Abstract
Bacteriophage λ integrase (λ-Int) is the prototypical member of a large family of enzymes that catalyze site-specific DNA recombination via single-strand cleavage and the formation of a Holliday junction intermediate. Crystallographic and biochemical evidence indicate that substantial conformational change (i.e., folding) in the catalytic domain of the protein is required for substrate recognition and catalysis. We have examined the solution conformation of the catalytic domain (C170) in the absence and presence of a cognate "half-site" DNA oligonucleotide by electrospray ionization mass spectrometry, and circular dichroism and fluorescence spectroscopy. The distribution of ions in the positive ion electrospray mass spectrum of the free protein reveals the presence of three distinct species in solution, one corresponding to the folded protein, one to the unfolded protein, and one to a dimer. In the presence of DNA, ions are observed only for the protein–DNA complex and the folded form of the free protein. We therefore conclude that DNA binding stabilizes the global fold of the protein in a manner that is consistent with folding-coupled target recognition as a mechanism to control site-specific recombination. Furthermore, we find that inspection of the charge state distribution of ions in electrospray mass spectra provides a quick and effective means to identify conformational heterogeneity of proteins in solution and to investigate dynamic protein–nucleic acid interactions.
Keywords: DNA binding protein, mass spectrometry, binding-induced folding, charge state distribution, λ integrase, C170
Bacteriophage λ integrase (λ-Int) is the prototypical member of a large family of enzymes that catalyze site-specific DNA recombination via single-strand cleavage and the formation of a Holliday junction intermediate (Argos et al. 1986; Landy 1989; Nunes-Duby et al. 1998). The crystal structure of C170-Y342F, a mutant of the catalytic domain of λ-Int in which the catalytic tyrosine nucleophile has been replaced by phenylalanine (Kwon et al. 1997; Tirumalai et al. 1997), revealed no density for a portion of the active site that had been proposed to be flexible based on proteolytic susceptibility (Tirumalai et al. 1997). Indeed, the structure implied that, in the absence of its substrate, the tyrosine nucleophile, adjacent to this disordered region, was located far (>18 Å) from the other conserved catalytic residues (Kwon et al. 1997). This arrangement and the implied flexibility indicated that, in order to form an intact active site, that portion of the protein must undergo substantial conformational changes (i.e., folding) on binding to a DNA substrate. An appealing aspect of the dynamic behavior of the protein is that via folding-coupled recognition of its cognate sequence (Spolar and Record 1994), it could achieve an important degree of control over the recombination reaction (Landy 1989; Kwon et al. 1997). Furthermore, protein dynamics could accommodate the paradoxically observed mechanistic duality in which protein active sites are entirely composed of residues from a single monomer (cis) (Nunes-Duby et al. 1994), or in which the tyrosine nucleophile is provided by a neighboring protein in the Holliday junction complex (trans) (Han et al. 1993).
However, high-resolution structures of λ-Int in complex with its DNA substrate and/or a recombination intermediate are not available, leaving uncertain the nature of the necessary conformational changes. In our laboratory, NMR structural studies of the catalytic domain of λ-Int in complex with its DNA substrate have been complicated by exchange broadening: Although the catalytic domain is capable of performing sequence-specific DNA cleavage, its affinity for its substrate is low (>1 μM; Tirumalai et al. 1998; S. Subramaniam and M. Foster, unpubl.) and thus, under normal solution conditions, C170 is in dynamic exchange between the free and bound states. Consequently, the very dynamic processes that are of most interest for understanding the protein’s function in substrate binding, dissociation and cleavage, have made their detailed investigation by NMR extremely difficult.
Because of its gentle ionization process, electrospray ionization mass spectrometry (ESI-MS) has emerged as a useful tool to study both changes in the three-dimensional conformational states of proteins and their interactions with ligands (for reviews, see Loo 1997; Last and Robinson 1999; Veenstra 1999; Hernandez and Robinson 2001). In particular, when combined with limited proteolysis and measurement of hydrogen/deuterium (H/D) exchange rates, MS can provide detailed insights into the dynamics of local binding and/or folding events. Although very powerful, performing these H/D exchange experiments by MS requires substantial amounts of sample, significant expertise, and nonroutine access to instrumentation.
However, valuable insights into the solution structure of proteins can be readily obtained from analysis of their ESI pattern in volatile buffers. For instance, changes in the distribution of ions in ESI-MS have been used to study pH and temperature denaturation of cytochrome C and ubiquitin (Chowdhury et al. 1990; Mirza et al. 1993), methanol-induced denaturation of myoglobin (Babu and Douglas 2000), assembly of the MtGimC chaperone complex (Fandrich et al. 2000), DNA binding by the gene V, trp repressor and Tus proteins (Cheng et al. 1996; Potier et al. 1998; Kapur et al. 2002), and to identify unfolded species and monomeric intermediates in the assembly of the dimeric DNA binding protein HU (Vis et al. 1998). We report here the results of ESI-MS and spectroscopic experiments to study DNA binding by the catalytic domain of λ-Int (C170). Based on analysis of the charge state distribution of protein ions obtained in the presence and absence of DNA, we conclude that the latter stabilizes a more compact, folded tertiary structure for the protein. Such behavior is consistent with a role for binding-coupled protein folding as a mechanism to control recognition and catalysis.
Results and discussion
ESI-MS of free C170
Positive ion ESI of proteins produces spectra containing ions that differ by charge and the mass of the added protons, distributed over an approximately Gaussian envelope (Fenn 1993; Siuzdak 1996; Dobo and Kaltashov 2001). The width and center of this distribution depend on the tertiary structure of the ionizing molecule in solution: Protein molecules that have been denatured prior to ionization give rise to a broad distribution of highly charged ions (low m/z), reflecting the accessibility of multiple protonation sites on the molecule (Chowdhury et al. 1990; Loo et al. 1991; Mirza and Chait 1997). Folded proteins, on the other hand, have fewer accessible sites of protonation and therefore produce ESI spectra that exhibit a narrow distribution of charge states at higher m/z (Chowdhury et al. 1990; Mirza and Chait 1997; Vis et al. 1998).
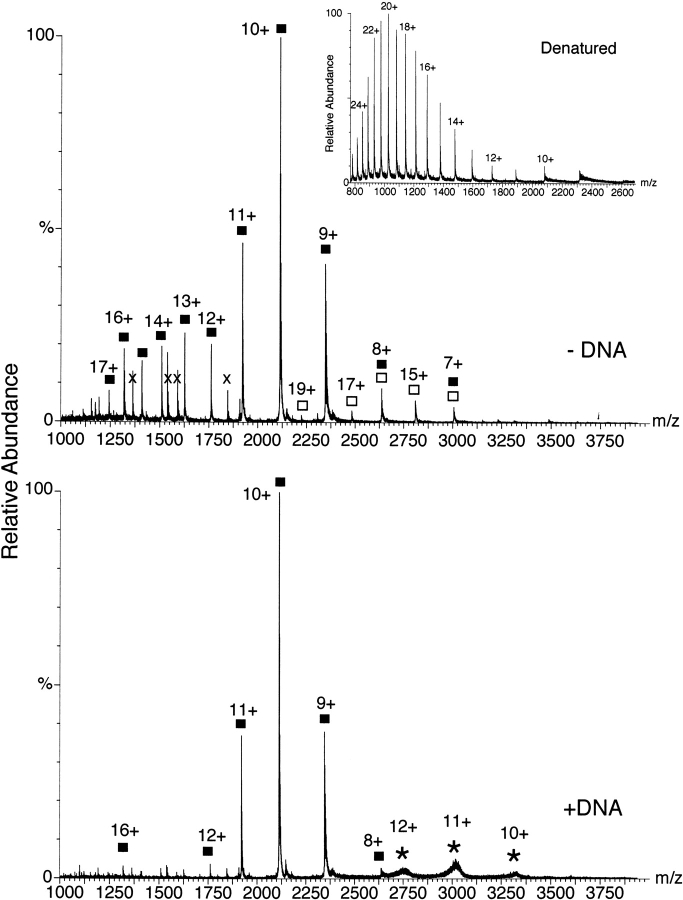
The molecular weight of the C170 protein (residues 170–356 of λ-Int) is 21,119 Da. The multiply-charged ions in the ESI mass spectrum of C170 recorded in the absence of DNA that match those expected for multiple protonation of the protein are indicated with boxes in Figure 1 ▶ (top). At least three species of ions are observed in the spectrum. The most abundant of these is a narrow distribution, with a maximum centered at the 10+ charge state (M+10H)10+, which is characteristic of that expected for a compact folded protein. Abundant ions were observed with m/z of 1920.6, 2112.1, and 2345.6 (1920.9, 2112.9, and 2347.6 expected), corresponding to charge states of 11+, 10+, and 9+, respectively. In addition, two other significant populations of ions are observed. The first of these is a broad distribution of more highly charged species of C170 (ranging from 17+ to 12+: m/z of 1243.5, 1319.2, 1409.2, 1509.7, 1625.9, and 1761.2); this broad distribution and higher charge are consistent with that observed from ionization of denatured proteins. Evidence that these ions correspond to the (partially) unfolded protein was obtained by recording ESI-MS of C170 under denaturing conditions (Fig. 1 ▶, inset).
Figure 1.
Electrospray mass spectra of C170 in the absence (top) and presence (bottom) of a half-site DNA substrate. The ions corresponding to the indicated protonated charge states (M + nH)+n of the free protein (Mr 21,119) are indicated with filled squares, the dimeric C170 (Mr 42,238) with open squares, and the C170–DNA complex (Mr 21,119 + 12,286 = 33,405) with asterisks. Ions marked with an "x" are from a minor contaminant. The spectrum of the free protein (top) exhibits a bimodal distribution of charge states, one corresponding to the folded protein (centered at the 10+ state), and a broader distribution at lower m/z (higher charge) centered at ∼ 15+; these signals result from ionization of unfolded or partially folded protein. The signals corresponding to the unfolded protein are absent in the spectra recorded in the presence of DNA (bottom). The presence of ions corresponding to dimeric C170 indicates that the protein is at least partially dimerized/oligomerized in solution. The spectrum of acid-denatured C170 is shown in the inset, exhibiting the characteristic broad distribution of highly charged species.
The third significant distribution of ions (open boxes in Fig. 1 ▶) corresponds to the mass of the dimeric protein (2 × 21,119 Da = 42,238 Da). Ions from a protein dimer that bear an even charge will overlap those of the monomer with half the charge: m/z = (M2+2nH)2n+ = (M+nH)n+. However, dimer ions bearing an odd charge are readily identified because they appear in a series between those of the protein monomer. Thus, unique (odd-charge) ions matching the protein dimer are observed at m/z of 2223.2, 2483.1, and 2809.7, corresponding to the 19+, 17+, and 15+ charge states, respectively (m/z 2224.1, 2485.6, and 2816.9 expected). Although size exclusion chromatography indicates that the protein is not predominantly dimeric in solution, dynamic light scattering and NMR experiments (S. Subramaniam and M. Foster, unpubl.) reveal that at 500 μM concentrations the protein exhibits a significantly larger effective mass than that expected for the monomeric protein, indicating it may transiently dimerize in solution. At the concentrations of the experiments reported here (10–50 μM), nonspecific aggregation would not be expected, suggesting that the ions reflect a real population of dimeric species in solution. Because the enzyme functions in vivo by assembling into a multimeric complex involving four protein molecules and two DNA strands, this observation could have mechanistic implications.
ESI-MS of C170 in the presence of DNA
Addition to C170 of a DNA hairpin containing a consensus half-site (Nunes-Duby et al. 1989) results in two significant changes in the ESI mass spectrum (Fig. 1 ▶, bottom). In particular, a new set of broad signals corresponding to ions from the intact protein–DNA complex (Mr 21,119 + 12,286 = 33,405) are observed at 12+ through 10+ charge states (m/z 2748, 3016, and 3325 observed; 2785, 3038, and 3342 expected); these ions are broadened because of the tight binding of trace amounts of cations to the anionic DNA (Siuzdak 1996; Crain and McCloskey 1998). These ions reflect the association and joint ionization of the intact protein–DNA complex. Thus, despite the relatively low affinity of the protein for the DNA substrate (>1 μM), the complex is observed in the mass spectrum, confirming the expected 1:1 stoichiometry of the interaction.
However, the most interesting observation was that, in the presence of DNA, the signals derived from ionization of the free protein exhibit only the narrow distribution of charge states expected for the folded protein (centered at 10+). This result is consistent with stabilization of the folded form of the protein by DNA, thereby preventing it from unfolding. Because most of the ions observed in the presence of DNA correspond to the mass of the free protein, we suggest that the DNA may be serving as a sort of “chaperone” in that it interacts transiently with the protein in a manner that stabilizes the folded structure. Stabilization of a protein structure by its interaction with nucleic acids has been observed for many nucleic acid binding proteins, including zinc finger proteins (Foster et al. 1997), the architectural transcription factor Lef-1 (Love et al. 1995), and the trp (Arrowsmith et al. 1991; Potier et al. 1998) and lac repressors (Kalodimos et al. 2002). Such “induced fit” binding has been argued to play an important role in sequence-specific recognition (Spolar and Record 1994) and in λ-Int could play a role in controlling the recombination (Landy 1989; Kwon et al. 1997).
Circular dichroism (CD)
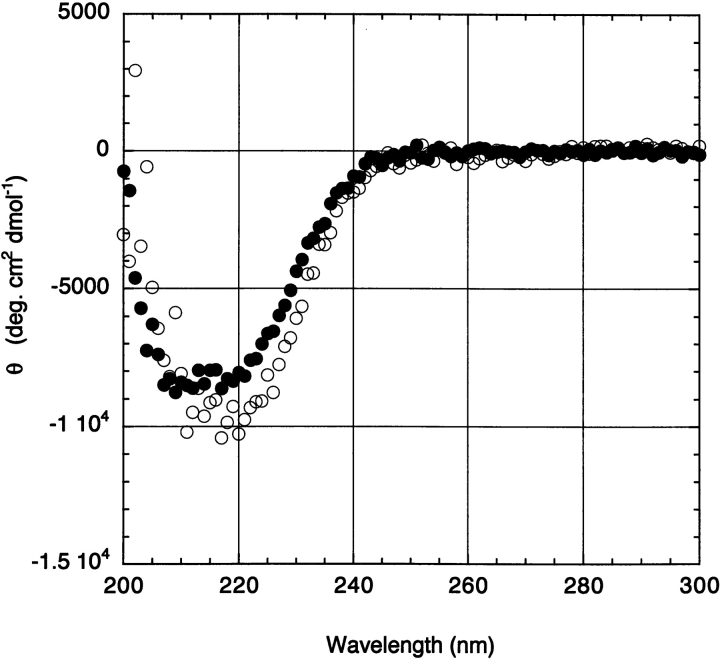
The effect of DNA on the solution structure of C170 was also investigated by far UV CD under identical conditions as the MS experiments (Fig. 2 ▶). The CD spectrum of C170 recorded in the presence of 1:1 DNA reveals a measurable and reproducible increase in ellipticity at 222 nm, consistent with stabilization of the protein’s helical secondary structure by DNA. No additional changes in the protein CD spectrum are observed at DNA stoichiometries greater than 1:1. Although far UV CD is only sensitive to changes in secondary structure, whereas ESI-MS is sensitive to tertiary structure, both measurements are consistent with stabilization of the protein fold by DNA.
Figure 2.
Circular dichroism spectra of the catalytic domain of λ-Int (C170) recorded in the absence (filled circles) and presence (open circles) of a (reversible) half-site DNA substrate. The increased ellipticity at 222 nm reflects a DNA-induced stabilization of α-helical structure in the protein.
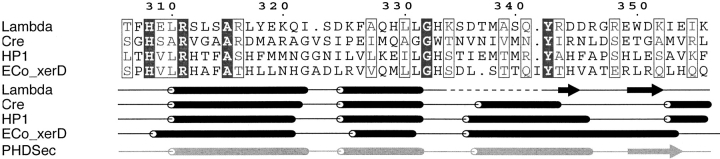
The apparent increased helicity is particularly interesting in the context of the available crystallographic data on λ-Int and related members of the tyrosine recombinase family. Namely, in addition to a conserved mechanism, each of these proteins exhibits substantial topological homology at the tertiary and secondary structural level (Fig. 3 ▶; Nunes-Duby et al. 1998). However, among the family members whose structures have been determined, including Cre (Guo et al. 1997), XerD (Subramanya et al. 1997), HP1 (Hickman et al. 1997), and Flp (Chen et al. 2000), λ-Int differs in that its tyrosine nucleophile was observed to be located on a β-hairpin adjacent to a disordered loop (Kwon et al. 1997), whereas it is present on a helix in the other family members (Fig. 3 ▶). Further, the position of the nucleophile, far removed from the other conserved residues, indicated that, in order for C170 to initiate strand cleavage, dramatic structural rearrangements in the catalytic loop are required.
Figure 3.
Partial sequence alignment of λ-Int and related family members illustrating the sequence and secondary structure conservation. (Lambda) λ-Int (Kwon et al. 1997); (Cre) Cre recombinase (Guo et al. 1997); (HP1) bacteriophage HP1 integrase (Hickman et al. 1997); (Eco_xerD) Escherichia coli XerD recombinase (Subramanya et al. 1997). The secondary structure predicted for C170 by PHDSec (Rost et al. 1994) is indicated. Alignment is from Nunes-Duby et al. (1998) and the figure was generated using ESPript (Gouet et al. 1999). The numbering is according to the λ-Int sequence; His308 and Arg311 are two critical catalytic residues, whereas Tyr342 is the nucleophile. The flexible loop observed in λ-Int (C170 Y342F) is a helix in the other family members.
Although the data presented here do not identify where in the protein the helical structure has been stabilized, it is tempting to speculate that the increase in helical structure in C170 reflects a conformational change that would enable it to recognize and cleave its DNA substrate (Kwon et al. 1997; Tekle et al. 2002). It should be noted that the CD signature reflects an ensemble average, so that it cannot distinguish between an increase in helicity for all of the protein molecules versus stabilization of a population of (partially folded) molecules, whereas the ESI-MS data indicate that there is indeed a population of species that is less compact.
It is also possible that the protein is partially destabilized in ammonium acetate buffer, as reported for the trp repressor (Potier et al. 1998). Indeed, although the CD spectrum recorded in ammonium acetate and phosphate buffer (50 mM sodium phosphate at pH 6.3, 100 mM NaCl, 1 mM dithiothreitol [DTT]) are indistinguishable, the tryptophan fluorescence spectrum recorded in ammonium acetate buffer exhibits reduced emission intensity when compared with that in phosphate (data not shown). Furthermore, as was seen in studies of the DNA binding protein HU (Vis et al. 1998), ESI mass spectra of C170 recorded in high concentrations of ammonium acetate (1 M) revealed only a single distribution of charges, corresponding to that of the folded protein (data not shown). Nevertheless, it is clear from the data in Figures 1 and 2 ▶ ▶ that DNA does indeed stabilize the structure of C170.
Conclusion
Ion suppression and interference from adducts generally precludes performing ESI-MS experiments under physiological conditions (salts, nonvolatile buffers). However, the pioneering work of several laboratories demonstrated that substantial insights into the native states of proteins and their noncovalent interactions with ligands could be obtained by recording ESI-MS spectra in volatile buffers (for reviews, see Loo 1997; Veenstra 1999; Hernandez and Robinson 2001).
The ESI-MS spectrum of C170 indicates that, in the absence of DNA, three distinct species are present in solution: a folded, compact globular species; a partially unfolded conformation; and a dimeric species. On addition of a cognate DNA oligonucleotide, the unfolded species disappears, indicating that the compact species has been stabilized by its interaction with the DNA. The fact that relatively few ions are observed for the ionized protein–DNA complex could reflect the relatively weak (and transient) nature of the C170–DNA interaction, coupled with the potential for dissociation during droplet formation and ion suppression due to the negative charge of the DNA phosphate backbone or the effect of the cations on the spectrum.
Although the ESI spectra do not provide the same detailed level of information obtainable from mass spectrometric measurements of H/D exchange coupled with proteolytic digestion, they can identify the presence of heterogeneous populations of protein conformations in solution, are easy to perform, and require very little sample and only routine access to instrumentation. In addition, it is worth emphasizing that, unlike in aqueous solution where hydrophobic effects dominate intermolecular interactions, electrostatic interactions are favored in the gas phase (Robinson et al. 1996). Thus, although it can be difficult in solution to characterize interactions between molecules that bind transiently through electrostatic interactions, these interactions may be easier to see in ESI-MS. In conclusion, we found that ESI-MS was able to identify DNA-induced stabilization folding of C170, a protein that is in rapid exchange between a free, partially denatured state and a bound, folded state.
Materials and methods
C170, the λ-Int catalytic domain
C170 consists of 187 residues (170–356) of the wild-type λ-Int enzyme. The amino acid sequence for the fragment is AAKSEVRRSR LTADEYLKIY QAAESSPCWL RLAMELAVVT GQRVGDLCEM KWSDIVDGYL YVEQSKTGVK IAIPTALHID ALGISMKETL DKCKEILGGE TIIASTRREP LSSGTVSRYF MRARKASGLS FEGDPPTFHE LRSLSARLYE KQISDKFAQH LLGHKSDTMA SQYRDDRGRE WDKIEIK. The recombinantly expressed protein additionally contains an amino-terminal methionine that is efficiently removed posttranslationally by methionine amino peptidase in vivo, yielding a polypeptide with an expected mass of 21,119 Da.
Expression of C170
The DNA plasmid encoding C170, the catalytic domain of λ-Int, under control of a T7 promoter, and carrying the gene for ampicillin resistance (Tirumalai et al. 1997, 1998), was provided by Arthur Landy (Brown University, Providence, RI). The C170 gene contains eight instances of the rare AGA/AGG arginine codons, three of which occur in the first 10 codons. The frequency of occurrence of these codons implied that overexpression would be highly dependent on the availability of the corresponding rare tRNAs (Brinkmann et al. 1989; Zahn and Landy 1996). Consequently, the argU gene product (also known as dnaY), encoding an arginyl tRNA that can decode these rare codons (Brinkmann et al. 1989; Saxena and Walker 1992), expressed on a vector encoding kanamycin resistance (pARG-U), was cotransformed into C170-expressing cells to boost protein production. C170 was overexpressed in doubly transformed BL21(DE3) cells grown in LB at 37°C in the presence of 50 mg/L carbenicillin and 30 mg/L kanamycin. Cultures were induced with 1 mM IPTG at an OD600 of 0.6 and were grown for 8 h after induction. The cells were harvested by centrifugation (5000 rpm, Sorvall SLA-3000 rotor, 10 min, 4°C) and stored at −20°C until lysed.
Purification of C170 and preparation of samples for mass analysis
C170 was purified from cell pellets as described in (Tirumalai et al. 1998) except that the last step in the purification protocol (a hydroxylapatite column) was replaced with a size exclusion column (Sephacryl SW-100 column, Pharmacia, 1 ml/min) in phosphate buffer (50 mM sodium phosphate (pH 6.3), 0.1 mM EDTA, 1 mM DTT and 100 mM NaCl). For MS experiments, C170 was further exchanged into ammonium acetate buffer (10 mM ammonium acetate (pH 6.3), 1 mM DTT) using a size exclusion column (PD-10, Pharmacia). An acid-denatured sample (pH 3) was prepared by adjusting the pH of the protein solution with acetic acid.
Oligonucleotides
The sequence of the reversible hairpin DNA substrate is 5′-d(CGC TCA AGT TA*G TAT ACG CTT GCG TAT ACT AAC TTG AGC G)-′3, in which the consensus (B′) recognition sequence (Nunes-Duby et al. 1989) is in italics, the site of cleavage is indicated by the asterisk, and the four-nucleotide hairpin is underlined. The molecular weight of the deoxyoligonucleotide hairpin is 12,286 Da. The synthetic oligonucleotide (Integrated DNA Technologies) was dissolved in deionized water (Waters MilliQ, 18 Ω) to make up a stock concentration of 4.2 mM. To favor formation of the hairpin structure, we diluted the stock with ammonium acetate buffer (pH 6.3), heated it to 95°C for 5 min, and cooled it on ice for 15 min.
Protein–DNA complex
The C170 protein–DNA complex was assembled at room temperature by incubating protein and DNA in equimolar ratios for 5 min in ammonium acetate buffer. The mixture was then injected directly into the mass analyzer. The protein concentration (8–50 μM) was maintained constant while recording mass spectra of free protein and complex. The molecular weight of the C170–DNA complex is 21,119 + 12,286 = 33,405 Da.
Mass spectrometry
ESI experiments were performed on a Micromass Q-Tof(tm) II (Micromass) mass spectrometer equipped with an orthogonal electrospray source (Z-spray) operated in positive ion mode. Sodium iodide was used for mass calibration for a calibration range of m/z 100–2500. The capillary potential was set to 3000 V and the cone voltage to 85 V; cone temperature was set to 90°C; the ESI gas was nitrogen. The charge-to-mass ratio of ions were scanned in the range of 1000 to 4000. Samples were infused into the electrospray source at a rate of 10 μL/min. Mass measurements were performed at protein concentrations of 47 and 10 μM, and DNA concentrations of 27 and 10 μM, respectively.
Circular dichroism
CD measurements were performed on an AVIV model 62A DS spectropolarimeter at 25 °C in a 1-mm path-length cell. Protein samples were 10 μM in 10 mM ammonium acetate buffer (pH 6.3) and 1 mM DTT. The effect of DNA on the protein was obtained by subtracting the CD spectrum from the free DNA in the same buffer. Experiments were repeated at least three times, using either 1:1 or 2:1 DNA:protein ratios. Titration curves could not be generated because addition of DNA at low stoichiometry (<0.5:1 DNA:protein) resulted in irreversible protein precipitation.
Tryptophan fluorescence spectroscopy
The structural integrity of the C170 protein in ammonium acetate buffer was examined by intrinsic tryptophan fluorescence emission spectroscopy. Protein samples were 0.5 μM. Samples were excited at 295 nm (5 nm bandwidth) and the emission spectrum recorded from 315 to 450 nm. Spectra were recorded at 25°C using a 1-cm path-length quartz cell using a FluoroMax-3 spectropolarimeter. Fluorescence spectra were also recorded at pH 3.0 to verify that the MS data recorded at this pH corresponded to the unfolded protein.
Acknowledgments
We acknowledge A. Landy and S. Nunes-Duby (Brown University) for reagents and enlightening discussions; G. Suizdak (The Scripps Research Institute) and M. Freitas (OSU) for helpful suggestions and assistance; and the ACS/Petroleum Research Fund, the National Science Foundation, The Ohio State University Research Foundation, and the Ohio Board of Regents for financial support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0234303.
References
- Argos, P., Landy, A., Abremski, K., Egan, J.B., Haggard-Ljungquist, E., Hoess, R.H., Kahn, M.L., Kalionis, B., Narayana, S.V., Pierson, III, L.S., et al. 1986. The integrase family of site-specific recombinases: Regional similarities and global diversity. EMBO J. 5: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith, C.H., Czaplicki, J., Iyer, S.B., and Jardetzky, O. 1991. Unusual dynamic features of the trp repressor from Escherichia coli. J. Am. Chem. Soc. 113: 4020–4022. [Google Scholar]
- Babu, K.R. and Douglas, D.J. 2000. Methanol-induced conformations of myoglobin at pH 4.0. Biochemistry 39: 14702–14710. [DOI] [PubMed] [Google Scholar]
- Brinkmann, U., Mattes, R.E., and Buckel, P. 1989. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene 85: 109–114. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Narendra, U., Iype, L.E., Cox, M.M., and Rice, P.A. 2000. Crystal structure of a Flp recombinase-Holliday junction complex: Assembly of an active oligomer by helix swapping. Mol. Cell 6: 885–897. [PubMed] [Google Scholar]
- Cheng, X., Harms, A.C., Goudreau, P.N., Terwilliger, T.C., and Smith, R.D. 1996. Direct measurement of oligonucleotide binding stoichiometry of gene V protein by mass spectrometry. Proc. Natl. Acad. Sci. 93: 7022–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, S.K., Katta, V., and Chait, B. 1990. Probing conformational changes in proteins by mass spectrometry. J. Am. Chem. Soc. 112: 9012–9013. [Google Scholar]
- Crain, P.F. and McCloskey, J.A. 1998. Applications of mass spectrometry to the characterization of oligonucleotides and nucleic acids. Curr. Opin. Biotechnol. 9: 25–34. [DOI] [PubMed] [Google Scholar]
- Dobo, A. and Kaltashov, I.A. 2001. Detection of multiple protein conformational ensembles in solution via deconvolution of charge-state distributions in ESI MS. Anal. Chem. 73: 4763–4773. [DOI] [PubMed] [Google Scholar]
- Fandrich, M., Tito, M.A., Leroux, M.R., Rostom, A.A., Hartl, F.U., Dobson, C.M., and Robinson, C.V. 2000. Observation of the noncovalent assembly and disassembly pathways of the chaperone complex MtGimC by mass spectrometry. Proc. Natl. Acad. Sci. 97: 14151–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn, J.B. 1993. Ion formation from charged droplets—Roles of geometry, energy, and time. J. Am. Soc. Mass Spectrom. 4: 524–535. [DOI] [PubMed] [Google Scholar]
- Foster, M.P., Wuttke, D.S., Radhakrishnan, I., Case, D.A., Gottesfeld, J.M., and Wright, P.E. 1997. Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA. Nat. Struct. Biol. 4: 605–608. [DOI] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. 1999. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- Guo, F., Gopaul, D.N., and van Duyne, G.D. 1997. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389: 40–46. [DOI] [PubMed] [Google Scholar]
- Han, Y.W., Gumport, R.I., and Gardner, J.F. 1993. Complementation of bacteriophage λ integrase mutants: Evidence for an intersubunit active site. EMBO J. 12: 4577–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, H. and Robinson, C.V. 2001. Dynamic protein complexes: Insights from mass spectrometry. J. Biol. Chem. 276: 46685–46688. [DOI] [PubMed] [Google Scholar]
- Hickman, A.B., Waninger, S., Scocca, J.J., and Dyda, F. 1997. Molecular organization in site-specific recombination: The catalytic domain of bacteriophage HP1 integrase at 2.7 Å resolution. Cell 89: 227–237. [DOI] [PubMed] [Google Scholar]
- Kalodimos, C.G., Boelens, R., and Kaptein, R. 2002. A residue-specific view of the association and dissociation pathway in protein–DNA recognition. Nat. Struct. Biol. 9: 193–197. [DOI] [PubMed] [Google Scholar]
- Kapur, A., Beck, J.L., Brown, S.E., Dixon, N.E., and Sheil, M.M. 2002. Use of electrospray ionization mass spectrometry to study binding interactions between a replication terminator protein and DNA. Protein Sci. 11: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, H.J., Tirumalai, R., Landy, A., and Ellenberger, T. 1997. Flexibility in DNA recombination: Structure of the λ integrase catalytic core. Science 276: 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy, A. 1989. Dynamic, structural, and regulatory aspects of λ site-specific recombination. Annu. Rev. Biochem. 58: 913–949. [DOI] [PubMed] [Google Scholar]
- Last, A.M. and Robinson, C.V. 1999. Protein folding and interactions revealed by mass spectrometry. Curr. Opin. Chem. Biol. 3: 564–570. [DOI] [PubMed] [Google Scholar]
- Loo, J.A. 1997. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 16: 1–23. [DOI] [PubMed] [Google Scholar]
- Loo, J.A., Loo, R.R., Udseth, H.R., Edmonds, C.G., and Smith, R.D. 1991. Solvent-induced conformational changes of polypeptides probed by electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 5: 101–105. [DOI] [PubMed] [Google Scholar]
- Love, J.J., Li, X., Case, D.A., Giese, K., Grosschedl, R., and Wright, P.E. 1995. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376: 791–795. [DOI] [PubMed] [Google Scholar]
- Mirza, U.A. and Chait, B.T. 1997. Do proteins denature during droplet evolution in electrospray ionization? Int. J. Mass Spectrom. Ion Process. 162: 173–181. [Google Scholar]
- Mirza, U.A., Cohen, S.L., and Chait, B.T. 1993. Heat-induced conformational changes in proteins studied by electrospray ionization mass spectrometry. Anal. Chem. 65: 1–6. [DOI] [PubMed] [Google Scholar]
- Nunes-Duby, S.E., Matsumoto, L., and Landy, A. 1989. Half-att site substrates reveal the homology independence and minimal protein requirements for productive synapsis in λ excisive recombination. Cell 59: 197–206. [DOI] [PubMed] [Google Scholar]
- Nunes-Duby, S.E., Tirumalai, R.S., Dorgai, L., Yagil, E., Weisberg, R.A., and Landy, A. 1994. λ Integrase cleaves DNA in cis. EMBO J. 13: 4421–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Duby, S.E., Kwon, H.J., Tirumalai, R.S., Ellenberger, T., and Landy, A. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26: 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier, N., Donald, L.J., Chernushevich, I., Ayed, A., Ens, W., Arrowsmith, C.H., Standing, K.G., and Duckworth, H.W. 1998. Study of a noncovalent trp repressor: DNA operator complex by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 7: 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C.V., Chung, E.W., Kragelund, B.B., Knudsen, J., Aplin, R.T., Poulsen, F.M., and Dobson, C.M. 1996. Probing the nature of noncovalent interactions by mass spectrometry. A study of protein-CoA ligand binding and assembly. J. Am. Chem. Soc. 118: 8646–8653. [Google Scholar]
- Rost, B., Sander, C., and Schneider, R. 1994. PHD—An automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 10: 53–60. [DOI] [PubMed] [Google Scholar]
- Saxena, P. and Walker, J.R. 1992. Expression of argU, the Escherichia coli gene coding for a rare arginine tRNA. J. Bacteriol. 174: 1956–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuzdak, G. 1996. Mass spectrometry for biotechnology, pp. xvi, 161. Academic Press, San Diego, CA.
- Spolar, R.S. and Record Jr., M.T. 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science 263: 777–784. [DOI] [PubMed] [Google Scholar]
- Subramanya, H.S., Arciszewska, L.K., Baker, R.A., Bird, L.E., Sherratt, D.J., and Wigley, D.B. 1997. Crystal structure of the site-specific recombinase, XerD. EMBO J. 16: 5178–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekle, M., Warren, D.J., Biswas, T., Ellenberger, T., Landy, A., and Nunes-Duby, S.E. 2002. Attenuating functions of the C terminus of λ integrase. J. Mol. Biol. 324: 649–665. [DOI] [PubMed] [Google Scholar]
- Tirumalai, R.S., Healey, E., and Landy, A. 1997. The catalytic domain of λ site-specific recombinase. Proc. Natl. Acad. Sci. 94: 6104–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumalai, R.S., Kwon, H.J., Cardente, E.H., Ellenberger, T., and Landy, A. 1998. Recognition of core-type DNA sites by λ integrase. J. Mol. Biol. 279: 513–527. [DOI] [PubMed] [Google Scholar]
- Veenstra, T.D. 1999. Electrospray ionization mass spectrometry: A promising new technique in the study of protein/DNA noncovalent complexes. Biochem. Biophys. Res. Commun. 257: 1–5. [DOI] [PubMed] [Google Scholar]
- Vis, H., Heinemann, U., Dobson, C.M., and Robinson, C.V. 1998. Detection of a monomeric intermediate associated with dimerization of protein Hu by mass spectrometry. J. Am. Chem. Soc. 120: 6427–6428. [Google Scholar]
- Zahn, K. and Landy, A. 1996. Modulation of λ integrase synthesis by rare arginine tRNA. Mol. Microbiol. 21: 69–76. [DOI] [PubMed] [Google Scholar]