Abstract
The apoptosis-associated Par-4 protein has been implicated in cancers of the prostate, colon, and kidney, and in Alzheimer’s and Huntington’s diseases, among other neurodegenerative disorders. Previously, we have shown that a peptide from the Par-4 C-terminus, which is responsible for Par-4 self-association as well as interaction with all currently identified effector molecules, is natively unfolded at neutral pH, but forms a tightly associated coiled coil at acidic pH and low temperature. Here, we have alternately mutated the two acidic residues predicted to participate in repulsive electrostatic interactions at the coiled coil interhelical interface. Analysis of circular dichroism spectra reveals that a dramatic alteration of the folding/unfolding equilibrium of this peptide can be effected through directed-point mutagenesis, confirming that the two acidic residues are indeed key to the pH-dependent folding behavior of the Par-4 coiled coil, and further suggesting that alleviation of charge repulsion through exposure to either a low pH microenvironment or an electrostatically complementary environment may be necessary for efficient folding of the Par-4 C-terminus.
Keywords: Coiled coil, leucine zipper, circular dichroism, apoptosis, protein folding, neurodegeneration, mutagenesis
Par-4 is a 38-kD protein that sensitizes certain cells to apoptotic stimuli (Sells et al. 1994). When expressed at high levels, Par-4 has been linked to neuron death in patients suffering from various neurodegenerative diseases, including Alzheimer’s (Guo et al. 1998) and Huntington’s (Duan et al. 2000) diseases, HIV encephalitis (Kruman et al. 1999), and amylotrophic lateral sclerosis (Pedersen et al. 1999). Low levels of Par-4 expression have been linked to cancers of the prostate (Sells et al. 1994, 1997), colon (Zhang and DuBois 2000), and kidney (Cook et al. 1999). More recently, Par-4 has been shown to selectively induce apoptosis in androgen-insensitive prostate cancer cell lines (Xu et al. 2001).
The C-terminal region of Par-4 is required for apoptotic activity (Sells et al. 1997), and is required for self-association and association with all currently identified potential effector molecules (Diaz-Meco et al. 1996; Johnstone et al. 1996; Page et al. 1999). The primary sequence of Par-4 suggests the presence of a C-terminal coiled coil. Coiled coils are amphipathic oligomerization motifs with a seven residue heptad repeat, denoted (abcdefg)n. A key driving force for coiled coil formation is interhelical association between hydrophobic residues at the a and d positions. In addition, interhelical electrostatic interactions between residues in the e and g′ positions (where the prime distinguishes between monomer units) can contribute significantly to folding propensities, depending upon charge complementary or non-complementary. It is still uncertain to what extent complementary e–g′ interactions contribute to stability, however, it is clear that repulsive e–g′ interactions can significantly destabilize coiled coil formation (Krylov et al. 1994, 1998; Zhou et al. 1994; Kohn et al. 1995, 1997; Jelesarov and Bosshard 1996; Lavigne et al. 1996; Durr et al. 1999).
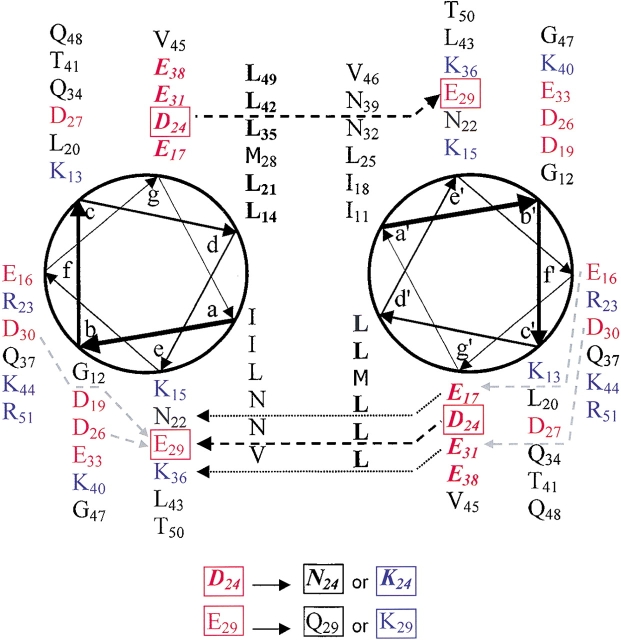
The existence of a coiled coil in a peptide containing the C-terminal 47 residues of Par-4 (Fig. 1 ▶) was recently experimentally established (Dutta et al. 2001). However, coil formation occurred efficiently only at acidic pH and low temperature. Electrostatic repulsion between side chains in the g position (D24) of one helix and the e′ position (E29′) of the interacting helix was hypothesized as the dominant factor preventing coil formation at high pH. It was proposed that complete or partial side chain protonation at low pH may be responsible for alleviation of this (and perhaps other) negative–negative charge clashes, permitting coiled coil formation under acidic conditions.
Figure 1.
Residues 11–51 of the Par-4 WT peptide in a helical wheel format. Positions in the helical wheel are denoted by abcdefg. The Leucine repeat at the d position is shown in bold black letters. Acidic residues are red, basic residues blue. The acidic repeat at the g position is in bold red italics. The black dashed arrows mark selected e–g′ and e′–g interhelical contacts. The gray dashed arrows mark several possible intrahelical charge repulsions involving e and g residues. The residues targeted for mutagenesis are boxed. The substitutions used to create the four mutants in this study are specified at the bottom of the figure.
Here we show that four separate point mutants, targeted to the acidic residues D24 or E29, each exert a substantial effect upon coiled coil formation. The charge-reversing or basic E29K point mutation is capable of converting the above Par-4 peptide from a strongly pH-dependent coiled coil to a pH-insensitive coiled coil throughout the range of pH values tested (5.5–8.0) at low temperature, although some pH dependence does return at significantly elevated temperatures. The alternative D24K mutation produces a similar effect, introducing slightly higher stability than does E29K. Charge neutralizing or polar mutants (D24N and E29Q) behave in an intermediate fashion, with stabilities lying between those of the WT and basic mutants. These results strongly suggest that charge–charge repulsion at the coiled coil interface, and specifically between D24 and E29′, is largely responsible for the pH-dependent folding behavior of the Par-4 C-terminal peptide, and helps to confirm that efficient coiled coil formation by the Par-4 C-terminus in a cellular environment would require exposure to a microenvironment that effectively reduces the local pH, or otherwise neutralizes or complements the charges found at the coiled coil interface.
Results
pH dependence
The D24N and E29Q mutants will be referred to as “polar,” and the D24K and E29K mutants are referred to as “basic.” The data depicted in Figures 2 ▶–7 ▶ was obtained in the presence of 12 mM NaPi and 20 mM NaCl. Figure 2 ▶ shows the ellipticity of the Par-4 WT LZ peptide and four mutant peptides at 5°C, each as a function of pH. As pH decreases from 8.0 to 6.0, the WT fractional helicity increases from 15% to 75% (Fig. 2A ▶). D24N increases in helicity from 33% to 74% (Fig. 2B ▶), and E29Q increases from 37% to 70% helicity (Fig. 2C ▶), between pH 8.0 and 6.0. Thus, the polar mutants increase in helicity by 33% and by 41% over this pH range, while the WT peptide increases by 60%, with all three peptides arriving at 70%–75% helicity at pH 6.0. The principal distinction is that the WT peptide is ∼20% less helical at high pH. In contrast, CD spectra of the basic mutants are much less sensitive to pH, with both D24K (Fig. 2D ▶) and E29K (Fig. 2E ▶) containing 72%–80% helix throughout the pH range tested, a helical percentage that WT and polar mutant peptides approach only near pH 6.0.
Figure 2.
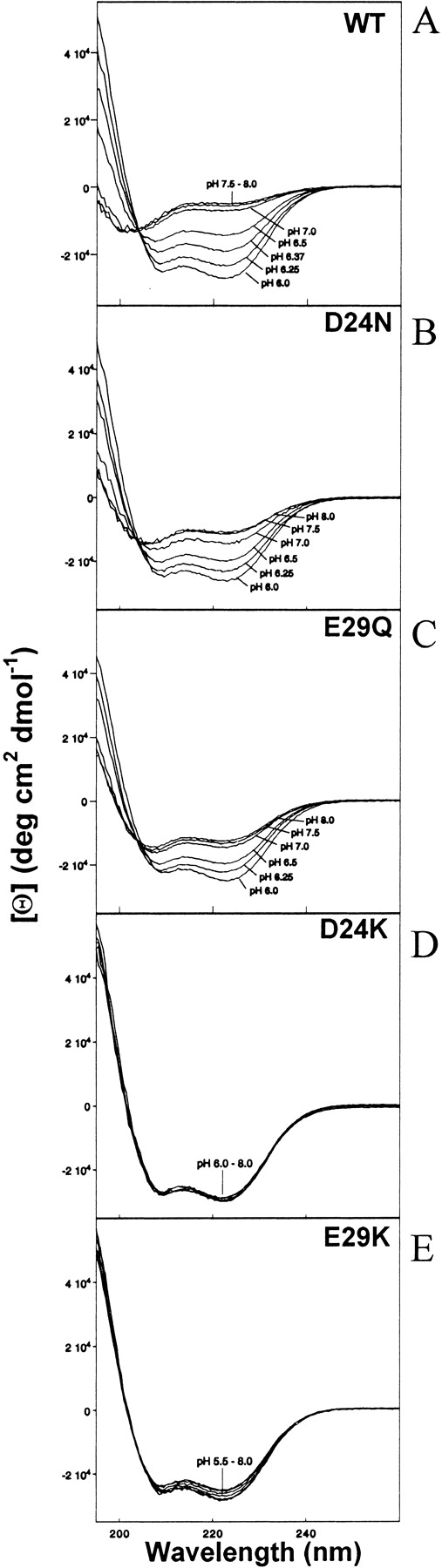
Molar ellipticity versus wavelength at various pH values for (A) 38 μM WT; (B) 34 μM D24N; (C) 40 μM E29Q; (D) 43 μM D24K; (E) 37 μM E29K peptides at 5 °C in 12 mM NaPi and 20 mM NaCl.
Figure 7.
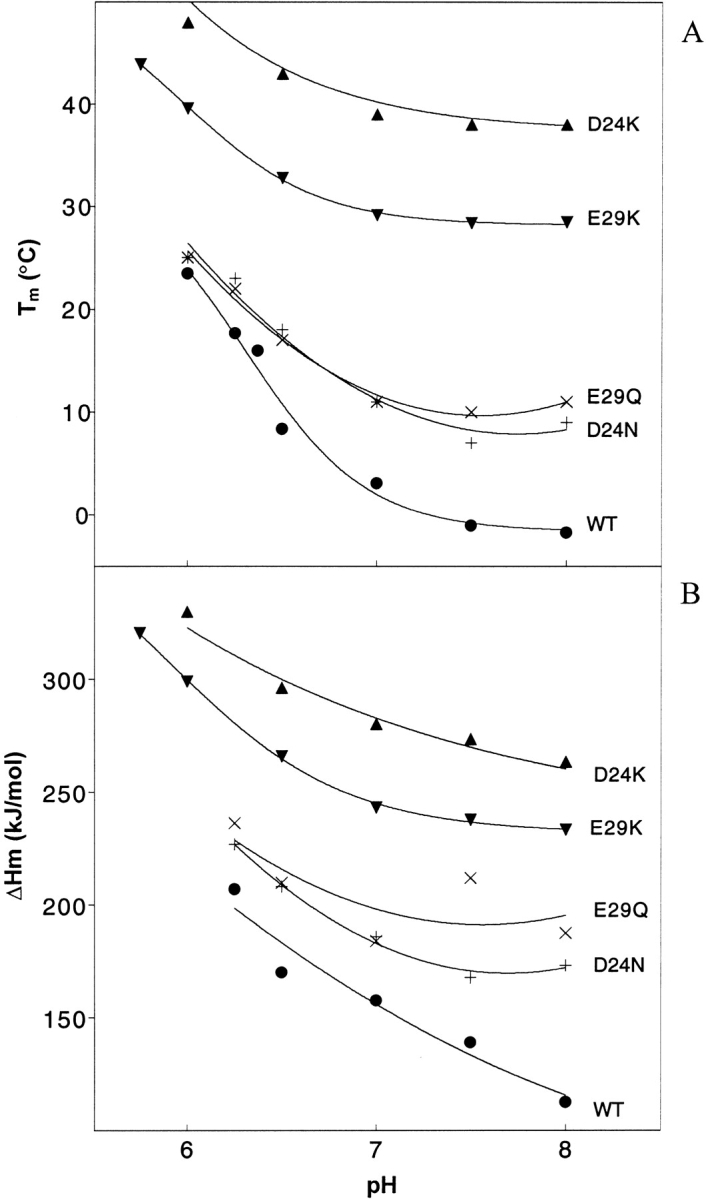
Graphs of (A) Tm and (B) ΔHm as a function of pH for WT and four mutants, derived from fitting of Figure 6 ▶ curves. The symbols and concentrations are as described in the Figure 2 ▶ legend.
Temperature dependence
For brevity, Figures 3 ▶, 4 ▶, and 6 ▶ contain raw data for only WT, D24N, and E29K. Figures 5 ▶ and 7 ▶ present compiled and extracted parameters for WT and all mutants. Figure 3 ▶ shows the change in ellipticity as a function of temperature at pH 6.0. In each panel, as the temperature is increased, the CD spectra reflect a transition from a predominately folded (low temperature) to a predominately unfolded (high temperature) state. The fractional helicity of Par-4 WT (Fig. 3A ▶) decreases from 78% to 16%, a 62% change, as the temperature is increased from 5°C to 40°C. The polar mutant (D24N) shows a similar decrease from 75% to 19%, a 55% change, over this temperature range, with only slight additional changes outside of this range (Fig. 3B ▶). From 5°C to 40°C, the E29K mutant decreases in helicity from 77% to 40%, a 37% change, and further decreases to 8% when the temperature is increased to 65°C (Fig. 3C ▶). By interpolation, the temperature at which 50% helix would be present is approximately 18°C, 23°C, and 37°C for WT, D24N, and E29K, respectively. Thus, the temperature stability is in the order WT < polar < basic mutants.
Figure 3.
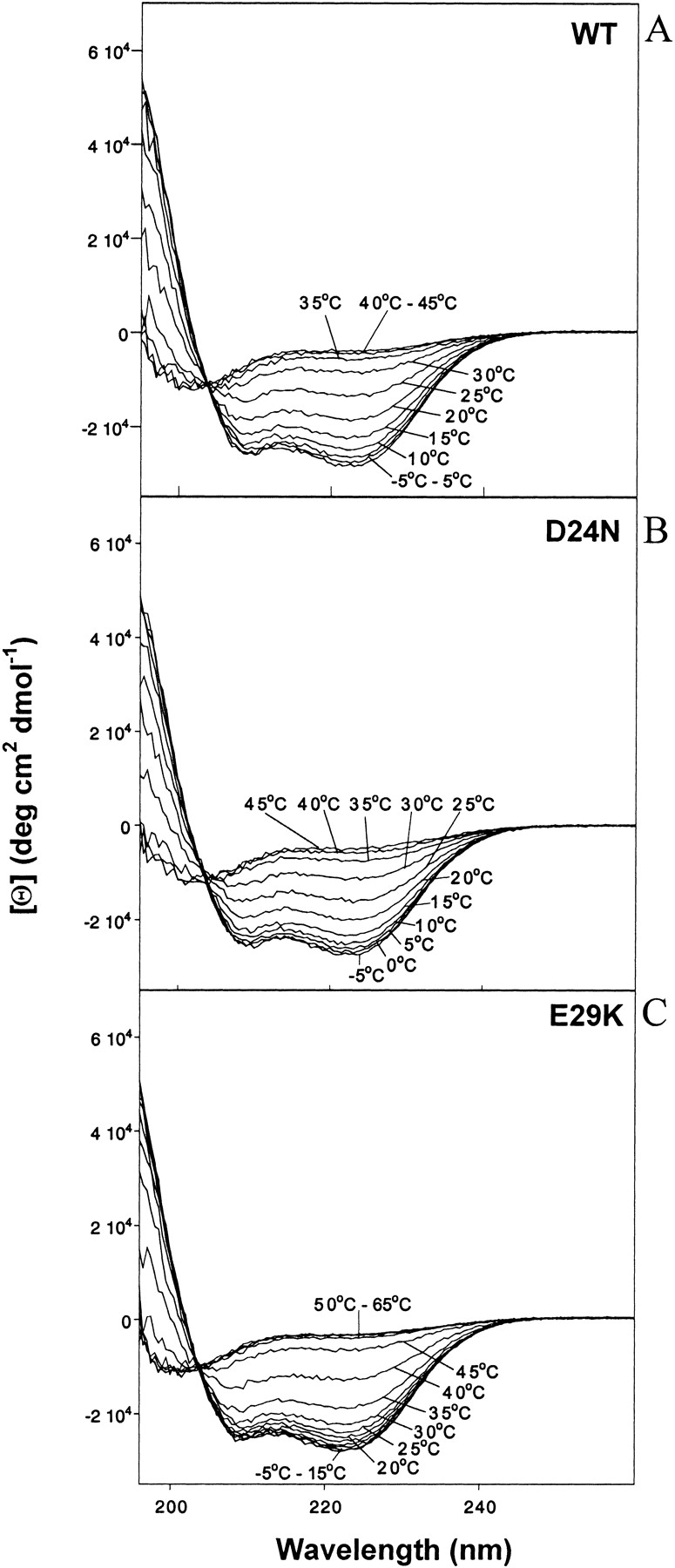
Molar ellipticity versus wavelength at various temperatures for (A) 40 μM WT; (B) 34 μM D24N; (C) 37 μM E29K peptides at pH 6.0 in 12 mM NaPi and 20 mM NaCl.
Figure 4.
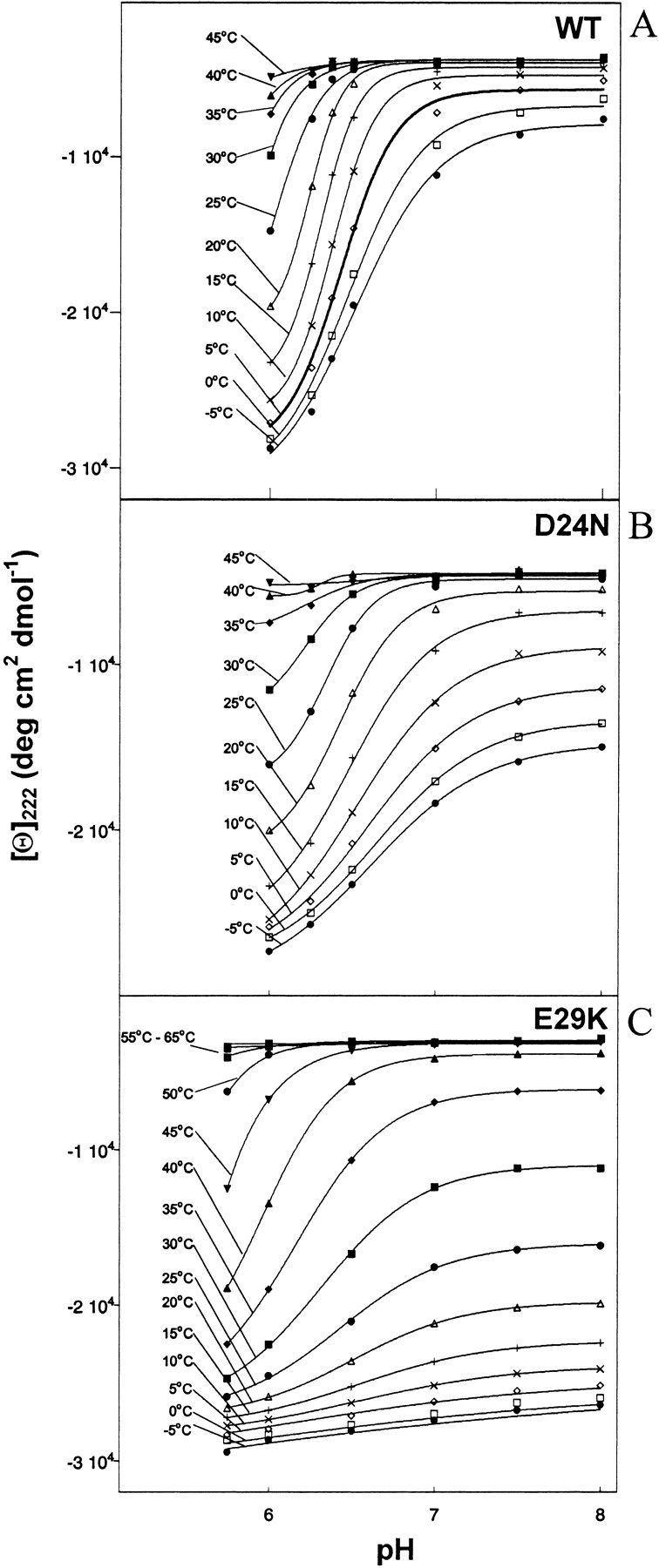
Molar ellipticity at 222 nm versus pH at various temperatures for (A) 38 μM WT; (B) 34 μM D24N; (C) 37 μM E29K peptides in 12 mM NaPi and 20 mM NaCl.
Figure 6.
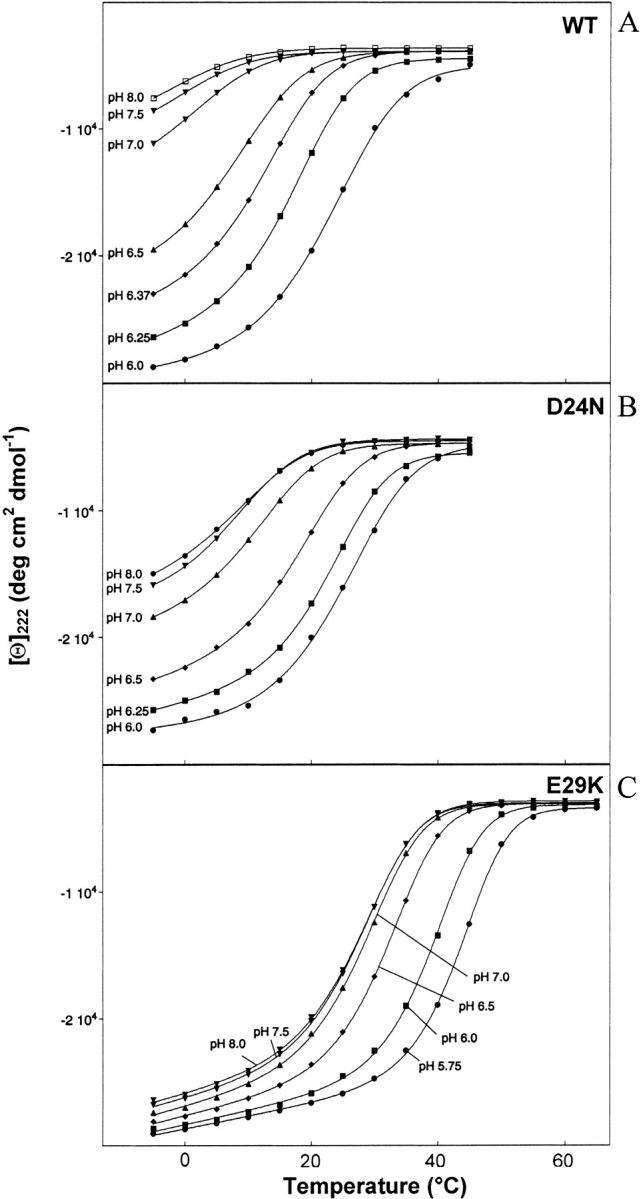
Molar ellipticity at 222 nm versus temperature at various pH values for (A) 38 μM WT; (B) 34 μM D24N; (C) 37 μM E29K peptides in 12 mM NaPi and 20 mM NaCl.
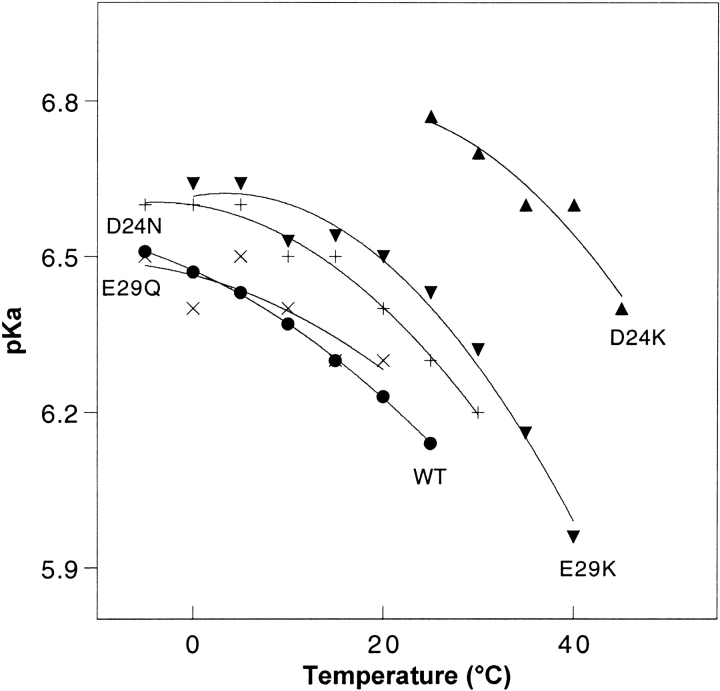
Figure 5.
Conformational pKa values as a function of temperature for WT and four mutants, derived from fitting of Figure 4 ▶ curves. Concentrations are as described in the Figure 1 ▶ legend. Symbols: filled circles, WT; +, D24N; ×, E29Q; filled up triangles, D24K; filled down triangles, E29K.
Conformational pKa
Figure 4 ▶ contains plots of [θ]222 versus pH at several temperatures. Each set of data points was fitted to a modified Henderson-Hasselbalch equation (Dutta et al. 2001) to extract values of what we have chosen to call the conformational pKa. In analogy with the standard use of the term pKa to represent the pH at which titration of a particular charge group is half complete, the conformational pKa represents the pH at which the conformational transition is half complete (Fig. 5 ▶). Extraction of conformational pKa values without large uncertainty is only possible below 25°C for WT and D24N, below 20°C for E29Q, between 0°C and 40°C for E29K, and between 25°C and 45°C for D24K. Outside of these respective ranges, the peptides remain largely insensitive to pH (unfolded at high temperature, or mostly folded at low temperature). Curves within these temperature ranges can be fit with a two-state model, consistent with a two state cooperative transition between what we will call the two end states. These two states represent a less helical (high pH) and more helical (low pH) conformation. Note that the degree to which each of the peptides folds/unfolds varies, and thus the end states for one peptide are not necessarily of the same percentage helicity as the end states for another peptide. Indeed, the two end states for each individual peptide vary as a function of temperature. For instance, it is clear that at low temperature, high pH does not increase the energy sufficiently to cause unfolding of a large percentage of the E29K coiled coil (Fig. 4C ▶, lower traces). This fact should be considered when interpreting the pH-sensitivity of the peptides (see Discussion).
For each peptide, the conformational pKa drops as the temperature is raised, indicating that when additional thermal energy is provided, a more acidic environment is necessary to induce helicity. Within the above temperature limits, the pKa values lie in this order: WT ≈ E29Q < D24N < E29K < D24K. Although a very acidic environment is required to induce folding of the WT and E29Q peptides, considerably less acidity is required by D24K, and the D24N and E29K mutants are intermediate. Thus, substitution of D24 appears to confer a greater resistance to pH denaturation than a similar substitution at E29. At either the 24 or 29 position, a basic substitution confers greater pH stability than a polar substitution. The D24K mutant is extremely stable presumably because it carries the combined characteristics of a D24 substitution with a basic residue. We will return to this observation in the Discussion section.
Tm and ΔHm
Figure 6 ▶ contains plots of [θ]222 versus temperature at various pH values. The melting curves of the WT (Fig. 6A ▶) are very sensitive to pH, while the E29K basic mutant curves (Fig. 6C ▶) are much less sensitive (more tightly grouped) particularly at low temperature. The D24N polar mutant (Fig. 6B ▶) displays pH sensitivity intermediate between WT and E29K. This is in agreement with the order of pKa values presented in Figure 5 ▶. Folding of the basic mutant appears to be somewhat more cooperative than folding of the WT or polar mutants (Fig. 6 ▶). In addition, when compared to the WT, the midpoint of the polar and especially the basic curves are shifted to the right, particularly at high pH (Fig. 6 ▶). This data can be further analyzed by extraction of the melting temperature, Tm (temperature at which the transition is half complete) and ΔHm (the enthalpy of melting) at each pH by fitting the data to a two state model (Dutta et al. 2001). Results are graphed in Figure 7 ▶. Both the Tm and ΔHm values follow the order: WT < polar < basic. As temperature increases, the WT unfolds first, followed by the polar mutants, and finally by the basic mutants. Correspondingly, more energy (enthalpy) is required to unfold the basic mutants. At pH 8.0, the Tm of the WT is −1.7, the polar mutants are 9 (D24N) and 11 (E29Q), and the basic mutants are 29 (E29K) and 38 (D24K) °C (Fig. 7A ▶). At pH 6.5, ΔHm of the WT is 170.3, the polar mutants are 208.1 (D24N) and 209.9 (E29Q), and the basic mutants are 265.9 (E29K) and 297.9 (D24K) kJ/mole (Fig. 7B ▶). At lower pH values, the gaps may narrow, but the order remains the same at least down to pH 6.0. Thus, the basic substitutions confer more resistance to thermal denaturation than the polar substitutions, particularly at high pH.
Discussion
Par-4 WT
Previous studies have demonstrated that the Par-4 WT peptide forms a coiled coil, but only at low pH and low temperature (Dutta et al. 2001). Isodichroic points near 203 nm (Figs. 2A ▶, 3A ▶) suggest a two-state transition between a largely unfolded and a largely helical conformation. The [Θ]222/ [Θ]208 ratios near 1.1 (Figs. 2A ▶, 3A ▶, bottom traces) and concentration dependence studies (Dutta et al. 2001) indicate that the helical conformation is an intermolecular coiled coil. In addition to the leucine repeat at the d position, an acidic repeat is present at the g position (Fig. 1 ▶). Interhelical interactions between acidic g side chains and residues in the e′ positions are either complementary (E31–K36′), neutral (E17–N22′ and E38–L43′) or repulsive (D24–E29′). Analysis of these interactions, together with the pH-dependent folding of the WT coiled coil, lead us to hypothesize that interhelical charge–charge repulsion between D24 and E29′ (also D24′–E29) is a key factor in destabilizing the coiled coil at high pH (Dutta et al. 2001). Increased acidity may partially protonate or electrostatically shield D24 and/or E29, favoring coiled coil formation.
Mutants
We have created D24 and E29 mutants to further examine the role of these residues in the pH and temperature-dependent folding behavior of the Par-4 coiled coil. As with the WT peptide, isodichroic points near 203 nm (Figs. 2B ▶, 2C ▶, 3B ▶, 3C ▶), [Θ]222/[Θ]208 ratios near 1.1 (Figs. 2B ▶–2E ▶, 3B ▶, 3C ▶, bottom traces) suggest that each of the mutants in this study forms a self-associated coiled coil similar in nature to that formed by the WT peptide. The principal effect of mutation appears to be in the degree to which the coil is induced to form under various conditions.
Elimination of the repulsive D24–E29′ interaction via point mutation to a polar (D24N or E29Q) or basic (D24K or E29K) residue indeed alters the folding behavior significantly. The basic mutations display the greatest stabilizing effect, which can be quantitated in a number of ways: (1) At pH 8.0 and 5°C, WT, polar and basic peptides contain ∼15%, 35%, and 75% helix, respectively (Fig. 2 ▶). (2) At pH 6.0 and 25°C, helical percentages are 42%, 46%, and 70%, respectively (Fig. 3 ▶). (3) Throughout the pH range tested, Tm and ΔHm values of the basic mutants are significantly increased (Fig. 7 ▶). (4) The basic mutants are more resistant to pH-induced denaturation. This last point appears to be evident from the curves of Figure 5 ▶. However, a more careful analysis must consider a few subtle points. As noted in the Results section, the pKa (as well as Tm) values specify transition midpoints, but do not specify the nature of the end states. It can be extrapolated from the data of Figure 4 ▶ that at 25°C, for instance, E29K transitions between end states containing 70% and 46% helix, while D24N converts between end states with 46% and 14% helix between pH 6.0 and 8.0. The E29K transition represents a smaller percentage change. In addition, the E29K end points are more helical than the D24N end points. Thus, when the end states are taken into account, it becomes even more evident that the basic mutants are both more helical and more resistant to helix loss through pH denaturation than the polar mutants.
Elimination of charge–charge clashes has previously been shown to stabilize coiled coil formation by several peptides, while the contribution to stability from charge–charge attraction is less well accepted (Krylov et al. 1994, 1998; Zhou et al. 1994; Kohn et al. 1995, 1997; Jelesarov and Bosshard 1996; Lavigne et al. 1996; Durr et al. 1999; see also Dutta et al. 2001, and discussions and references therein). Here, polar mutants were designed to eliminate repulsion without introducing charge attraction, except those involving partial charges of the introduced amino side-chain group (N24 or Q29). The fact that the basic mutations introduce more stability to pH and temperature denaturation seems to suggest that, at least in this case, charge–charge attraction may also contribute to stability. The difference in ΔHm (ΔΔHm) for basic versus polar mutants, as well as for polar mutants versus WT, is ∼60–70 kJ/mole at high pH (Fig. 7 ▶). By comparison, the strength of a Glu–Lys salt bridge in the low dielectric core of a protein has been estimated at 86 kJ/mole (Voet and Voet 1995), assuming a dielectric constant of 4. The similarity in magnitude to ΔΔHm is intriguing, because stabilization is likely to be related to the loss of two negative–negative repulsions in the polar mutants (D24–E29′ and D24′–E29) and perhaps to the conversion of these into two new salt bridges in the basic mutants (e.g., K24–E29′ and K24′–E29). The decrease in ΔΔHm (Fig. 7B ▶) and in Tm differences (Fig. 7A ▶) as the pH becomes acidic suggests that as charges become neutralized or shielded, the stabilities also converge. However, the location of these side chains on the surface of the coiled coil implies that the relevant dielectric constant is quite high, and hence, the energy of the salt bridges (or charge repulsions) should be reduced accordingly.
D24K versus E29K
Upon WT coiled coil formation, one interhelical and four intrahelical charge–charge interactions are predicted to involve residues D24 or E29 (twice this number when considering the dimer; see Fig. 1 ▶). Of these, one is complementary (D24–R23) and four noncomplementary (E29–D26, E29–D27, E29–D30, and E29–D24′) (Dutta et al. 2001). The E29K mutation eliminates the interhelical (E29–D24′) and three of the four intrahelical (E29–D26, E29–D27, and E29–D30) noncomplementary interactions, whereas the D24K mutant eliminates only the interhelical noncomplementary (E29–D24′) and the complementary (D24–R23) interactions. It might therefore be anticipated that the E29K mutation would be more stabilizing to coiled coil formation than D24K. However, the data shows that D24K is more resistant to temperature denaturation than E29K (Fig. 7A ▶), and that the conformational pKa values for D24K are elevated relative to E29K (Fig. 5 ▶). A detailed analysis of the end states during pH titration (at fixed temperature) or thermal melting (at fixed pH) further supports the superior stability of D24K versus E29K, as the transitions represented in the D24K curves are uniformly between states of higher coiled coil content than are the transitions represented in the E29K curves (see Electronic Supplemental Material). It therefore appears that, at least in this case, elimination of the interhelical repulsive interaction is much more influential to folding than elimination of the intrahelical repulsive interactions.
This hypothesis is consistent with the fact that E29K is not more stable than D24K, but the actual reversal of the predicted relative stabilities of these two mutants suggests that other factors are contributing as well. It is certainly possible that size and shape considerations in the D24K mutant, perhaps involving the K24–E29′ salt bridge or other contacts, may be more favorable than in the case of the E29K mutant. In terms of geometry, the polar substitutions are relatively isosteric, while the basic mutants alter both the shape and the size of the side chain. The D24K mutant results in the largest increase in side chain length, introducing three additional methylene groups, whereas E29K introduces two new methylene. These additional hydrophobic groups may interact with the Leucine-rich hydrophobic core (residues in “a” and “d” positions). The difference in the number of added methylenes could account for the greater stability of D24K versus E29K, and may also contribute to the elevated stability of the basic mutants versus the polar mutants, in which no new methylene groups are introduced. Because hydrophobic interactions are largely entropic in nature, the curves of Figure 7B ▶ suggest that other (enthalpic) factors may also play a role. Additional mutational and thermodynamic analysis may provide further information regarding these enthalpic factors and their magnitude relative to entropic factors.
Site-directed mutagenesis has confirmed that D24 and E29 are key residues in the pH-dependent folding of the Par-4 C-terminal coiled coil. Analysis of isosteric D24N and E29Q mutants suggests that elimination of electrostatic repulsion is critical in the pH-dependent behavior of the WT peptide. Results using charge reversing D24K and E29K mutants suggest that introduction of charge attraction and an increase in hydrophobic surface across the interhelical interface may also play roles in stabilization of these mutant coiled coils. The intermolecular g–e′ electrostatic interaction (between residues 24 and 29′) appears to be a much larger factor in the stability of the coiled coil than are intrahelical electrostatic interactions involving these residues. Taken together, these results suggest that efficient coiled coil formation by the Par-4 C-terminus may require exposure either to a low pH microenvironment or to an electrostatic surface or environment capable of complementing the large number of negative charges present, particularly those at the interhelical interface. In this light, it is interesting to note that a cellular binding partner of Par-4, the atypical isoforms of protein kinase C, localize preferentially to late endosomal compartments (Sanchez et al. 1998), the interior of which are highly acidic. However, it remains to be determined whether Par-4 interacts with atypical protein kinase C isoforms within the endosomal interior.
Materials and methods
Mutant constructs
E29K
Using the wild-type construct as a template (Alexandrov et al. 2001), two separate PCR reactions were performed, the first of which amplified the DNA from 5′ of the MCS to 3′ of the mutation site (primer I: malE forward primer, New England Biolabs, primer II: 5′-GTTGCTCGTTTTCGTCCTTCATGTCATCGAG GTCTC-3′), and the second of which amplified DNA from 5′ of the mutation site to 3′ of the MCS (primer III: 5′-AAGGAAGA GATTGATTTGTTAAATAGAGACCTCGATGACATGAAGG ACGAAAACGAGCAACTAAAG-3′, primer IV: malE reverse primer, New England Biolabs). The two fragments were then separately gel purified, mixed to allow cross hybridization, and extended by PCR to create a small proportion of double stranded DNA spanning the entire MCS region containing the complete Par-4 insertion with the point mutation. This product was further amplified using primers I and IV, digested with BamH1 and Sal1, and incorporated into the H-MBP-3C vector (Alexandrov et al. 2001). The D24K, E29Q, and D24N mutants were constructed similarly using variants of primers II and III.
Expression and purification
Expression and purification of peptides was essentially as previously described for the wild-type Par-4 LZ peptide (Alexandrov et al. 2001). Briefly, mutant peptides were over expressed in BL21 (DE3) codon plus cells as H-MBP-3C fusion proteins. Initial purification was performed via His-tag affinity chromatography. The fusion proteins were subsequently cleaved with recombinant 3C protease, and further purified using a Hi-Trap Q Sepharose column (Amersham-Pharmacia). The eluted proteins were exchanged into aqueous buffer containing 20 mM NaCl and 12 mM NaPi at pH 7.5. The purity as determined via MALDI mass spectroscopy and SDS-PAGE was > 99%. Solutions of 0.1 mM HCl and 0.1 mM NaOH were used to adjust the pH for CD analysis. Because peptides did not contain aromatic residues, sample concentrations were determined via amino acid analysis.
Circular dichroism
An AVIV model 202 CD spectrometer equipped with a thermoelectric sample temperature controller and a 0.1-cm path length cuvette was used to record CD spectra. Data were collected from 260 to 190 nm at an interval of 0.5 nm with an integration time of 2 sec at each wavelength. Each spectrum was baseline corrected using a cuvette containing buffer. Whenever a complete CD spectrum was available (190–260 nm), helical content was estimated using the SELCON program (Sreerama and Woody 1993). When only [θ]222 was available as in some melting or pH-denaturation curves, [θ]222/−35,000 was used (Chen et al. 1974). In our experience with these peptides, these two methods generally agree to within 1% or 2%. Thermal stability was determined by monitoring [θ]222 as a function of temperature at 5°C intervals with 5 min equilibration and a collection time of 30 sec at each temperature. No appreciable deviation from reversibility was observed during pH and temperature variation. At a variety of pH values, the thermodynamic parameters Tm (melting temperature) and ΔHm (enthalpy of melting) were extracted from the temperature dependence of CD spectra (Dutta et al. 2001). Similarly, at a variety of temperatures, the pH dependence of CD spectra was used to extract the conformational pKa values and Hill coefficient by fitting to a modified Henderson-Hasselbalch equation (Dutta et al. 2001), using a nonlinear least-squares regression method.
Electronic supplemental materials
Tables showing the conformational pKa and thermodynamic parameters extracted from the thermal denaturation curves and pH denaturation curves for WT and the four mutants at several Par-4 peptide concentrations. Thermal denaturation data at several NaCl concentrations is also included.
Acknowledgments
This work was supported by U.S. Army Prostate Cancer Research Program award DAMD17-98-1-8551 (to S.M.P.). We thank Stephen Sells, Vivek Rangnekar, Maria Diaz-Meco, Jorge Moscat, Satoshi Ohnishi, Yaqiong Lin, and Huang He.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Par-4, prostate apoptosis response protein number 4
LZ, leucine zipper
MBP, maltose-binding protein
Pi, inorganic phosphate (PO4−)
CD, circular dichroism
[Θ], molar ellipticity
[Θ222], molar ellipticity at 222 nm
[Θ208], molar ellipticity at 208 nm
Tm, characteristic melting temperature
ΔHm, enthalpy of melting
ΔΔHm, difference in enthalpy of melting for the two specified peptides
WT, 51 residue wild type Par-4 peptide corresponding to Par-4 residues 286–332 preceded by a four residue cloning artifact
D24N, E29Q, D24K, E29K: Four point mutants of WT peptide
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0223903.
References
- Alexandrov, A., Dutta, K., and Pascal, S.M. 2001. MBP-fusion with a viral protease cleavage site: One-step cleavage/purification of insoluble proteins. Biotechniques 30 1194–1198. [DOI] [PubMed] [Google Scholar]
- Chen, Y.J., Yang, K., and Chan, K. 1974. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13 3350–3359. [DOI] [PubMed] [Google Scholar]
- Cook, J., Krishnan, S., Ananth, S., Sells, S.F., Shi, Y., Walther, M.M., Linehan, W.M., Sukhatme, V.P., Weinstein, M.H., and Rangnekar, V.M. 1999. Decreased expression of the pro-apoptotic protein Par-4 in renal cell carcinoma. Oncogene 18 1205–1208. [DOI] [PubMed] [Google Scholar]
- Diaz-Meco, M.T., Municio, M.M., Frutos, S., Sanchez, P., Lozano, J., Sanz, L., and Moscat, J. 1996. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell 86 777–786. [DOI] [PubMed] [Google Scholar]
- Duan, W., Guo, Z., and Mattson, M.P. 2000. Participation of par-4 in the degeneration of striatal neurons induced by metabolic compromise with 3-nitropropionic acid. Exp. Neurol. 165 1–11. [DOI] [PubMed] [Google Scholar]
- Durr, E., Jelesarov, I., and Bosshard, H.R. 1999. Extremely fast folding of a very stable leucine zipper with a strengthened hydrophobic core and lacking electrostatic interactions between helices. Biochemistry 38 870–880. [DOI] [PubMed] [Google Scholar]
- Dutta, K., Alexandrov, A., Huang, H., and Pascal, S.M. 2001. pH-induced folding of an apoptotic coiled coil. Protein Sci. 10 2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Q., Fu, W., Xie, J., Luo, H., Sells, S.F., Geddes, J.W., Bondada, V., Rangnekar, V.M., and Mattson, M.P. 1998. Par-4 is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer disease [see comments]. Nat. Med. 4 957–962. [DOI] [PubMed] [Google Scholar]
- Jelesarov, I. and Bosshard, H.R. 1996. Thermodynamic characterization of the coupled folding and association of heterodimeric coiled coils (leucine zippers). J. Mol. Biol. 263 344–358. [DOI] [PubMed] [Google Scholar]
- Johnstone, R.W., See, R.H., Sells, S.F., Wang, J., Muthukkumar, S., Englert, C., Haber, D.A., Licht, J.D., Sugrue, S.P., Roberts, T., et al. 1996. A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms’ tumor suppressor WT1. Mol. Cell. Biol. 16 6945–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn, W.D., Monera, O.D., Kay, C.M., and Hodges, R.S. 1995. The effects of interhelical electrostatic repulsions between glutamic acid residues in controlling the dimerization and stability of two-stranded α-helical coiled-coils. J. Biol. Chem. 270 25495–25506. [DOI] [PubMed] [Google Scholar]
- Kohn, W.D., Kay, C.M., and Hodges, R.S. 1997. Salt effects on protein stability: Two-stranded α-helical coiled-coils containing inter- or intrahelical ion pairs. J. Mol. Biol. 267 1039–1052. [DOI] [PubMed] [Google Scholar]
- Kruman, I.I., Nath, A., Maragos, W.F., Chan, S.L., Jones, M., Rangnekar, V.M., Jakel, R.J., and Mattson, M.P. 1999. Evidence that Par-4 participates in the pathogenesis of HIV encephalitis. Am. J. Pathol. 155 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov, D., Mikhailenko, I., and Vinson, C. 1994. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 13 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov, D., Barchi, J., and Vinson, C. 1998. Inter-helical interactions in the leucine zipper coiled coil dimer: pH and salt dependence of coupling energy between charged amino acids. J. Mol. Biol. 279 959–972. [DOI] [PubMed] [Google Scholar]
- Lavigne, P., Sonnichsen, F.D., Kay, C.M., and Hodges, R.S. 1996. Interhelical salt bridges, coiled-coil stability, and specificity of dimerization [letter; comment]. Science 271 1136–1138. [DOI] [PubMed] [Google Scholar]
- Page, G., Kogel, D., Rangnekar, V., and Scheidtmann, K.H. 1999. Interaction partners of Dlk/ZIP kinase: Co-expression of Dlk/ZIP kinase and Par-4 results in cytoplasmic retention and apoptosis. Oncogene 18 7265–7273. [DOI] [PubMed] [Google Scholar]
- Pedersen, W.A., Luo, H., Kruman, I., Kasarskis, E., and Mattson, M.P. 1999. Involvement of the prostate apoptosis response-4 (Par-4) protein in motor neuron degeneration in ALS. Soc. Neurosci. Abstr. 25 49. [DOI] [PubMed] [Google Scholar]
- Sanchez, P., De Carcer, G., Sandoval, I.V., Moscat, J., and Diaz-Meco, M.T. 1998. Localization of atypical protein kinase C isoforms into lysosome-targetted endosomes through interaction with p62. Mol. Cell. Biol. 18 3069–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama, N. and Woody, R. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209 32–44. [DOI] [PubMed] [Google Scholar]
- Sells, S.F., Wood, Jr., D.P., Joshi-Barve, S.S., Muthukumar, S., Jacob, R.J., Crist, S.A., Humphreys, S., and Rangnekar, V.M. 1994. Commonality of the gene programs induced by effectors of apoptosis in androgen-dependent and -independent prostate cells. Cell Growth Diff. 5 457–466. [PubMed] [Google Scholar]
- Sells, S.F., Han, S.S., Muthukkumar, S., Maddiwar, N., Johnstone, R., Boghaert, E., Gillis, D., Liu, G., Nair, P., Monnig, S., et al. 1997. Expression and function of the leucine zipper protein Par-4 in apoptosis. Mol. Cell. Biol. 17 3823–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet, D. and Voet, J.G. 1995. Biochemistry. John Wiley amd Sons, Inc., New York.
- Xu, Y., Kiningham, K., Rangnekar, V.M., Prachayasitikul, V., St. Clair, D.K., and Chakraborty, M. 2001. Par-4 drives trafficking and activation of Fas and Fasl to induce prostate cancer cell apoptosis and tumor regression. DNA Cell Biol. 20 473–481. [PubMed] [Google Scholar]
- Zhang, Z. and DuBois, R.N. 2000. Par-4, a proapoptotic gene, is regulated by NSAIDs in human colon carcinoma cells. Gastroenterology 118 1012–1017. [DOI] [PubMed] [Google Scholar]
- Zhou, N.E., Kay, C.M., and Hodges, R.S. 1994. The net energetic contribution of interhelical electrostatic attractions to coiled-coil stability. Protein Eng. 7 1365–1372. [DOI] [PubMed] [Google Scholar]