Abstract
The domain organization of calretinin (CR) was predicted to involve all six EF-hand motifs (labeled I to VI) condensed into a single domain, as characterized for calbindin D28k (Calb), the closest homolog of calretinin. Unperturbed 1H,15N HSQC NMR spectra of a 15N-labeled calretinin fragment (CR III–VI, residues 100–271) in the presence of the unlabeled complimentary fragment (CR I–II, residues 1–100) show that these fragments do not interact. Size exclusion chromatography and affinity chromatography data support this conclusion. The HSQC spectrum of 15N-labeled CR is similar to the overlaid spectra of individual 15N-labeled CR fragments (CR I–II and CR III–VI), also suggesting that these regions do not interact within intact CR. In contrast to these observations, but in accordance with the Calb studies, we observed interactions between other CR fragments: CR I (1–60) with CR II–VI (61–271), and CR I–III (1–142) with CR IV–VI (145–271). We conclude that CR is formed from at least two independent domains consisting of CR I–II and CR III–VI. The differences in domain organization of Calb and CR may explain the specific target interaction of Calb with caspase-3. Most importantly, the comparison of CR and Calb domain organizations questions the value of homologous modeling of EF-hand proteins, and perhaps of other protein families.
Keywords: Calretinin, calbindin D28k, hexa EF-hand, calcium, domain organization
Calretinin (CR) and calbindin D28k (Calb) are two closely related hexa EF-hand proteins with 58% amino acid identity (Celio 1996). They consist of six EF-hand motifs and have predominant and distinct neuronal distributions (Rogers and Resibois 1992; Celio 1996). The proteins are used as markers of neuronal cell types and several disease states (Celio 1996). Both proteins have been classified as calcium buffer proteins, protecting cells against insults of high intracellular calcium concentration. However, the interactions of Calb with caspase-3 (Bellido et al. 2000) and CR with cytoskeleton components (Marilley and Schwaller 2000) and basic helix-loop-helix transcription factors (Zimmermann and Schwaller 2002) suggest that both proteins may also participate in calcium signaling pathways. Here, we show that the domain organization of EF-hand motifs in CR is different from that in Calb (Linse et al. 1997; Berggard et al. 2000).
In the past, the general rule was that EF-hand proteins contain an even number of EF-hand motifs that are organized into paired domains separated by linkers allowing a high degree of relative domain orientation. Structures of calmodulin (two paired domains; Ikura et al. 1992), troponin C (two paired domains; Slupsky and Sykes 1995), and calbindin D9k (one paired domain; Kördel et al. 1989) provided model examples. The structure of parvalbumin (a three-EF-hand domain, from which the term ‘EF-hand’ was derived; Kretsinger and Nockolds 1973) is a notable exception to this rule. Since the mid-1990s, exceptions to the paired EF-hand domain rule have come to predominate: sorcin, with five EF-hands, forms dimers that do not appear to contain any truly independent paired EF-hand domains (Xie et al. 2001); the majority of S100 proteins (with two EF-hands) form integrated homodimers (Drohat et al. 1998, 1999) or heterodimers (Hessian and Fisher 2001); recoverin consists of a single, rigid four EF-hand domain, although two EF-hand pairs are discerned within this structure (Tanaka et al. 1995); and a number of bacterial proteins contain a single EF-hand motif integrated into a larger domain structure (e.g., van Asselt et al. 1999). A mutant of calbindin D9k, a protein with two EF-hands that is normally found in a monomeric state, was found to produce stable, intertwined dimers (Hakansson et al. 2001), thus further breaking down the original structural rule. Linse and coworkers made no assumptions when studying the domain organization of Calb as a prelude towards more detailed structural and functional analyses of the protein. They concluded that Calb consists of a single domain of six EF-hands (Linse et al. 1997; Berggard et al. 2000). This rigid domain structure might confer a high degree of target specificity to the protein or better adapt Calb to a multiple-target scaffolding role.
We decided to define the domain organization of CR in order to support our high-resolution nuclear magnetic resonance (NMR) structural studies of CR. To achieve this, we applied NMR, size exclusion, and affinity chromatography methods to study the interactions between CR fragments.
The heteronuclear single-quantum correlation (HSQC) spectrum of a C-terminal fragment of CR (EF-hands III–VI, CR III–VI) does not change in the presence of its complementary, unlabeled N-terminal fragment consisting of the first two EF-hands (CR I–II; Fig. 1A ▶5). If CR III–VI interacted with CR I–II, then a shift in some of the CR III–VI resonances should be detected. For example, the interaction of a 12-amino acid polypeptide derived from actin capping protein CapZ with S100B (a homodimeric S100 protein with two EF-hand motifs) resulted in the visible shift of almost all of the ∼100 S100B resonances in a similar HSQC-based experiment (Kilby et al. 1997). Figure 1B ▶ indicates that the overlaid spectra of 15N-labeled CR I–II and CR III–VI resemble that of CR (Fig. 1C ▶). This suggests the CR I–II and CR III–VI regions do not interact within intact CR. The conclusion from the NMR data presented in Figure 1 ▶ is that CR I–II forms a domain that is independent of the CR III–VI fragment of CR.
Figure 1.

NMR analysis of CR and CR fragments in 50 mM Tris, 10 mM CaCl2, 20 mM NaCl, pH 7.7 buffer. (A) 1H,15N HSQC spectra of 50 μM 15N-labeled CR III–VI in the absence (black) and presence (red) of an equivalent concentration of unlabeled CR I–II. (B) Overlaid 1H,15N HSQC spectra of CR I–II (black) and CR III–VI (red). (C) 1H,15N HSQC spectrum of CR. The pattern of peaks from CR resembles the summed spectra of CR I–II and CR III–VI, particularly in well dispersed areas of the spectra. All spectra in panels B and C were obtained on separate samples.
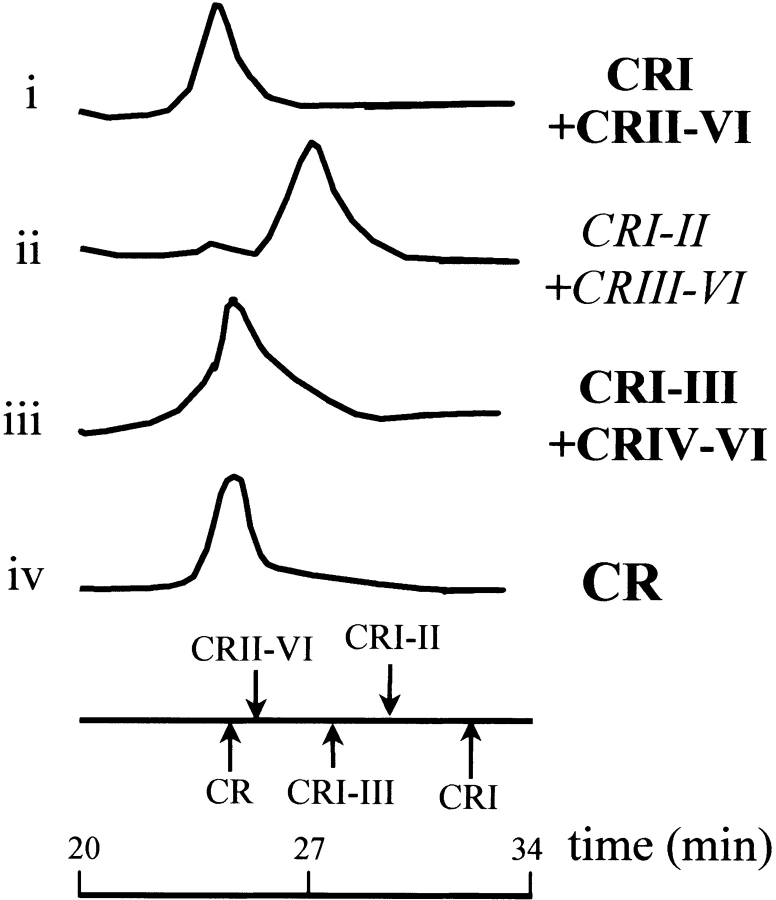
Size exclusion chromatography (SEC) supports our NMR data (Fig. 2 ▶). No interaction between CR I–II and CR III–VI was observed; the mixture of protein fragments elutes as an overlapping peak corresponding to the individual components. Other interactions between CR fragments were detected. For example, we observed an interaction between EF-hand I and the complementary CR II–VI region that had been produced by limited trypsinolysis of CR, and also between the CR I–III and CR IV–VI regions. A distinct shift from the component elution profiles was observed in these cases, and the mixtures eluted as a single peak with similar elution times as the intact protein (Fig. 2 ▶).
Figure 2.
Size exclusion chromatography analysis of complementary mixtures of CR fragments in calcium-containing buffer. (i) Mixture of CR I and CR II–VI (obtained by limited trypsinolysis of CR). (ii) Mixture of CR I–II and CR III–VI. (iii) Mixture of CR I–III and CR IV–VI. The elution times of CR and some CR fragments are marked below. Positive results are in bold, negative results in italics.
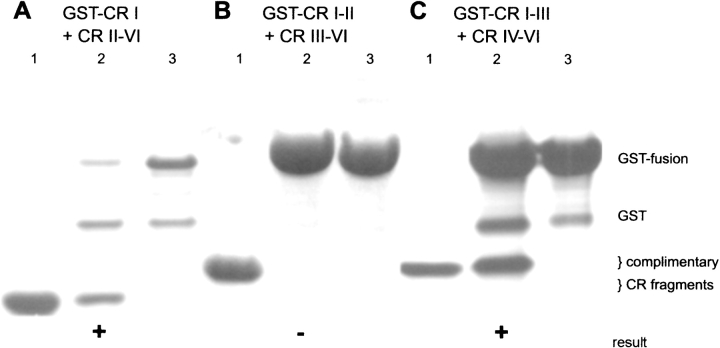
Figure 3 ▶ presents affinity chromatography data that confirmed the interactions between glutathione-S-transferase (GST)-CR I and CR II–VI, and between GST-CR I–III and CR IV–VI. CR IV–VI did not interact with GST-CR I–II in a control experiment, supporting the conclusion that CR IV–VI specifically interacts with EF-hand III and not with the Sepharose support, GST, or the CR I–II region of CR I–III (data not shown). CR III–VI did not interact with GST-CR I–II, which is in agreement with our other results.
Figure 3.
Affinity chromatography analysis of CR motif interactions. (A) Interaction of CR II–VI with glutathione-attached GST-CR I. (B) Interaction of CR III–VI with glutathione-attached GST-CRI–II. (C) Interaction of CR IV–VI with glutathione-attached GST-CRI–III. Lane 1, total applied complementary CR fragment; lane 2, SDS-eluted, specifically bound fraction; lane 3, GST-fusion-loaded resin eluted with SDS as a control. Wash fractions were analyzed but are omitted from this figure. The relative positions of the proteins are indicated to the right. Positive and negative results are marked by + or − signs below the appropriate lanes.
The data presented here suggest that CR I–II is independent of CR III–VI. Structural studies of CR I–II showed that it is capable of forming a regular two EF-hand domain (Palczewska et al. 2001). The exact nature of the interaction between the CR I–III and CR IV–VI fragments and the organization of the CR III–VI fragment are undetermined due to poor solubility of smaller C-terminal fragments (Palczewska et al. 1999).
The conditions under which our experiments were performed represent physiologically relevant conditions that overlap those used for Calb (Linse et al. 1997; Berggard et al. 2000). No interaction between CR I–II and CR III–VI was observed using three different experimental techniques at protein concentrations up to 50 μM. In contrast, SEC and affinity chromatography provided positive data for the CR I/CR II–VI and CR I–III/CR IV–VI interactions at ≤10 μM fragment concentrations (Figs. 2 ▶,3 ▶). Linse et al. (1997) reconstituted a native-like Calb from six polypeptides, constituting individual EF-hand sequences, at micromolar concentrations. This reconstitution required the presence of all six Calb EF-hand polypeptides. In a follow-up study using larger fragments, Calb was confirmed to reconstitute into a single domain (Berggard et al. 2000). The latter study removes any doubt that the reconstitution of Calb was facilitated by the ∼30% excised residues in the earlier study. We have already commented on the monomeric state of CR I–II (Palczewska et al. 2001) compared to the homodimeric Calb I–II domain (Klaus et al. 1999). We think that the homodimeric interface of Calb I–II may represent a heterodimeric interface between the highly identical Calb I–II and Calb III–IV segments. Clearly, considering fragment concentrations and the degree of reconstitution in the two sets of studies, our results distinguish the domain organizations of CR and Calb.
CR and Calb have different domain organizations, perhaps pointing to different structures and potential functions. This is supported by the results of Bellido et al. (2000) showing that caspase-3 is a specific binding partner for Calb. Secretagogin is the newest member of the hexa-EF-hand family (Wagner et al. 2000) with ∼40% identity to CR and Calb. Clues to the structure of secretagogin would be difficult to accurately predict on the basis of the different domain organizations of Calb and CR.
Our results might also have wider implications for studies involving homologous modeling. For example, DREAM has unique (for an EF-hand protein) DNA-binding properties (Carrion et al. 1999), and the structure of recoverin (Tanaka et al. 1995) would be expected to provide a suitable structural model (both proteins belong to the neuronal calcium sensor subgroup of four EF-hand proteins). DREAM and recoverin share only 35% identity. On the basis of our results on more identical proteins, how accurate is a threaded homologous model of DREAM based on the recoverin structure? Could a theoretical DREAM structure provide clues to the interactions between DREAM and DNA? Our work indicates that the links between structure and function within the EF-hand protein family are still unclear and diverse. In addition, our observations highlight the potential dangers of homologous modeling, although it can be successful in many applications.
Materials and methods
Protein expression and purification
The proteins and fragments used in this study include CR (full-length, residues 1–271), CR I (1–60), CR I–II (1–100), CR I–III (1–142), CR II–VI (61–271), CR III–VI (100–271), CR IV–VI (145–271), and, for affinity chromatography, their GST fusions. All protein sequences (except CR II–VI) are preceded by a GlySer dipeptide as a result of the thrombin cleavage site engineered into the GST fusions. CR and its fragments were prepared as described (Kuźnicki et al. 1995a; Palczewska et al. 2001) with one modification: overnight cultures of bacteria were transferred from Luria Broth to 15N-labeled Martek-9 media (Martek) for large-scale culture and expression of 15N-labeled protein. Cell extracts containing the glutathione-S-transferase (GST) fusion proteins were applied to glutathione-Sepharose columns and cleaved with thrombin. Protein solutions, after ion exchange chromatography, were dialyzed against Milli-Q grade water and lyophilized.
Limited proteolysis
A mixture of CR I (residues 1–60) and CR II–VI (61–271) was obtained by limited trypsinolysis of CR in 50 mM Tris, 150 mM NaCl, pH 8.0 buffer for 10 min (Kuźnicki et al. 1995b), using immobilized trypsin (trypsin-agarose, Sigma). The reaction was terminated by centrifugation on a 0.45 μm filter microspin column.
NMR spectroscopy
HSQC spectra of 0.25 mM 15N-labeled CR I–II and 50 μM CR III–VI (Bruker DRX 500), and 1 mM CR (Varian UNITYINOVA 750) in 50 mM Tris, 10 mM CaCl2, 20 mM NaCl, pH 7.7 buffer were acquired using standard pulse sequences. The CR III–VI spectrum was acquired once more after the addition of an equivalent of unlabeled CR I–II. The integrated volumes of peaks in the 6–11 ppm region of 1D 1H NMR spectra were used to confirm the 1:1 stoichiometry of 15N-labeled CR III–VI and unlabeled CR I–II in the sample. Processing was performed using standard Spectrometer software, and spectra were manipulated in SPARKY 3 (Goddard and Kneller 2001).
Size exclusion chromatography
Pharmacia FPLC equipment fitted with an HR10–30 SEC column was used to obtain SEC data at room temperature in 50 mM Tris, 1 mM CaCl2, 20 mM NaCl, pH 8.0 buffer. Approximately 50 μg protein was injected into a 25 μL loop, and the elution times were noted.
Affinity chromatography
Ten μL of glutathione-Sepharose was applied to a microspin column and equilibrated in PBS containing 1% triton. Aliquots of cell extract containing fusion proteins of CR fragments, stored at −70°C, were then applied to the gel. The gel was washed with PBS-T (4 × 100 μL) and then with binding buffer (50 mM Tris, 20 mM NaCl, 1 mM CaCl2, 2 × 200 μL). An aliquot of complementary CR fragment (or mixture of CR I and CR II–VI obtained by limited trypsinolysis) was applied to the column and the flowthrough collected, as were 2 × 100 and 1 × 20 μL washes with binding buffer. The bound proteins were eluted with 1% SDS, which also removed the attached GST fusion proteins. All fractions, together with total applied complementary protein and GST-tagged protein washed and eluted with SDS as controls, were subjected to 10% Tris-tricine PAGE (Schagger and von Jagow 1987) and stained with Coomassie Blue.
Acknowledgments
We thank Sara Linse (Lund University, Sweden) for useful discussions and Barbara Zarzycka (Warsaw) for technical assistance. Access to the CDSB facilities was funded under the 5th Framework Program of the European Union. Other funding was provided by International Center for Genetic Engineering and Biotechnology grants CRP/Pol97-01(t1) and CRP/Hun97-01(t1) and by Hungarian Grant OTKA T 029089.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0215303.
The proteins and fragments used in this study include: CR (full-length, residues 1–271); CR I (1–60); CR 1–II (1–100); CRI–III (1–142); CR II–VI (61–271); CR III–VI (100–271); CR IV–VI (145–271) and, for affinity chromatography, their GST fusions. All protein sequences (except CR II–VI) are preceded by a GlySer dipeptide as a result of the thrombin cleavage site engineered into the GST fusions.
References
- Bellido, T., Huening, M., Raval-Pandya, M., Manolagas, S.C., and Christakos, S. 2000. Calbindin-D28k is expressed in osteoblastic cells and suppresses their apoptosis by inhibiting caspase-3 activity. J. Biol. Chem. 275 26328–26332. [DOI] [PubMed] [Google Scholar]
- Berggard, T., Thulin, E., Åkerfeldt, K.S., and Linse, S. 2000. Fragment complementation of calbindin D28k. Protein Sci. 9 1094–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion, A.M., Link, W.A., Ledo, F., Mellstrom, B., and Naranjo, J.R. 1999. DREAM is a Ca2+-regulated transcriptional repressor. Nature 398 80–84. [DOI] [PubMed] [Google Scholar]
- Celio, M.R. (ed.), 1996. Guidebook to the calcium-binding proteins, 1st ed., pp. 23–28. Sambrook and Tooze with Oxford University Press, Oxford, UK.
- Drohat, A.C., Baldisseri, D.M., Rustandi, R.R., and Weber, D.J. 1998. Solution structure of calcium-bound rat S100B (ββ) as determined by nuclear magnetic resonance spectroscopy. Biochemistry 37 2729–2740. [DOI] [PubMed] [Google Scholar]
- Drohat, A.C., Tjandra, N., Baldisseri, D.M., and Weber, D.J. 1999. The use of dipolar couplings for determining the solution structure of rat apo-S100B(ββ). Protein Sci. 8 800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, T.D. and Kneller, D.G. 2001. SPARKY 3 NMR software, University of California, San Francisco.
- Hakansson, M., Svensson, A., Fast, J., and Linse, S. 2001. An extended hydrophobic core induces EF-hand swapping. Protein Sci. 10 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessian, P.A. and Fisher, L. 2001. The heterodimeric complex of MRP-8 (S100A8) and MRP-14 (S100A9). Antibody recognition, epitope definition and the implications for structure. Eur. J. Biochem. 268 353–363. [DOI] [PubMed] [Google Scholar]
- Ikura, M., Clore, G.M., Gronenborn, A.M., Zhu, G., Klee, C.B., and Bax, A. 1992. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256 632–638. [DOI] [PubMed] [Google Scholar]
- Kilby, P.M., Van Eldik, L.J., and Roberts, G.C. 1997. Identification of the binding site on S100B protein for the actin capping protein CapZ. Protein Sci. 6 2494–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus, W., Grzesiek, S., Labhardt, A.M., Buchwald, P., Hunziker, W., Gross, M.D., and Kallick, D.A. 1999. NMR investigation and secondary structure of domains I and II of rat brain calbindin D28k (1–93). Eur. J. Biochem. 262 933–938. [DOI] [PubMed] [Google Scholar]
- Kördel, J., Forsén, S., and Chazin, W.J. 1989. 1H NMR Sequential resonance assignments, secondary structure, and global fold in solution of the major (trans-Pro43) form of bovine calbindin D9k. Biochemistry 28 7065–7074. [DOI] [PubMed] [Google Scholar]
- Kretsinger, R.H. and Nockolds, C.E. 1973. Carp muscle calcium-binding protein. II. Structure determination and general description. J. Biol. Chem. 248 3313–3326. [PubMed] [Google Scholar]
- Kuźnicki, J., Strauss, K.I., and Jacobowitz, D.M. 1995a. Conformational changes and calcium binding by calretinin and its recombinant fragments containing different sets of EF hand motifs. Biochemistry 34 15389–15394. [DOI] [PubMed] [Google Scholar]
- Kuźnicki, J., Wang, T.L., Martin, B.M., Winsky, L., and Jacobowitz, D.M. 1995b. Localization of Ca(2+)-dependent conformational changes of calretinin by limited tryptic proteolysis. Biochem. J. 308 607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linse, S., Thulin, E., Gifford, L.K., Radzewsky, D., Hagan, J., Wilk, R.R., and Åkerfeldt, K.S. 1997. Domain organization of calbindin D28k as determined from the association of six synthetic EF-hand fragments. Protein. Sci. 6 2385–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marilley, D. and Schwaller, B. 2000. Association between the calcium-binding protein calretinin and cytoskeletal components in the human colon adenocarcinoma cell line WiDr. Exp. Cell Res. 259 12–22. [DOI] [PubMed] [Google Scholar]
- Palczewska, M., Groves, P., and Kuźnicki, J. 1999. Use of Pichia pastoris for the expression, purification and characterization of rat calretinin “EF-hand” domains. Protein Expr. Purif. 17 465–476. [DOI] [PubMed] [Google Scholar]
- Palczewska, M., Groves, P., Ambrus, A., Kaleta, A., Kövér, K.E., Batta, G., and Kuźnicki, J. 2001. Structural and biochemical characterization of neuronal calretinin domain I-II (residues 1–100); comparison to homologous calbindin D28k domain I-II (residues 1–93). Eur. J. Biochem. 268 6229–6237. [DOI] [PubMed] [Google Scholar]
- Rogers, J.H. and Resibois, A. 1992. Calretinin and calbindin-D28k in rat brain: Patterns of partial co-localization. Neuroscience 51 843–865. [DOI] [PubMed] [Google Scholar]
- Schagger, H. and von Jagow, G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166 368–379. [DOI] [PubMed] [Google Scholar]
- Slupsky, C.M. and Sykes, B.D. 1995. NMR solution structure of calcium-saturated skeletal muscle troponin C. Biochemistry 34 15953–15964. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Ames, J.B., Harvey, T.S., Stryer, L., and Ikura, M. 1995. Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376 444–447. [DOI] [PubMed] [Google Scholar]
- van Asselt, E.J., Dijkstra, A.J., Kalk, K.H., Takacs, B., Keck, W., and Dijkstra, B.W. 1999. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure Fold. Des. 7 1167–1180. [DOI] [PubMed] [Google Scholar]
- Wagner, L., Oliyarnyk, O., Gartner, W., Nowotny, P., Groeger, M., Kaserer, K., Waldhausl, W., and Pasternack, M.S. 2000. Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J. Biol. Chem. 275 24740–24751. [DOI] [PubMed] [Google Scholar]
- Xie, X., Dwyer, M.D., Swenson, L., Parker, M.H., and Botfield, M.C. 2001. Crystal structure of calcium-free human sorcin: A member of the penta-EF-hand protein family. Protein Sci. 10 2419–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, L. and Schwaller, B. 2002. Monoclonal antibodies recognizing epitopes of calretinins: Dependence on Ca2+-binding status and differences in antigen accessibility in colon cancer cells. Cell Calcium 31 13–25. [DOI] [PubMed] [Google Scholar]