Abstract
Increasing resistance of malaria parasites to conventional antimalarial drugs is an important factor contributing to the persistence of the disease as a major health threat. The ongoing search for novel targets has resulted in identification and expression of several enzymes including cysteine proteases that are implicated in hemoglobin degradation. Falcipain-2 and falcipain-3 are considered to be the two principal cysteine proteases in this degradation, and hence, are potential drug targets. A homology model of falcipain-3 was built and validated by various structure/geometry verification tools as well as docking studies of known substrates. The correlation coefficient of 0.975 between interaction energies and Km values of these substrates provided additional support for the model. On comparison with the previously reported falcipain-2 homology model, the currently constructed falcipain-3 structure showed important differences between the S2 pockets that might explain the variations in the Km values of various substrates for these enzymes. Further, docking studies also provided insight into possible binding modes and interactions of ligands with falcipain-3. Results of the current study could be employed in de novo drug design leading to development of new antimalarial agents.
Keywords: Plasmodium falciparum, falcipain, homology modeling, docking
Malaria is one of the most important infectious diseases in the world, is prevalent in more than 90 countries, and affects about 40% of the world’s population (Breman 2001). It is estimated that there are some 300–500 million cases, and between 1.5 to 2.7 million deaths from malaria each year (Butler et al. 1997). The increasing resistance of malaria parasites, in particular Plasmodium falciparum, to antimalarial drugs is a key factor in the persistence of this disease as a major worldwide public health threat. Various potential biochemical targets have been proposed and are being pursued for the de novo design of novel antimalarials (Olliaro and Yuthavong 1999). Among these targets are proteases that hydrolyze hemoglobin and are known to play vital roles at various stages of the parasite life cycle (Klemba and Goldberg 2002; Rosenthal 2002). The intraerythrocytic cycle of infection is responsible for the clinical manifestations of malaria in humans. Intraerythrocytic Plasmodium falciparum trophozoites hydrolyze hemoglobin into amino acids (Rosenthal and Meshnick 1996) in the food vacuole, essential to the survival of the parasite. Proteases like metallo (Eggleson et al. 1999), aspartic (Francis et al. 1994), and cysteine protease (Sajid and McKerrow 2002) are the major reported hemoglobinases present in the food vacuole.
In P. falciparum, three papain-like cysteine proteases have been identified, characterized, and isolated thus far. Falcipain-1 was identified in erythrocytic parasites and found to hydrolyze hemoglobin (Salas et al. 1995). However, its low abundance and difficulties in developing expression systems have limited its study. Recently, two closely related cysteine proteases were identified and expressed. Falcipain-2 was shown to be one of the principal trophozoite cysteine proteases and hemoglobinases (Shenai et al. 2000), and more recently, falcipain-3 was identified in the acidic food vacuole of the parasite (Sijwali et al. 2001). Both proteases require a reducing environment and acidic pH for optimal activity. They differ, however, in that falcipain-3 undergoes efficient transformation into an active enzyme only at acidic pH. It is more active and stable at this pH with greater activity against native hemoglobin. Thus, falcipain-3 is the second P. falciparum hemoglobinase well suited for the hydrolysis of native hemoglobin in the food vacuole. It has been estimated that the concentration of falcipain-2 in trophozoites is 1.8 times that of falcipain-3; however, the latter appears to cleave native hemoglobin about twice as rapidly as the former. Thus, the relative contribution of the two enzymes to the hydrolysis of native hemoglobin is essentially equivalent, making falcipain-2 and falcipain-3 equally important targets for inhibition of hemoglobin degradation (Sijwali et al. 2001).
These findings imply that inhibitors of these enzymes could serve as potential leads for malarial chemotherapy. In fact, various studies have shown that inhibitors of falcipain-1 blocked the hydrolysis of globin and development of cultured parasites (Rosenthal et al. 1991, 1993, 1996; Ring et al. 1993; Olson et al. 1999). A few inhibitors of falcipain-2 have also been designed based on its homology model and shown to be active in vitro in the low micromolar range (Sabnis et al. 2002). Like falcipain-2, the crystal structure of falcipain-3 has yet to be elucidated. Homology modeling, based on sequence similarity among various proteins of the same family, offers a reasonable alternative for structure based drug design in such cases. Thus, to obtain a rational three-dimensional (3D) structure of falcipain-3, comparative protein structure modeling was employed. The resulting model was validated with various structure/geometry verification tools as well as docking studies of known substrates. The docking studies also provided insight into the possible binding modes and interactions of ligands with the enzyme.
Results and discussion
Homology modeling
A homology model for falcipain-3 was derived based on the multiple sequence alignment of the falcipain-3 sequence with homologs as shown in Figure 1 ▶. The average sequence homology of falcipain-3 with the six homologs was 35%. Interestingly, the average pairwise RMS fit of the C-α coordinates of these homologs was 0.72, indicating a strong structural conservation (Fig. 1 ▶) across the cysteine protease family as has been previously reported (Sajid and McKerrow 2002). A total of 11 structurally conserved regions (SCRs) were conceived, which constituted a major segment (about 83%) of the total sequence. The remaining trivial parts of the sequence, that is, the structurally variable regions (SVRs), were not in close proximity to the binding pocket (vide infra). Several key residues of this family of enzymes such as the catalytic cysteine (Cys51), histidine (His183), and asparagine (Asn213) are conserved in falcipain-3 as well. Moreover, glutamine (Gln45) and tryptophan (Trp215) forming the "oxyanion hole" (Sajid and McKerrow 2002) are also preserved.
Figure 1.

Sequence alignment of falcipain-3 (FP3) with homologs of the cysteine protease family. Residues in box represent the structurally conserved regions (SCRs). The sequences are labeled with PDB codes. Vertical lines in the sequences indicate missing residues in the PDB structures. Only the mature sequence of falcipain-3 starting from Thr243 (original numbering) is presented here. Unless otherwise stated, the numbering of the residues in this study reflects the ones shown in this figure.
Model evaluation
Refinement of the homology model resulted in a structure with RMSD of 1.55 for the heavy atoms and only 1.22 for the backbone, compared to the unrefined structure. An average value of −0.17 kT of the MatchMaker (Godzic et al. 1992) score suggested a reasonable 3D model. The Ramachandran plot (Edsall et al. 1966) showed a normal distribution of points with Phi (φ) angles mostly restricted to negative values and Psi (ξ) values clustered in a few distinct regions with 98% of residues occupying the allowed region. ProStat check for bond lengths, C-α chirality, amide torsion (ω), Phi and Psi torsions for helices, Phi for Prolines and side chain torsions (χ1 and χ2) showed no major deviation from the corresponding allowed values. Only 3.2% of the total residues showed somewhat higher values (≤5.0 standard deviations) for bond angles. However, none of these residues were part of the binding site. Profiles-3D (Lüthy et al. 1992) analysis suggested only one misfold across residues 73–77. On further investigation it was realized that the average C-α distance of these residues from that of active site Cys51 was 16 Å and the minimum distance was 11 Å. Hence, this misfold was considered trivial. The overall self-compatibility score for the model was 86.99, as against a score of 45.55 or less, the latter would indicate an almost certainly incorrect structure. The secondary structure prediction by PSIPRED (Jones 1999; McGuffin et al. 2000) suggested five helices and six major strands with the remaining structure being predicted as coil. These results coincided with the secondary structure of the final model (Table 1). In summary, the above-mentioned analyses indicate that the model structure is consistent with current understanding of protein structure.
Table 1.
Secondary structural motifs of the falcipain-3 model
| Structural motifs | Homology model |
| Helix-1 | W33-H36 |
| Helix-2 | W52-K68 |
| Helix-3 | Q77-C82 |
| Helix-4 | I94-D103 |
| Helis-5 | K146-Y151 |
| Strand-1 | Y137-S139 |
| Strand-2 | I155-I159 |
| Strand-3 | H183-G191 |
| Strand-4 | Y208-K212 |
| Strand-5 | Y224-E228 |
| Strand-6 | A244-P247 |
| Turn-1 | L104-G105 |
| Turn-2 | Q110-D112 |
| Turn-3 | D164-A166 |
| Turn-4 | G216-D218 |
| Turn-5 | G220-G223 |
| Turn-6 | T237-S239 |
Structure of falcipain-3
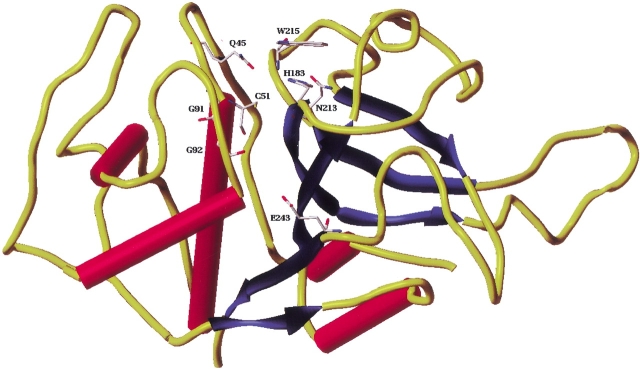
As expected, the model showed structural properties characteristic of the cysteine protease family of enzymes (Maes et al. 1996; Gillmor et al. 1997; Selzer et al. 1997; Zhao et al. 1997; Choi et al. 1999). The important secondary structural motifs comprising the five helices, six strands, and six turns are described in Table 1. Residues that constitute the binding pocket surround the catalytic Cys51, located at the N-terminus of helix-2, as shown in Figure 2 ▶. In proximity to Cys51 is His183 (N-terminus of Sheet-3), which may be involved in thiolate/imidazolium ion pair formation as has been observed in the case of other cysteine proteases (Leung et al. 2000). The appropriate orientation for this ion pair formation may be facilitated by an asparagine (Asn213). One of the key events in the catalytic hydrolysis of the peptide bonds is the nucleophilic attack of the thiolate anion on the appropriate electron deficient carbonyl of the substrate. This 1,2-addition results in the formation of a tetrahedral adduct bearing a negative charge, which is reported to be stabilized by the "oxyanion hole" formed by side-chain amide protons of glutamine and indole hydrogen of tryptophan (Sajid and McKerrow 2002). In falcipain-3, as in other enzymes of the family, Gln45 and Trp215 are in a similar orientation (Fig. 2 ▶) in the active pocket, and hence, can be proposed to play a parallel role in the stabilization of this hemithioketal. Furthermore, the glycine rich region (Brinen et al. 2000) comprising mainly of Gly91 and Gly92 may provide additional stability to the complex by forming a constellation of hydrogen bonds with the substrates/inhibitors.
Figure 2.
Structure of falcipain-3 model. Red cylinders show the helix; blue ribbons, strands; yellow tubes, turns/coils. Important residues are represented as solid sticks and colored by atom types. In this and in the following figures hydrogens are not shown for clarity.
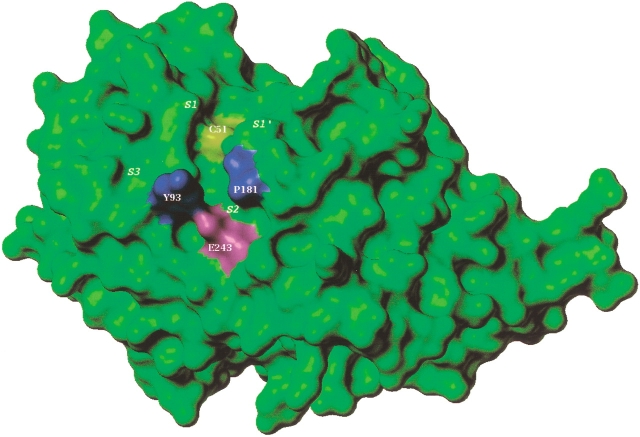
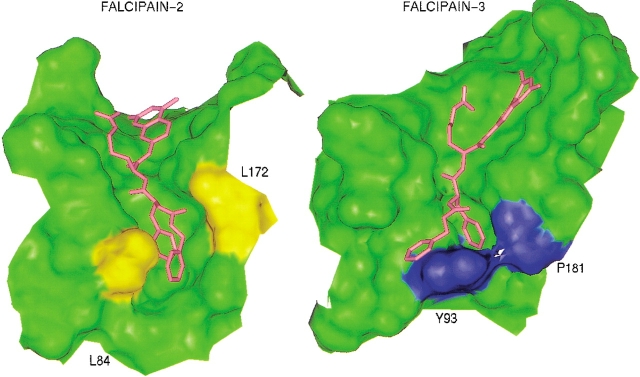
Residues within 6 Å of the active site Cys51 line the binding pocket and are listed in Table 2. The active site of the cysteine protease family of enzymes is generally considered to be constituted by four pockets as shown in Figure 3 ▶, viz. S1, S2, S1‘, and S3 (Sajid and McKerrow 2002). The S1 pocket is the least defined pocket in cysteine proteases, which usually holds the glutamine of the "oxyanion hole." The most well-defined pocket governing ligand specificity is the S2 pocket. Although most of the residues here are hydrophobic, occasionally a polar residue like glutamic or aspartic acid is present at the hollow end of the pocket, for example, falcipain-2 and cruzain. One of the highly conserved residues in the S1‘ pocket is tryptophan, which is known to participate in hydrophobic interactions with substrates. The glycine rich region of the binding site represents the S3 pocket. In falcipain-3, the overall topology of the active site is similar to that of other family members because the majority of the binding site residues are conserved. Interestingly, there are significant differences observed which might lead to diverse ligand specificity. These variations are vital when compared to falcipain-2 because these two enzymes are key hemoglobinases in P. falciparum. The important differences between falcipain-2 and falcipain-3 are given in Table 2. The major alterations observed in the S2 specificity pocket might explain trends in the experimentally observed Km values for various substrates (Sijwali et al. 2001). The two leucine residues, Leu84 and 172, present on either end of the S2 pocket in falcipain-2, are replaced by tyrosine (Tyr93) and proline (Pro181), respectively, in falcipain-3. A detailed examination of the binding pockets reveals that the distal end of the S2 pocket in falcipain-3 is narrower than that in falcipain-2. As seen in Figure 4 ▶, this important steric difference appears to be mainly the result of bulkier tyrosine and proline residues in falcipain-3. This might also explain the smaller relative volume (324 Å3) of the binding pocket in falcipain-3 compared to 538 Å3 in falcipain-2, providing that the correct rotamers for both Tyr93 and Pro181 had been determined. A study of the various rotamers of these residues revealed that the current structure was indeed the one with the lowest overall nonbonded energy and minimum number of steric overlaps. Another important difference was replacement of aspartic acid in falcipain-2 (Asp234) with glutamic acid in falcipain-3 (Glu243). However, this appears to be a minor change as this residue lies at the end of the pocket and does not significantly contribute to the shape and volume of the cavity (Fig. 4 ▶). Moreover, both residues provide a similar negative electrostatic environment for ligand interactions. It is noteworthy that the S2 pocket of the human cysteine protease cathepsin K (Zhao et al. 1997) is completely hydrophobic as the residues lining the pocket are nonpolar (Table 2). In addition, there are other differences between the binding pockets of human versus plasmodial cysteine proteases, as shown in Table 2. These differences may allow development of selective inhibitors.
Table 2.
Important residues lining the binding pockets of falcipain-3, falcipain-2, and cathepsin Ka
| Subsites | Falcipain-3 | Falcipain-2b | Cathepsin Kc |
| S1 | Q45-G49-C89-Y90 | Q36-G40-C80-N81 | Q19-G23-C63-G64 |
| S2 | Y93-I94-S158-P181-E243 | L84-I85-S149-L172-D234 | Y67-M68-A134-L160-L209 |
| S1′ | A160-A161-S162-A166-H183-W215 | V150-V152-S153-A157-H174-W206 | D136-A137-S138-F142-H162-W184 |
| S3 | K85-N86-N87-G91-G92 | K76-N77-Y78-G82-G83 | E59-N60 -D61-G65-G66 |
a The altered residues compared to falcipain-3 are in bold.
bBased on the homology model of falcipain-2 (Sabnis et al. 2002).
c The residues are numbered as in 1ATK.pdb.
Figure 3.
Connolly surface of the binding pocket of falcipain-3 model. Subsites are labeled as S1, S2, S1‘, and S3.
Figure 4.
A close view of the S2 pockets of falcipain-3 (green) and falcipain-2 (blue; Sabnis et al. 2002). Differences in residues are presented as a space-filled model, whereas other important residues are shown as solid sticks. Cysteines are colored yellow while Glu243 (in falcipain-3) and Asp234 (in falcipain-2) are shown in red.
Docking studies
Docking studies not only provide an understanding of the binding mode of the ligands but have also been employed to validate homology models (Folkers 1998). The task of docking known substrates of falcipain-3 was undertaken to authenticate the model by calculating the correlation between the Km values of the substrates (Sijwali et al. 2001) and their respective docking scores. Also, the possible binding mode of these ligands was exploited and compared with that in falcipain-2. The interaction energies calculated for the docked substrates are shown in Table 3. An excellent correlation coefficient of 0.975 between the Km values and the interaction energies was observed. Thus, the model can explain variations in the Km values, which in turn, is related to binding affinities of the substrates, implying that it had been constructed with reasonable accuracy.
Table 3.
Interaction energies and Kmvalues for substrates in falcipain-3
| Substratesa | Kmb (μM) | Interaction energies (kcal/mole)c |
| Z-Phe-Arg-AMC | 71.3 | −21.9 |
| Z-Leu-Arg-AMC | 72.0 | −16.5 |
| Z-Val-Arg-AMC | 185.0 | −7.2 |
| Z-Val-Leu-Arg-AMC | 27.4 | −29.8 |
| Z-Val-Val-Arg-AMC | 39.8 | −27.1 |
| Boc-Val-Leu-Lys-AMC | 10.9 | −32.4 |
a Abbreviations used: Z-benzyloxycarbonyl; AMC-7-amino-4-methyl coumarin; Boc-tertiary butyloxy carbonyl.
c Correlation coefficient between interaction energies and Km was calculated to be 0.975.
Comparison of the best docking modes of various substrates exhibited common binding characteristics among them. As an example, in Figure 5 ▶, the docking of Z-Phe-Arg-AMC in falcipain-3 reveals that the AMC moiety occupies the S1‘ pocket where it is involved in a hydrophobic interaction with Trp215; the benzyloxycarbonyl group is positioned between the S2 and S3 pockets with more inclination toward the latter and the phenyl ring lies in the S2 pocket. Although the arginine in this case occupies the S1 pocket, in other substrates it is either positioned between the S1 and the S1‘ pockets or in the S1‘ pocket. In case of substrates with three amino acid residues, the central residue is observed to be in vicinity of S1‘ pocket. Table 4 describes residues involved in hydrogen bonding with various docked substrates. Residues corresponding to Gln45, Gly92, His183, and Trp215 in falcipain-3 have been implicated in ligand interactions in other cysteine proteases (Sajid and McKerrow 2002). These residues are indeed observed to form H-bonds with the substrates in the current study.
Figure 5.

Docked orientation of Z-Phe-Arg-AMC (red) in falcipain-3. Only the crucial binding site residues are shown and colored by atom type. Substructures of the substrate are labeled blue. Z-benzyloxycarbonyl, AMC-7-amino-4-methyl coumarin.
Table 4.
Residues involved in H-bond formation with the substrates for falcipain-3
| Substratesa | Residues |
| Z-Phe-Arg-AMC | Q45, H183, W215 |
| Z-Leu-Arg-AMC | G92, N182 |
| Z-Val-Arg-AMC | G92 |
| Z-Val-Leu-Arg-AMC | Q45, G92, N182, H183 |
| Z-Val-Val-Arg-AMC | G92 |
| Boc-Val-Leu-Lys-AMC | Q45, G92 |
a For abbreviations, refer to footnotes in Table 3.
A correlation coefficient of 0.904 was calculated between the Km values for the substrates in falcipain-2 (Sijwali et al. 2001) and the corresponding interaction energies. When compared to the docking of these substrates in falcipain-2, it was observed that the overall binding mode in both the enzymes was similar (Fig. 6 ▶). Both in falcipain-2 and falcipain-3, the distance between sulfur of catalytic Cys51 and the point of cleavage of the substrate (carbonyl carbon of arginine) was observed to be in the range of 3–5 Å. This is in accordance with the experimentally observed binding modes of various ligands in other cysteine proteases like cruzain from Trypanosoma cruzi (Brinen et al. 2000). However, important differences were noted in regards to the position of the benzyloxycarbonyl group. In falcipain-2, this group fits into the S2 pocket along with the phenyl group whereas in falcipain-3 it is displaced from the center of the S2 pocket. On close inspection, the displacement of this group in falcipain-3 appeared to be mainly due to the narrower S2 pocket, clearly illustrated in Figure 6 ▶. This finding might explain the lower Km values (Sijwali et al. 2001) for all substrates in falcipain-2 compared to falcipain-3.
Figure 6.
Docked structure of Z-Phe-Arg-AMC (red) in falcipain-3 and falcipain-2. Connolly surfaces of only binding pockets are shown. L84 and L172 in falcipain-2 are colored yellow while Y93 and P181 in falcipain-3 are rendered blue. Z-benzyloxycarbonyl; AMC-7-amino-4-methyl coumarin.
Materials and methods
Computational methods
Computational studies were performed on a Silicon Graphics Octane 2 workstation, equipped with two parallel R12000 processors, V6 graphics board, and 512 MB memory. Homology modeling was performed with the COMPOSER module of SYBYL 6.7 (Tripos Associates Inc.). Energy minimizations and molecular dynamics were accomplished in the DISCOVER module of InsightII (Accelrys Inc.). The geometrical and local environmental consistency of the model was evaluated with the PROSTAT and Profiles-3D modules of InsightII. Secondary structure prediction was performed by a PSIPRED protein structure prediction server, and the AFFINITY (Luty et al. 1995) module of InsightII was employed for substrate docking.
Homology modeling
The falcipain-3 sequence (GenBank accession no. Q9NAW4) was obtained from the SWISS-PROT and TrEMBL databases of the ExPASy Molecular Biology Server (Bairoch and Apweiler 2000). Only the mature sequence (Fig. 1 ▶) starting from residue Thr243, as per the numbering in the complete sequence (Sijwali et al. 2001), was considered in deriving the homology model. A WU-BLAST 2.0 (Gish 1996–2002) PDB search was performed on the mature falcipain-3 sequence with default parameters of BLASTP gapped alignment. We employed WU-BLAST 2.0 because the searches were more rapid than the ungapped version 1.4 programs, while using identical ungapped parameters. Crystal structures of cysteine proteases from various sources were identified as homologs for the query search. Only those sequences that passed the identity filter (>30%) and had a significance score of 21 or more were chosen as closely homologous to the query sequence and were retained for building the 3D model. This included homologs belonging to the hydrolase family of enzymes: cathepsin K (1ATK), cysteine protease from Zingiber officinale (1CQD), chymopapain from Carica papaya (1YAL), cruzain (1AIM and 1EWP) from Trypanosoma cruzi, and two theoretical models: leishmania cysteine protease from Leishmania major (1BMJ) and falcipain-2 from Plasmodium falciparum (Sabnis et al. 2002). Three different models were constructed. One based only on the crystal structures (Model 1) whereas, 1BMJ and falcipain-2 models were utilized in addition to the crystal structures to get the second (Model 2) and the third (Model 3) model, respectively. Profiles-3D predicted six misfolds for Model 1, one for Model 2 and five for Model 3. It was surprising to notice five misfolds for Model 3 because falcipain-2 model itself was not shown to have any misfolds. Further comparison of Model 1 and Model 2 revealed several important differences. Model 1 was shown to have 18 residues in the misfolded region compared to five residues in Model 2. This was also reflected in a lower Profiles-3D score (80.5) for Model 1 against 86.99 for Model 2. The phi-psi occupancy for Model 1 was 80% versus 98% for Model 2. Also, the energy as calculated by cvff forcefield for Model 1 was 490.01 kcal/mole compared to 410.9 kcal/mole for Model 2. RMSD of backbone atoms of the residues lining the binding pockets of Model 1 and Model 2 was found to be 0.6, whereas that for all backbone atoms was 2.02. These observations suggested that the conformation of binding pockets in both the models was quite similar, and hence, either of the models could be used for further studies. However, based on the overall folding and energy values as discussed above, it was decided to use Model 2 for all further studies.
The multiple sequence alignment among the template structures and the target sequence was performed against each of the homolog sequences using the method of Needleman and Wunsch (1970). Gaps were inserted into the sequences to find an optimal alignment, based on the length-independent gap penalty of 6. Initial sets of topologically equivalent residues in the reference proteins were used to generate an optimal structural alignment for the family of homologs. The resulting alignment was manually refined to optimize the matching of several characteristics including (a) conserved protease hydrophobic side chains and buried positions in the template structures, (b) functionally important sites such as the active site cysteine, and (c) insertions/deletions. Because the alignment and superposition of the six structures showed that the common fold began at Arg28 of the falcipain-3 sequence, the 27-residue N-terminus was not considered for building the model. Thus, the final model was built for 223 residues. Eleven SCRs were conceived based on this alignment and are shown in Figure 1 ▶. Cruzain, having an overall percent identity of 35% and more than 80% in the SCRs with falcipain-3 (Fig. 1 ▶), was chosen as a major contributor to derive coordinates for building the SCRs.
The last step in the generation of the 3D model was building the structurally variable regions (SVRs) or the loops onto the SCRs. The Tweak Loop (Shenkin et al. 1986,Shenkin et al. 1987) approach was used to build loops onto the SCRs using the default parameters in COMPOSER. Different rotamers for the residues that line the active pocket were also studied and the most energy stable rotamer was retained.
Refinement
At this point, a theoretically reasonable model was constructed with a few trivial abnormalities, which were further corrected by minimization using the DISCOVER module of InsightII. The model was subjected to minimization for 2000 iterations of steepest descents followed by conjugate gradients, in two stages to a gradient of 0.001 kcal/mole/Å or less. In the first stage, the heavy atoms were tethered with a force constant of 100 kcal/Å, while in the second stage, the force constant was reduced to 75 kcal/Å. This protocol resolved all major geometrical defects with minimum deviation (RMSD 1.55 for heavy atoms) from the coarse model, as desired.
Docking
In the absence of reported inhibitors against falcipain-3, the homology model was validated by docking studies of various substrates for which the Km values were available. The substrates were built and minimized for 1000 steps each of steepest descents followed by conjugate gradients and finally by the BFGS method to a gradient of 0.001 kcal/mole/Å or less. The starting orientations were obtained by placing the substrates in the pocket surrounding Cys51 and intermolecular interaction energies were reduced to reasonable values by manual adjustments. The affinity docking was then performed by AFFINITY employing the cvff force field. Residues within 6 Å of the sulfur atom of the catalytic Cys51 were defined as the binding subset. Bulk of the protein, defined as atoms not in the binding subset, was held rigid during the docking process. Effect of solvent was introduced implicitly by solvation model of Stouten et al. (1993). First, 20 distinct docking poses of the substrates were collected using the Monte Carlo Minimization method, during which the ligands were subjected to random combinations of translational and rotational motions followed by minimization with 2500 iterations of conjugate gradients. Only those docking modes were retained wherein the ligands differed by a minimum RMS distance of 1 Å (RMS tolerance) and having an energy within 200 kcal (energy tolerance) of that of the lowest energy structure. During this stage the Quartic_vdw_no_Coul method was used to calculate energy. This choice employs a purely repulsive bounded quartic potential for modeling VDW interactions, and Coulombic interactions are set to zero. In the second stage, a more refined Cell-multipole method (Greengard and Rokhlin 1987) for calculation of nonbonded interactions was utilized. Here the structures so obtained were refined by minimization for 100 steps of conjugate gradients followed by a dynamics run for 5000 iterations of equilibration at 300 K. The Newton’s equations of motion were integrated using the Verlet Algorithm (Verlet 1967) with a time step of 1 fs using NVT ensemble. Temperature control was achieved by direct scaling of atom velocities. Finally, the structures were minimized to a gradient of 0.001 kcal/mole/Å or less using conjugate gradients. The same protocol was also employed for the docking of these substrates in the falcipain-2 homology model. The various docking poses of each substrate were ranked based on the empirical interaction energies between the substrate and the enzyme. The one with the lowest interaction energy was selected for further analysis.
Conclusions
One of the promising approaches in antimalarial drug discovery is the development of protease inhibitors for interfering with the organisms’ ability to metabolize human hemoglobin as a source of nutrients (Rosenthal 1998). Identification and isolation of two major cysteine proteases viz. falcipain-2 and falcipain-3 has highlighted and unraveled the unforeseen level of complexity involved in hemoglobin hydrolysis by the parasite. A homology model of falcipain-3 was built and validated to arrive at a reliable model for structure based drug design. Further, an excellent correlation between the interaction energies for various docked substrates and their respective Km values proven to be an endorsement for the model. In addition, these studies also provided insight into the nature of binding and interaction of ligands with the enzyme.
The falcipain-3 model offers a promising opportunity to design and develop novel inhibitors that could block one of the major mediators of hemoglobin denaturation. However, effective blockade of parasite development by suppression of parasite hemoglobin hydrolysis would likely require a potent inhibition of both falcipain-3 and falcipain-2. Comparison of the binding pockets of falcipain-3 with that of falcipain-2 model suggested some interesting features relevant to the drug design process. Some differences in the residues lining the S2 pockets of these enzymes were observed leading to a narrower S2 pocket in falcipain-3. Although, this difference may affect the ligand interaction to some extent, overall similarity of the binding pockets of these enzymes does not rule out the possibility of development of ligands with dual inhibition of falcipain-2 and falcipain-3. In any event, the proposed falcipain-3 model could be used to direct structure-based drug design studies leading to development of chemotherapeutic agents to combat malaria.
Acknowledgments
This work was supported by a CDC Cooperative agreement number U50/CCU418839.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0228103.
References
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acid Res. 28 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breman, J.G. 2001. The ears of the hippopotamus: Manifestations, determinants, and estimates of the malaria burden. Am. J. Trop. Med. Hyg. 64 1–11. [DOI] [PubMed] [Google Scholar]
- Brinen, L.S., Hansell, E., Cheng, J., Roush, W.R., McKerrow, J.H., and Fletterick, R.J. 2000. A target within the target: Probing cruzain’s P1‘ site to define structural determinants for the Chagas’ disease protease. Structure 8 831–840. [DOI] [PubMed] [Google Scholar]
- Butler, D., Maurice, J., and O’Brien, C. 1997. Time to put malaria control on the global agenda. Nature 386 535–536. [DOI] [PubMed] [Google Scholar]
- Choi, H.K., Laursen, R.A., and Allen, K.N. 1999. The 2.1 Å structure of a cysteine protease with proline specificity from ginger rhizome, Zingiber officinale. Biochemistry 38 11624–11633. [DOI] [PubMed] [Google Scholar]
- Edsall, J.T., Flory, P.J., Liquori, A.M., Kendrew, J.C., Nemethy, G., Ramachandran, G.N., and Scheraga, H.A. 1966. A proposal of standard conventions and nomenclature for the description of polypeptide conformation. Biopolymers 4 121–129. [PubMed] [Google Scholar]
- Eggleson, K.K., Duffin, K.L., and Goldberg, D.E. 1999. Identification and characterization of falcilysin, a metallopeptidase involved in hemoglobin catabolism within the malaria parasite Plasmodium falciparum. J. Biol. Chem. 274 32411–32417. [DOI] [PubMed] [Google Scholar]
- Folkers, G. 1998. Integrated homology modeling and X-ray study of herpes simplex virus I thymidine kinase. In Structure-based rug design experimental and computational approaches (ed. P.W. Codding), pp. 271–283. Kluwer Academic Publishers, Norwell, MA, USA.
- Francis, S.E., Gluzman, I.Y., Oksman, A., Knickerbocker, A., Mueller, R., Bryant, M.L., Sherman, D.R., Russell, D.G., and Goldberg, D.E. 1994. Molecular characterization and inhibition of a Plasmodium falciparum aspartic hemoglobinase. EMBO J. 13 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor, S.A., Craik, C.S., and Fletterick, R.J. 1997. Structural determinants of specificity in the cysteine protease cruzain. Protein Sci. 6 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish, W. WU-BLAST archives. 1996–2002. http://blast.wustl.edu.
- Godzik, A., Kolinski, A., and Skolnick, J. 1992. Topology finger print approach to the inverse protein folding problem. J. Mol. Biol. 227 227–238. [DOI] [PubMed] [Google Scholar]
- Goldberg, D.E. 2002. Parasitology: When the host is smarter than the parasite. Science 296 482–483. [DOI] [PubMed] [Google Scholar]
- Greengard, L. and Rokhlin, V.I. 1987. A fast algorithm for particle simulations. J. Comp. Phys. 73 325–348. [Google Scholar]
- Jones, D.T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Klemba, M. and Goldberg, D.E. 2002. Biological roles of proteases in parasitic protozoa. Annu. Rev. Biochem. 71 275–305. [DOI] [PubMed] [Google Scholar]
- Leung, D., Abbenante, G., and Fairlie, D.P. 2000. Protease inhibitors: Current status and future prospects. J. Med. Chem. 43 305–341. [DOI] [PubMed] [Google Scholar]
- Lüthy, R., Bowie, J.U., and Eisenberg, D. 1992. Assessment of protein models with three-dimensional profiles. Nature 356 83–85. [DOI] [PubMed] [Google Scholar]
- Luty, B.A., Wasserman, Z.R., Stouten, P.F.W., Hodge, C.N., Zacharias, M., and McCammon, J.A. 1995. A molecular mechanics/grid method for evaluation of ligand-receptor interactions. J. Comp. Chem. 16 454–464. [Google Scholar]
- Maes, D., Bouckaert, J., Poortmans, F., Wyns, L., and Looze, Y. 1996. Structure of chymopapain at 1.7 A resolution. Biochemistry 35 16292–16298. [DOI] [PubMed] [Google Scholar]
- McGuffin, L.J., Bryson, K., and Jones, D.T. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16 404–405. [DOI] [PubMed] [Google Scholar]
- Needleman, S. and Wunsch, C. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48 443–453. [DOI] [PubMed] [Google Scholar]
- Olliaro, P.L. and Yuthavong, Y. 1999. An overview of chemotherapeutic targets for antimalarial drug discovery. Pharm. Ther. 81 91–110. [DOI] [PubMed] [Google Scholar]
- Olson, J.E., Lee, G.K., Semenov, A., and Rosenthal, P.J. 1999. Antimalarial effects in mice of orally administered peptidyl cysteine protease inhibitors. Bioorg. Med. Chem. 7 633–638. [DOI] [PubMed] [Google Scholar]
- Ring, C.S., Sun, E., McKerrow, J.H., Lee, G.K., Rosenthal, P.J., Kuntz, I.D., and Cohen, F.E. 1993. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc. Natl. Acad. Sci. 90 3583–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, P. 1998. Proteases of malaria parasites: New targets for chemotherapy. Emerg. Infect. Dis. 4 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, P.J. 2002. Hydrolysis of erythrocyte proteins by proteases of malaria parasites. Curr. Opin. Hematol. 9 140–145. [DOI] [PubMed] [Google Scholar]
- Rosenthal, P.J. and Meshnick, S.R. 1996. Hemoglobin catabolism and iron utilization by malaria parasites. Mol. Biochem. Parasitol. 83 131–139. [DOI] [PubMed] [Google Scholar]
- Rosenthal, P.J., Wollish, W.S., Palmer, J.T., and Rasnick, D. 1991. Antimalarial effects of peptide inhibitors of a Plasmodium falciparum cysteine proteinase. J. Clin. Invest. 88 1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, P.J., Lee, G.K., and Smith, R.E. 1993. Inhibition of a Plasmodium vinckei cysteine proteinase cures murine malaria. J. Clin. Invest. 91 1052–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, P.J., Olson, J.E., Lee, G.K., Palmer, J.T., Klaus, J.L., and Rasnick, D. 1996. Antimalarial effects of vinyl sulfone cysteine proteinase inhibitors. Antimicrob. Agents Chemother. 40 1600–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis, Y., Rosenthal, P.J., Desai, P., and Avery, M.A. 2002. Homology modeling of falcipain-2: Validation, de novo ligand design and synthesis of novel inhibitors. J. Biomol. Struct. Dyn. 19 765–774. [DOI] [PubMed] [Google Scholar]
- Sajid, M. and McKerrow, J.H. 2002. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 120 1–21. [DOI] [PubMed] [Google Scholar]
- Salas, F., Fichmann, J., Lee, G.K., Scott, M.D., and Rosenthal, P.J. 1995. Functional expression of falcipain, a Plasmodium falciparum cysteine proteinase, supports its role as a malarial hemoglobinase. Infect. Immun. 63 2120–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer, P.M., Chen, X., Chan, V.J., Cheng, M., Kenyon, G.L., Kuntz, I.D., Sakanari, J.A., Cohen, F.E., and McKerrow, J.H. 1997. Leishmania major: Molecular modeling of cysteine proteases and prediction of new nonpeptide inhibitors. Exp. Parasitol. 87 212–221. [DOI] [PubMed] [Google Scholar]
- Shenai, B.R., Sijwali, P.S., Singh, A., and Rosenthal, P.J. 2000. Characterization of native and recombinant falcipain-2, a principal trophozoite cysteine protease and essential hemoglobinase of Plasmodium falciparum. J. Biol. Chem. 275 29000–29010. [DOI] [PubMed] [Google Scholar]
- Shenkin, P.S., Yarmush, D.L., Fine, R.M., Wang, H., and Levinthal, C. 1986. Predicting antibody hypervariable loop conformations II: Minimization and molecular dynamics studies of MCPC603 from many randomly generated loop conformations. Proteins Struct. Funct. Genet. 1 342–362. [DOI] [PubMed] [Google Scholar]
- ———. 1987. Predicting antibody hypervariable loop conformation. I. Ensembles of random conformations for ringlike structures. Biopolymers 26 2053–2085. [DOI] [PubMed] [Google Scholar]
- Sijwali, P.S., Shenai, B.R., Gut, J., Singh, A., and Rosenthal, P.J. 2001. Expression and characterization of the Plasmodium falciparum hemoglobinase falcipain-3. Biochem. J. 360 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouten, P.F.W., Froemmel, C., Nakamura, H., and Sander, C. 1993. An effective solvation term based on atomic occupancies for use in protein simulations. Mol. Simul. 10 97–120. [Google Scholar]
- Verlet, L. 1967. Computer "experiments" on classical fluids. I. Thermodynamical properties of Lennard-Jones molecules. Phys. Rev. 159 98–103. [Google Scholar]
- Zhao, B., Janson, C.A., Amegadzie, B.Y., D’Alessio, K., Griffin, C., Hanning, C.R., Jones, C., Kurdyla, .J., McQueney, M., Qiu, X., et al. 1997. Crystal structure of human osteoclast cathepsin K complex with E-64. Nat. Struct. Biol. 4 109–111. [DOI] [PubMed] [Google Scholar]