Abstract
The Yeast MATa1 and MATα2 are homeodomain proteins that bind DNA cooperatively to repress transcription of cell type specific genes. The DNA affinity and specificity of MATa1 in the absence of MATα2, however, is very low. MATa1 is converted to a higher affinity DNA-binding protein by its interaction with the C-terminal tail of MATα2. To understand why MATa1 binds DNA weakly by itself, and how the MATα2 tail affects the affinity of MATa1 for DNA, we determined the crystal structure of a maltose-binding protein (MBP)-a1 chimera whose DNA binding behavior is similar to MATa1. The overall MATa1 conformation in the MBP-a1 structure, which was determined in the absence of α2 and DNA, is similar to that in the a1/α2/DNA structure. The sole difference is in the C-terminal portion of the DNA recognition helix of MATa1, which is flexible in the present structure. However, these residues are not in a location likely to be affected by binding of the MATα2 tail. The results argue against conformational changes in a1 induced by the tail of MATα2, suggesting instead that the MATα2 tail energetically couples the DNA binding of MATα2 and MATa1.
Keywords: Homeodomain, MATa1, transcription, crystal structure, protein–DNA interactions, maltose binding protein, protein chimera
The MATa1 protein (which we shall call a1), plays an important role in yeast mating-type regulation (Johnson 1995). In diploid a/α cells, a1 binds DNA cooperatively with MATα2 (which we shall call α2), leading to the repression of haploid-specific genes (hsg). Previous genetic, biochemical, and structural studies have characterized the minimum functional fragments of both proteins necessary for DNA binding and heterodimerization (Goutte and Johnson 1993; Phillips et al. 1994; Li et al. 1995; Vershon et al. 1995). The α2 protein binds DNA with a homeodomain located near the carboxyl-terminus of the protein and contacts the a1 homeodomain with an 18-amino acid carboxyl-terminal tail that is unstructured in the absence of a1 (Wolberger et al. 1991; Li et al. 1995). The a1 protein, which also contains a homeodomain at its carboxyl-terminus portion, shows no reproducible sequence-specific DNA binding. However, in combination with α2, the a1/α2 heterodimer binds DNA with high affinity, and the sequence specificity of the heterodimer increases 3000-fold over that of α2 alone (Goutte and Johnson 1993; Phillips et al. 1994).
Structures of the a1/α2/DNA and the α2/DNA complex have been determined by X-ray crystallography (Wolberger et al. 1991; Li et al. 1995). In the a1/α2/DNA structure, the a1 and α2 homeodomains bind DNA in a head-to-tail orientation, with heterodimer contacts mediated by the 18-residue carboxyl-terminal tail of α2 (Fig. 1 ▶). The α2 tail becomes ordered only in the presence of a1, forming a short amphipathic helix that packs against the a1 homeodomain between helices 1 and 2. An evenly distributed 60° bend in the DNA is induced by the binding of the a1/α2 heterodimer. The conformation of α2 and its DNA contacts are virtually the same in both the a1/α2/DNA and the α2/DNA structures. The docking of the a1 homeodomain on the DNA is similar to that of the α2 protein. a1 contacts DNA bases in the major groove with five residues in helix 3: Val47, Ile50, Asn51, Met54, and Arg55 (Li et al. 1995). Unlike many other homeodomain proteins, the N-terminal arm of a1 is mostly unstructured, does not contact the DNA minor groove, and was shown to be dispensable for DNA binding (M. Stark and A.D. Johnson, pers. comm.).
Figure 1.
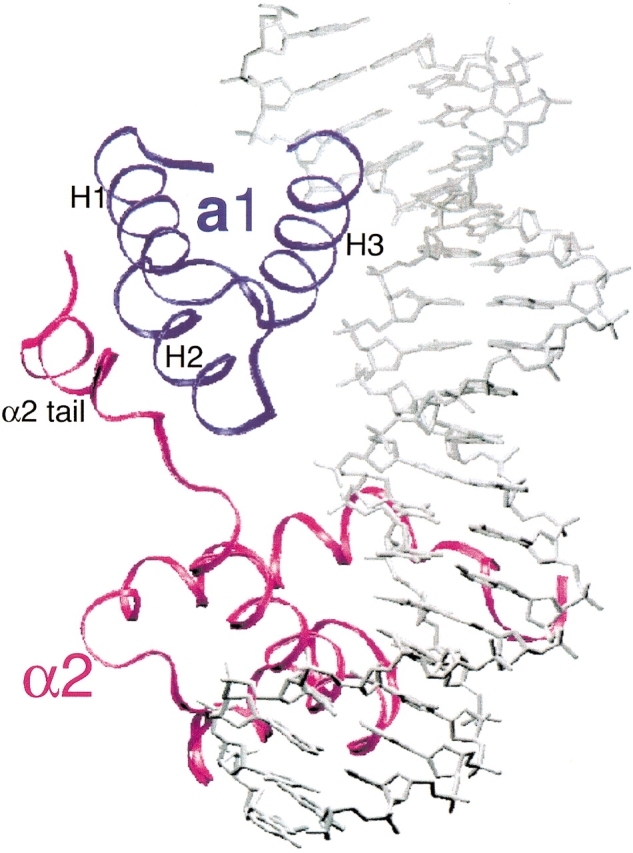
Overview of the crystal structure of the a1/α2/DNA ternary complex. The a1 homeodomain is shown in red, α2 in blue, and the DNA in silver. The three helices of a1 and the tail of α2 have been noted. The α2 tail folds into a short amphipathic helix and packs against the groove formed by Helix 1 and 2 of a1.
Given the similar docking of a1 and α2 to DNA and the extensive a1-DNA contacts, the prior structures have not explained why a1 binds DNA so weakly in the absence of α2. Also unclear is how the binding of the α2 tail to the non-DNA binding face of a1 dramatically increases the DNA binding affinity and specificity of a1. Two hypotheses have been proposed concerning the role of the α2 tail: (1) the α2 tail couples the DNA binding of a1 and α2, making the binding of a1 to DNA energetically more favorable than it would be in the absence of cooperativity; (2) upon heterodimarization, contacts with the α2 tail causes a1 to bind DNA with higher affinity and specificity. In support of the latter hypothesis, Stark et al. (1999) demonstrated that when the α2 tail is covalently linked to the homeodomain of a1, the engineered a1 can bind DNA tightly and specifically as a monomer (Stark et al. 1999). They also showed that an α2 peptide supplied in trans can induce tighter DNA binding by the a1 homeodomain. NMR studies of the free a1 protein further suggested that the α2 tail induces changes in the loop 1 region between helices 1 and 2 of a1 that push it towards a properly folded DNA binding conformation (Anderson et al. 2000).
To understand fully why a1 binds DNA so weakly, and how binding of the α2 tail to the a1 homeodomain improves the affinity of a1 for DNA, we set out to determine the crystal structure of the a1 homeodomain in the absence of DNA or the α2 protein. To overcome our inability to obtain crystals of free a1, we took an alternative approach and determined the structure of a chimeric protein consisting of the free a1 homeodomain fused to the maltose binding protein (MBP). The DNA binding properties of the MBP-a1 chimera are identical to that of the a1 homeodomain. The 2.1-Å crystal structure shows that the C-terminal portion of the DNA recognition helix of a1 is unstructured in the absence of the α2 tail and DNA, providing an explanation for the weak affinity of a1 alone for DNA. Other than the four C-terminal residues, the structure of the a1 homeodomain in the MBP-a1 structure is identical to that in the a1/α2/DNA structures. The absence of a conformational change leads us to conclude that the α2 tail probably increases the DNA specificity and affinity of a1 by coupling the DNA binding of a1 and α2 to minimize the overall energy cost. We discuss the possible sources of discrepancies between our crystal structures and the previous NMR structure.
Results
The MBP/aI chimera facilitates the crystallization and structure determination of the a1 homeodomain
All of our previous attempts to crystallize the a1 homeodomain in the absence of DNA and α2 protein failed. We therefore used MBP as a crystallization tag (Center et al. 1998; Liu et al. 2001). MBP is a well-behaved protein whose affinity for amylose has been widely utilized in protein purification and whose structure is known (Quiocho et al. 1997). It was first used by Center et. al. (1998) to promote crystallization of a trimeric human T cell leukemia virus type I gp21 ectodomain fragment by fusing MBP to its N-terminus. The MBP fusion vector in that study was engineered by replacing the flexible linker and protease cleavage site between the two fusion partners with a three-residue poly-alanine linker to increase the chance of crystallization. We adopted this system with modifications (Materials and Methods) to create a MBP-a1 chimera. In addition, part of the N-terminal arm of the a1 homeodomain was deleted because it is not well structured in the a1/α2/DNA complex structure (Li et al. 1995), and has been shown to be dispensable for a1 function (M. Stark and A.D. Johnson, pers. comm.). The fusion of the MBP molecule to the a1 homeodomain dramatically increased the expression level from 0.8 mg/L for the a1 homeodomain to ∼30 mg/L for the MBP-a1 chimera. The purification of MBP-a1 was a simple two-step process, with affinity purification on an amylose agarose column followed by either size-exclusion or anion exchange chromatography. The purified MBP-a1 protein behaves as a monomer on a size-exclusion column (data not shown).
The fusion of MBP with a1, unlike the isolated a1 protein, greatly increased the chance of crystallization. Among the 98 conditions in the Hampton crystal screen I and II kits, around one-fifth produced MBP-a1 crystals. At least two crystal forms were observed. One of them diffracted to 2.1-Å resolution and the other crystal form diffracted to 2.3 Å. The structure of the MBP-a1 fusion in the two crystal forms was solved by molecular replacement. To avoid any model bias in the determination of the a1 structure, only the coordinates of the maltose binding protein (PDB code: 4MBP) (Quiocho et al. 1997) were used as the search model. The density corresponding to the a1 protein was easily identifiable in electron density maps calculated with phases from the MBP model. The a1 portion of the structure was traced and the entire structure of the fusion protein was refined. Statistics for the models determined from both crystal forms are shown in Table 1. In crystal form I, the model of the MBP-a1 chimera contains residues 1 to 366 of the engineered MBP protein and residues 77 to 126 of the a1 protein. The amino acid residues of the a1 homeodomain were renumbered according to the homeodomain numbering convention (Qian et al. 1989; Kissinger et al. 1990; Otting et al. 1990; Phillips et al. 1991; Wolberger et al. 1991; Klemm et al. 1994) such that a1 residues 77–126 are renumbered 8–57. The last three amino acid residues of the a1 homeodomain in crystal form II are missing from the electron density map.
Table 1.
Data collection and refinement statistics
| Data processing | Crystal form I | Crystal form II |
| Space group | P43212 | P212121 |
| Resolution (Å) | 60–2.1 | 50–2.3 |
| Data redundancy | 6.8 | 9.97 |
| Unique reflections | 42612 | 19118 |
| Completeness (overall/outer shell) | 99.2/97.1 | 96.9/83.2 |
| I/σ (overall/outer shell) | 30.1/4.9 | 22.5/3.58 |
| Rsym (%, overall/outer shell) | 5.7/21.6 | 5.8/24.0 |
| Refinement | ||
| Rcrystal(%) | 23.1 | 26.4 |
| Rfree(%) | 26.2 | 29.2 |
| rms deviation of bond lengths (Å) | 0.0059 | 0.0064 |
| rms deviation of bond angles (°) | 1.16 | 1.33 |
The overall structure of the MBP-a1 chimera in crystal form I, with space group P43212, is shown in Figure 2 ▶. The five-residue poly-alanine linker between MBP and a1 that was designed to form an α-helical conformation adopts a turn conformation instead, thereby positioning a1 to avoid steric clashes with MBP. The only intramolecular contacts are between a few a1 residues at the C-terminal end of helix 2 and MBP. The orientation of MBP and a1 is such that both the DNA recognition helix (helix 3) and the α2 tail binding site located between helices 1 and 2 of a1 are fully exposed (Fig. 2 ▶). The exact same overall MBP-a1 structure is observed in crystal form II, which forms in space group P212121. The C-terminus of a1 adopts a different conformation due to the changed packing environment in this crystal form, as discussed below.
Figure 2.
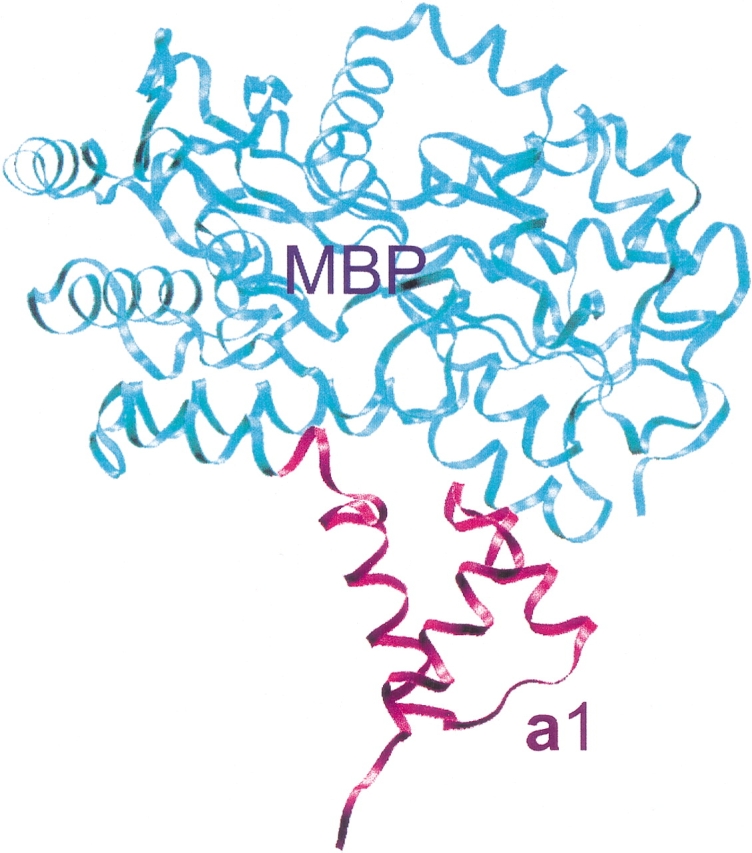
Overview of the MBP-a1 chimeric protein structure. The MBP moiety is shown in cyan; the five-residue polyalanine linker and the a1 homeodomain are shown in magenta. Note the short alanine linker that adopts a turn conformation to avoid steric clashes between a1 and MBP molecule. The α2 tail binding site on the a1 homeodomain is not in contact with MBP.
The MBP-a1 structure contains a distorted a1 C-terminus in the absence of DNA and α2
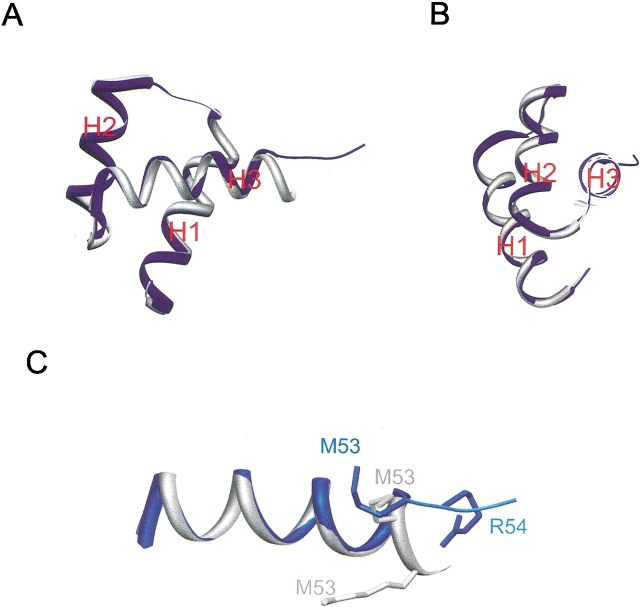
The structure of the uncomplexed a1 protein in the MBP-a1 fusion determined from both crystal forms superimposes well with the DNA-bound a1 protein in the a1/α2/DNA structure, with an r.m.s.d. in Cα positions (residues 9–53, excluding four residues from the C-terminus) of 0.4 Å (Fig. 3A,B ▶). We find no evidence for global conformational changes in the a1 homeodomain induced by the binding of the α2 tail or DNA. To further investigate whether there are any small but significant changes in the orientation of the three homeodomain helices in DNA-bound versus free a1, we carried out various alignments of bound and free a1 using only two of the three α-helices at a time and examined the orientation of the third helix. No significant change of the helix orientation was revealed by any of the suppositions.
Figure 3.
(A) Comparison of the free a1 conformation (in blue) in the MBP-a1 structure (crystal form I) and the DNA-bound conformation (in gray) in the a1/α2/DNA structure. The two a1 homeodomain structures are aligned with the Cα atoms of residues 9 –53. The same alignment rotated roughly 90° is shown in (B). No significant global conformational changes are observed in the aligned region of the a1 homeodomain. However, a large local conformational change is observed for the last four amino acid residues at the C-terminus of a1 (residues 54–57), as shown in more detail in (C). Instead of folding into α-helix in the DNA-bound a1, the C-terminal portion of the DNA recognition helix in the free a1 adopts an extended conformation and contacts the surface of a symmetry-related MBP molecule. Two important DNA major groove contacting residues, Met54 and Arg55, are in different conformation in the free a1 homeodomain, and would result in stetic clashes if docked onto the DNA.
The sole significant local conformational difference between the bound a1 in the a1/α2/DNA ternary complex and the free a1 homeodomain in the MBP-a1 fusion lies in the last four amino acid residues at the C-terminus of a1 (residues 54–57). Instead of the helical conformation adopted by the DNA-bound a1, the C-terminal portion of the DNA recognition helix (Helix 3) of free a1 in crystal form I adopts an extended conformation (Fig. 3C ▶) and contacts the surface of a symmetry-related MBP molecule. The Cα backbone displacements between the two structures for the four C-terminal residues are as high as 10 Å. The different backbone conformation causes the side chains of Met54 and Arg55, both of which make crucial DNA major groove contacts in the a1/α2/DNA structure, to deviate completely from their DNA-binding conformation. Adopting the free a1 conformation in the MBP-a1 structure would cause severe steric clashes when a1 binds DNA. The B-factors for the last four residues, which indicates how well ordered the atom positions are, rise steadily from the structure average of 24 Å2 to 55 Å2 at residue 57. This is in contrast to the B-factors of the same region of a1 in the a1/α2/DNA structure, in which the B-factor is 45 Å2 at the last residue, as compared to the structure average of 39 Å2. An unstructured C-terminus is also found in the free a1 homeodomain determined from MBP-a1 crystal form II. Without the crystal contact that stabilizes the extended a1 c-terminus conformation found in crystal form I, the a1 C-terminus begins to unfold at Met54, with no electron density visible for residues Arg55 to Lys57.
The DNA binding behavior of the MBP-a1 chimera is the same as the a1 homeodomain
To test whether fusion to MBP alters the DNA binding properties of the a1 homeodomain, we performed a series of electrophoretic mobility shift assays (EMSA) in which the DNA binding of both MBP-a1 and a1 was assayed in the presence or absence of the α2 protein. As the results show in Figure 4 ▶, the a1 homeodomain and the MBP-a1 chimera have the same DNA-binding behavior. Neither a1 nor MBP-a1 has detectable affinity for a DNA fragment containing an a1/α2 binding site in the absence of α2. In the presence of α2, however, both proteins show specific high-affinity DNA binding. The shifted bands contain the heterodimer bound to DNA (Fig. 4 ▶). The same behavior is observed in assays of complex formation with α2–3A, an α2 mutant that does not bind DNA by itself but still binds DNA cooperatively with a1 (Vershon et al. 1995; Jin et al. 1999). The affinity of a1 and MBP-a1 for DNA in the presence of α2 proteins is comparable, with an estimated Kd for the heterodimer of 4 × 10−13 M2.
Figure 4.
DNA binding behavior of the MBP-a1 and a1 homeodomain proteins. The DNA binding affinity of the MBP-a1 and a1 proteins was measured in electrophoretic mobility shift assays in the presence or absence of either wild-type α2 or α2–3A mutant protein. A series of fivefold dilution of a1 or MBP-a1, starting from 10−6 M, was used. The wild-type and the mutant α2 protein concentrations were kept constant at 10−7 M. The lanes containing DNA alone (lanes 3,19), the wild-type α2 alone bound to DNA (lanes 1,18), and the α2–3A bound to DNA (lanes 2,17) are also shown here.
Discussion
The reason the a1 homeodomain contributes high DNA binding affinity and specificity to the a1/α2/DNA complex, while having little intrinsic affinity and specificity for DNA in the absence of α2, has remained an open question. Previous results (Stark et al. 1999) had suggested that the tail of α2, which mediates all protein–protein contacts with a1 in the a1/α2/DNA structure, may play an allosteric role in inducing a1 to adopt a high-affinity DNA-binding conformation. A report (Anderson et al. 2000) comparing the solution NMR structure of the free a1 homeodomain with the (Anderson et al. 2000) crystal structure of a1 in a ternary complex with α2 and DNA (Li et al. 1995) identified structural differences located in the loop 1 region connecting helices 1 and 2 and in the C-terminus of a1. The authors proposed that changes in loop 1 of a1 induced by the α2 tail cause van der Waals stacking changes leading to the ordering of a final turn in the DNA-binding helix of a1. We took a different approach by determining the crystal structure of the free a1 homeodomain. The 2.1- and 2.3-Å resolution crystal structures from two crystal forms allow us to observe crystal structure of the free a1 and to compare bound and free structures determined by the same experimental technique. The fused MBP in the present crystal structure is unlikely to have altered the a1 conformation, because the DNA binding properties of the fusion protein are the same as the a1 homeodomain, and MBP does not contact the α2 tail binding site on a1. Interestingly, we draw different conclusions from our study.
The comparison of bound and free crystal structure of a1 suggests that the flexibility of the C-terminus of the a1 recognition helix can explain why the a1 homeodomain binds DNA weakly in the absence of α2. The free a1 conformation determined from the MBP-a1 structures clearly shows that the carboxyl-terminal portion of the DNA recognition helix (helix3) of a1 is destabilized in the absence of α2 and DNA. The conformation of two important DNA recognition amino acid residues is completely different from their DNA-bound conformation, and would occlude DNA binding unless they underwent a conformational change docked onto DNA. This part of our observation is in agreement with that from the solution NMR structure of the a1 homeodomain suggesting a poorly folded a1 C-terminus (Phillips et al. 1991). We note that an unstructured C-terminus of helix 3 has been found in several other free homeodomain protein structures. The last four residues of the free engrailed homeodomain crystal structure are disordered (Clarke et al. 1994), as are the last 10 and 8 residues of the free VND-NK2 and Antp, respectively (Billeter et al. 1990; Tsao et al. 1995). However, the other homeodomain proteins with an unstable helix 3 C-terminus retain a functional N-terminal arm that makes DNA minor groove contacts. Because the N-terminal arm of a1 does not appear to participate in DNA binding, the requirement for reordering the C-terminal residues of helix 3 may have proportionally greater effect on the affinity of a1 for DNA.
We did not observe any conformational differences in the remainder of the a1 homeodomain that would suggest an allosteric role for the binding of the α2 tail. This observation disagrees with the conclusion from the NMR structural analysis (Baxter et al. 1994; Anderson et al. 2000), which links van der Waals stacking changes in the a1 loop 1 region caused by the binding of the α2 tail to the ordering of a final turn in the DNA-binding helix. There are several discrepancies between the two studies. The r.m.s.d. of Cα positions (residues 10–52, excluding the flexible N- and C-terminus) in a superposition of the NMR structure of free a1 and the crystal structure of DNA-bound a1 is 1.83Å (Anderson et al. 2000), significantly higher than the 0.4 Å r.m.s.d. we observe comparing the crystal structures of the free and DNA-bound a1 protein. Most of the side-chain conformation changes in the loop 1 region observed in the NMR study are not supported by our crystal structure. Moreover, the relative orientations of the three homeodomain helices remain the same in the crystal structures of the free and DNA-bound a1. The apparent discrepancy is unlikely due to differences in experimental conditions such as pH and temperature at which the NMR and X-ray diffraction data were collected. We note that the free a1 conformation does not change when the MBP-a1 crystals are transferred from pH 5.0 to pH 8.0 (data not shown). The room temperature NMR structure is expected to be more dynamic than the crystal structures determined at 90 K, but this is unlikely to account for a significant part of the discrepancy in the Cα alignments.
We think it most likely that the apparent discrepancy between the two studies is the result of the different structure determination methods used. The conclusions of the previous study relied upon the assumption that differences between the NMR structure of free a1 and the crystal structure of bound a1 were attributable to the effects of binding to α2 and DNA. However, it has previously been observed that structures of a given protein determined by NMR and crystallographic methods do not necessarily superimpose well. For example, when the core Antennapedia homeodomain (residues 5–60) in the crystal structure of Antennapedia homeodomain–DNA complex are aligned with the 16 NMR models of the same complex, the average r.m.s.d. in Cα positions is 1.13 Å ( Billeter et al. 1993; Qian et al. 1993; Fraenkel and Pabo 1998). In the case of Ets-1 bound to DNA, the NMR (Werner et al. 1997) and crystal structures (Garvie et al. 2001) of the Ets domain superimpose with an r.m.s.d. of 2.3 Å. The previously observed discrepancies are of the same order of magnitude as the differences between the NMR and crystal structures of a1. Moreover, we note that the Ramachandran plot of a randomly picked free a1 NMR structure (Anderson et al. 2000) out of 20 in the ensemble has only 67% of the amino acid residues in the most favored regions, with one residue (2%) in the disallowed region. In contrast, the crystal structures we report here have 96% of the residues in the most favored regions and no residues in the disallowed or generously allowed regions. We also note that the NMR structure determination of a1 (Anderson et al. 2000) did not include measurements of hydrogen-bonding patterns (Grzesiek et al. 2001), or distance-independent residual dipolar coupling (Tjandra and Bax 1997), which have the potential to reduce the discrepancy between NMR and crystal structures. Although none of the aforementioned caveats rule out the possibility of a global conformational change, these considerations lead us to favor a model in which conformational changes are likely to be due to DNA binding alone and are localized to the C-terminus of the a1 homeodomain helix 3.
Because α2-induced conformational changes in a1 are unlikely according to our data, we instead favor an energetic coupling model, in which the DNA binding of a1 and α2 are strictly coupled to the heterodimerization of a1 and α2 mediated by the α2 tail. The a1 protein cannot dissociate from DNA without breaking the heterodimer contacts with the α2 protein. In such circumstances, the dissociation constant of the a1/α2 heterodimer will be the product of the dissociation constant of the two individual proteins. The dissociation constant of a1 from DNA is estimated to be from 10−5 to 10−6 M and that of α2 is ∼10−8 M. The strict coupling model predicts a 10−13 to 10−14 M2 dissociation constant of the a1/α2 heterodimer from DNA, which is exactly what was observed in the DNA-binding assays. Besides, the heterodimerization of a1/α2 obviously provides the free energy to compensate for the folding of the a1 C-terminus, which occurs upon binding to DNA. The seemingly weak Kd of the heterodimerization of a1 and α2 in solution, estimated to be 2 × 10−4 M (Phillips et al. 1994), is expected to be tighter in the a1/α2/DNA ternary complex, because the nearby DNA and a1 molecules further restrict the conformational space of the α2 tail, dramatically reducing the entropic cost of folding the α2 tail during heterodimerization. The above rationales, rather than an allosteric mechanism, are the more likely explanations for the role of the α2 tail in recruiting a weakly binding partner, a1, to the DNA.
Materials and methods
Protein purification
A plasmid (pMBP-a1) encoding a chimera of MBP, a five alanine linker, and a1 residues 77–126 was constructed as described (Center et al. 1998), with modifications. A gene fragment of MBP was PCR amplified using the primer pair p1: 5′-CTGATTTATAACAAAGATCTGCTGCCG (contains a Bgl II site) and p2: 5′-TGCCCGTGCTTGGGGTGACGCTGCAGCATTAGTCTGCGC GGCTGCCAGGGCTGCATCGACAGT (contains a Pst I site and part of the a1 N-terminal sequence) and pMAL-c2 as the template. A gene fragment of a1 (residues 77–126) was PCR amplified using the primer pair p3: 5′-GACTAATGCTGCAGCGTCACCCCAAGCA (contains a Pst I site) and p4: 5′-CGAGCACAGGATCCGTCATTATTTAGATCTCAT (contains a BamH I site) and pMSK66 encoding a1 homeodomain as the template. A gene fragment containing the chimera of MBP and a1 was generated by mixing two PCR products mentioned above and doing 5 PCR cycles, then adding primer p1 and p4 to the same tube and continuing PCR for additional 25 cycles. The PCR product was inserted into pMAL-c2 vector cut with Bgl II and BamH I to produce pMAL-a1. The protein sequence around the link between MBP and a1 is shown below:
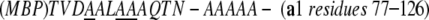 |
The underlined residues correspond to mutations E359A, K362A, and D363A. The fusion protein was expressed in E. coli BL21 codon plus cells, The cells were resuspended and lysed in a buffer containing 50 mM HEPES pH8.0, 200 mM NaCl, 1 mM EDTA by passing through microfluidizer twice. After centrifugation, the MBP-a1 in the supernatant was affinity-purified with an amylose-agarase column (New England Biolabs), yielding MBP-a1 that was 90% pure. The chimeric protein was further purified on a Mono S column (Pharmacia) by running a NaCl gradient from 50 to 800 mM. The peak fractions containing MBP-a1 were >99% pure as judged by SDS-PAGE and mass spectrometry.
A fragment of the wild-type a1 homeodomain (residues 74–126) was synthesized by standard solid-state peptide synthesis method using a MilliGen 9050 PepSynthesizer with FMOC chemistry. The peptide was cleaved from the resin surface and purified on reverse-phase chromatography C4 followed by C18 column. N-terminal sequencing and mass spectrometry confirmed the purity to be ∼99%. This a1 fragment was used in the DNA binding assays as the positive control.
Crystallization and structure determination
Crystallization conditions of MBP-a1 protein were screened using the hanging drop vapor diffusion method with Hampton Research screens I and II. The concentration of MBP-a1 was 15 mg/mL. Crystals appeared in around one-fifth of the conditions tested. Crystal form I with space group P43212 symmetry was grown in 100 mM MES, pH 5.0, and 2.4 M ammonium sulfate. Crystal form II, with space group P212121, was grown in 100 mM MES, pH 6.0, and 20% PEG 6000. Crystals were prepared for freezing by transferring stepwise to mother liquor plus 15% glycerol and flash frozen at −173°C. Diffraction data were collected with an R-Axis IV image plate detector mounted on a Rigaku rotating anode X-ray generator. The data were processed with DENZO and SCALEPACK (Table 1).
Electrophoretic mobility shift assay for DNA binding
Proteins were incubated for 1 h on ice with a 32P-labeled 34 base-pair DNA fragment containing either an a1/α2 binding site or an a1/a1 site and electrophoresed through an 8% native Tris-borate-EDTA polyacrylamide gel as described previously (Stark et al. 1999). Electrophoretic mobility shift assays were performed in an assay buffer containing 25 mM Tris (pH 7.5), 1 mM EDTA, 100 mM NaCl, 5 mg/mL bovine serum albumin, 5% glycerol, 0.1% Nonidet P-40, 10 μg/mL of sheared salmon sperm DNA, and 1 mM DTT. Dried gels were exposed to phosphor screens, and the images were scanned on a Molecular Dynamics model 425 phosphorimager.
Acknowledgments
We thank members of the Wolberger lab for assistance and David Shortle for discussions. This work was funded by NSF Grant MCB-9808412. The coordinates of the crystal form I and II of the MBP-a1 chimera protein structures have been deposited to the Protein Data Bank at Brookhaven with accession number of 1MH3 and 1MH4, respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0219103.
References
- Anderson, J.S., Forman, M.D., Modleski, S., Dahlquist, F.W., and Baxter, S.M. 2000. Cooperative ordering in homeodomain–DNA recognition: Solution structure and dynamics of the MATa1 homeodomain. Biochemistry 39 10045–10054. [DOI] [PubMed] [Google Scholar]
- Baxter, S.M., Gontrum, D.M., Phillips, C.L., Roth, A.F., and Dahlquist, F.W. 1994. Heterodimerization of the yeast homeodomain transcriptional regulators α 2 and a1: Secondary structure determination of the a1 homeodomain and changes produced by α 2 interactions. Biochemistry 33 15309–15320. [DOI] [PubMed] [Google Scholar]
- Billeter, M., Qian, Y., Otting, G., Muller, M., Gehring, W.J., and Wuthrich, K. 1990. Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy. J. Mol. Biol. 214 183–197. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain–DNA complex. J. Mol. Biol. 234 1084–1093. [DOI] [PubMed] [Google Scholar]
- Center, R.J., Kobe, B., Wilson, K.A., Teh, T., Howlett, G.J., Kemp, B.E., and Poumbourios, P. 1998. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 7 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, N.D., Kissinger, C.R., Desjarlais, J., Gilliland, G.L., and Pabo, C.O. 1994. Structural studies of the engrailed homeodomain. Protein Sci. 3 1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel, E. and Pabo, C.O. 1998. Comparison of X-ray and NMR structures for the Antennapedia homeodomain–DNA complex. Nat. Struct. Biol. 5 692–697. [DOI] [PubMed] [Google Scholar]
- Garvie, C.W., Hagman, J., and Wolberger, C. 2001. Structural studies of Ets–1/Pax5 complex formation on DNA. Mol. Cell 8 1267–1276. [DOI] [PubMed] [Google Scholar]
- Goutte, C. and Johnson, A.D. 1993. Yeast a1 and α 2 homeodomain proteins form a DNA-binding activity with properties distinct from those of either protein. J. Mol. Biol. 233 359–371. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S., Cordier, F., and Dingley, A.J. 2001. Scalar couplings across hydrogen bonds. Methods Enzymol. 338 111–133. [DOI] [PubMed] [Google Scholar]
- Jin, Y., Zhong, H., and Vershon, A.K. 1999. The yeast a1 and α2 homeodomain proteins do not contribute equally to heterodimeric DNA binding. Mol. Cell. Biol. 19 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A.D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5 552–558. [DOI] [PubMed] [Google Scholar]
- Kissinger, C.R., Liu, B.S., Martin-Blanco, E., Kornberg, T.B., and Pabo, C.O. 1990. Crystal structure of an engrailed homeodomain–DNA complex at 2.8 Å resolution: A framework for understanding homeodomain–DNA interactions. Cell 63 579–590. [DOI] [PubMed] [Google Scholar]
- Klemm, J.D., Rould, M.A., Aurora, R., Herr, W., and Pabo, C.O. 1994. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77 21–32. [DOI] [PubMed] [Google Scholar]
- Li, T., Stark, M.R., Johnson, A.D., and Wolberger, C. 1995. Crystal structure of the MATa1/MAT α 2 homeodomain heterodimer bound to DNA. Science 270 262–269. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Manna, A., Li, R., Martin, W.E., Murphy, R.C., Cheung, A.L., and Zhang, G. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. 98 6877–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting, G., Qian, Y.Q., Billeter, M., Muller, M., Affolter, M., Gehring, W.J., and Wuthrich, K. 1990. Protein–DNA contacts in the structure of a homeodomain–DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 9 3085–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, C.L., Vershon, A.K., Johnson, A.D., and Dahlquist, F.W. 1991. Secondary structure of the homeo domain of yeast α 2 repressor determined by NMR spectroscopy. Genes & Dev. 5 764–772. [DOI] [PubMed] [Google Scholar]
- Phillips, C.L., Stark, M.R., Johnson, A.D., and Dahlquist, F.W. 1994. Heterodimerization of the yeast homeodomain transcriptional regulators α2 and a1 induces an interfacial helix in α2. Biochemistry 33 9294–9302. [DOI] [PubMed] [Google Scholar]
- Qian, Y.Q., Billeter, M., Otting, G., Muller, M., Gehring, W.J., and Wuthrich, K. 1989. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: Comparison with prokaryotic repressors. Cell 59 573–580. [DOI] [PubMed] [Google Scholar]
- Qian, Y.Q., Otting, G., Billeter, M., Muller, M., Gehring, W., and Wuthrich, K. 1993. Nuclear magnetic resonance spectroscopy of a DNA complex with the uniformly 13C-labeled Antennapedia homeodomain and structure determination of the DNA-bound homeodomain. J. Mol. Biol. 234 1070–1083. [DOI] [PubMed] [Google Scholar]
- Quiocho, F.A., Spurlino, J.C., and Rodseth, L.E. 1997. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure 5 997–1015. [DOI] [PubMed] [Google Scholar]
- Stark, M.R., Escher, D., and Johnson, A.D. 1999. A trans-acting peptide activates the yeast a1 repressor by raising its DNA-binding affinity. EMBO J. 18 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra, N. and Bax, A. 1997. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science 278 1111–1114. [DOI] [PubMed] [Google Scholar]
- Tsao, D.H., Gruschus, J.M., Wang, L.H., Nirenberg, M., and Ferretti, J.A. 1995. The three-dimensional solution structure of the NK-2 homeodomain from Drosophila. J. Mol. Biol. 251 297–307. [DOI] [PubMed] [Google Scholar]
- Vershon, A.K., Jin, Y., and Johnson, A.D. 1995. A homeo domain protein lacking specific side chains of helix 3 can still bind DNA and direct transcriptional repression. Genes & Dev. 9 182–192. [DOI] [PubMed] [Google Scholar]
- Werner, M.H., Clore, G.M., Fisher, C.L., Fisher, R.J., Trinh, L., Shiloach, J., and Gronenborn, A.M. 1997. Correction of the NMR structure of the ETS1/DNA complex. J. Biomol. NMR 10 317–328. [DOI] [PubMed] [Google Scholar]
- Wolberger, C., Vershon, A.K., Liu, B., Johnson, A.D., and Pabo, C.O. 1991. Crystal structure of a MAT α 2 homeodomain–operator complex suggests a general model for homeodomain–DNA interactions. Cell 67 517–528. [DOI] [PubMed] [Google Scholar]