Abstract
The genetic basis of heterosis was investigated in an elite rice hybrid by using a molecular linkage map with 150 segregating loci covering the entire rice genome. Data for yield and three traits that were components of yield were collected over 2 years from replicated field trials of 250 F2:3 families. Genotypic variations explained from about 50% to more than 80% of the total variation. Interactions between genotypes and years were small compared with the main effects. A total of 32 quantitative trait loci (QTLs) were detected for the four traits; 12 were observed in both years and the remaining 20 were detected in only one year. Overdominance was observed for most of the QTLs for yield and also for a few QTLs for the component traits. Correlations between marker heterozygosity and trait expression were low, indicating that the overall heterozygosity made little contribution to heterosis. Digenic interactions, including additive by additive, additive by dominance, and dominance by dominance, were frequent and widespread in this population. The interactions involved large numbers of marker loci, most of which individually were not detectable on single-locus basis; many interactions among loci were detected in both years. The results provide strong evidence that epistasis plays a major role as the genetic basis of heterosis.
Keywords: hybrid vigor, molecular markers, quantitative trait loci, interaction between loci
The two earliest hypotheses regarding heterosis, both proposed in 1908—i.e., the dominance hypothesis (1) and the overdominance hypothesis (2, 3)—have competed for most of this century (4). However, pertinent data allowing for critical assessments of these hypotheses remained largely unavailable until very recently with the advent of discretely recognizable mendelian markers such as allozymes, restriction fragment length polymorphisms (RFLPs), and more recently high-density molecular linkage maps. Stuber et al. (5), who analyzed the genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines, observed that heterozygotes of almost all the quantitative trait loci (QTLs) for yield had higher phenotypic values than the respective homozygotes. They suggested that both overdominance and QTLs detected by single-locus analysis play a significant role in heterosis. On the other hand, Xiao et al. (6), who conducted an inheritance study of quantitative traits in an intersubspecific cross of rice, suggested that dominance may be the genetic basis of heterosis in rice. Both of the dominance and overdominance hypotheses are based only on single-locus theory.
Wright (7) visualized a “net-like” structure of population genotypes such that the variations of most characters are affected by many loci and that each gene replacement may have effects on many characters. Based on this perspective, epistasis should be one of the most important genetic components in the inheritance of quantitative characters. Using morphological markers marking small chromosome segments, Fasoulas and Allard (8) showed that epistasis was responsible for a major part of the total genetic variance for seven of the eight characters studied in crosses between near isogenic lines of cultivated barley. Population genetic analyses have also established that epistasis, functioning to assemble and maintain favorable multilocus genotypes, is a major mechanism of adaptation in various plant species (9). Surprisingly, however, little evidence for epistasis has been indicated in the recent data from molecular marker-based studies of yield and yield-associated characters, although epistasis has been observed in characters such as plant height in soybean, nitrogen-fixing ability in rice, and seed size in legumes (10–13). Thus, the significance of epistasis in the expression of heterosis in relation to hybrid performance has yet to be established.
Rice is the staple food for a large segment of the world population. The success of hybrid rice breeding, together with saturated molecular linkage maps (14, 15) and relatively small genome size of this species, has provided a rare opportunity for dissecting the genetic basis of heterosis in elite rice hybrids. In the study reported in this paper, we investigated, at both single-locus and two-locus levels, the genetic components conditioning the inheritance of yield and yield component traits in one of the most heterotic crosses. The main objective was to characterize the genetic basis of heterosis in this hybrid with the long-term goal of manipulating the genome for hybrid rice improvement.
MATERIALS AND METHODS
Experimental Population and Field Data Collection.
The experimental population consisted of 250 F3 families, each derived from bagged seeds of a single F2 plant, from a cross between Zhenshan 97 and Minghui 63. These two lines are the parents of Shanyou 63, which is the best hybrid in China, with an annual acreage of 6.7 million hectares in the last decade and accounting for approximately 25% of the rice production of China. The F3 families, two parents, and F1 were transplanted to a bird-net-equipped field in the experimental farm of Huazhong Agricultural University in the 1994 and 1995 rice-growing seasons in Wuhan, China. The field planting followed a randomized complete block design with three replications. For each family within a replication, seedlings approximately 35 days old were transplanted to a single-row plot, with distance of 17 cm between plants within a row, and the rows were 27 cm apart. The field management followed essentially the normal agricultural practice. Each plant was harvested individually at maturity to prevent loss from over-ripening. Only the 15 plants in the middle of each row were used for scoring. Traits examined included yield per plant, which was converted to metric tons/hectare (t/ha), tillers per plant scored as the number of seed-setting tillers per plant, grain weight as the weight (g) of 1,000 seeds, and grains per panicle.
DNA Markers and Laboratory Assay.
Leaf tissues were harvested either from single F2 plants or from bulks of at least 20 plants from each of the F3 families. Exactly 8 g of leaf tissue per sample was ground to fine powder under liquid nitrogen. Total cellular DNA was extracted using the protocol of Murray and Thompson (16).
Two classes of markers were employed to assay DNA polymorphisms in this population: RFLPs and simple sequence repeats. RFLP analysis followed the method described by Liu et al. (17). For surveying parental polymorphisms, DNA samples from each of the two parents were digested with six restriction enzymes and probed with a total of 537 clones from Cornell University and the Japanese Rice Genome Research Project (14, 15). Fourteen enzymes were added to the survey for genomic regions in which polymorphisms could not be detected using the six restriction enzymes.
The 54 primer pairs used for surveying simple sequence repeat polymorphisms included 29 from published data (18, 19) and 25 prepared by our group mostly from published sequences in GenBank. PCR detection followed essentially the methods by Wu and Tanksley (18). The DNA markers detecting polymorphisms between the parents were used to assay the entire population of 250 individuals.
Data Processing and Statistical Analyses.
A molecular marker linkage map was constructed using Mapmaker 3.0 (20). QTLs were determined using Mapmaker/QTL 1.1. (21). The entire genome was searched for digenic interactions for each trait with two-way analyses of variance using all possible two-locus combinations of marker genotypes. The calculation was based on unweighted cell means (22), and the sums of squares were multiplied by the harmonic means of the cell sizes to form the test criteria. There are eight degrees of freedom among the nine genotypes formed of two codominant loci, two degrees of freedom for each of the additive and dominance effects within each locus, and four degrees of freedom for interaction between loci. The interaction, often referred to as epistasis, can be further partitioned into four terms each specified by a single degree of freedom: additive (first locus) × additive (second locus) (AA), additive × dominance (AD), dominance × additive (DA), and dominance × dominance (DD). Statistical significance for each term was assessed using an orthogonal contrast test provided by the statistical package Statistica (23).
RESULTS
Measurements of the Traits.
The performance and heterosis of yield and yield component traits in parents, F1, and F3 grown in 1994 and 1995 are given in Table 1. The F1 yield was approximately the same in the two years tested, and is on the same order of magnitude as the average yielding level of this hybrid under normal agricultural conditions (24). The measurements of the three yield component traits were also similar in the two years. F1 heterosis, measured as the percentage of deviation of the F1 from the parental mean, is large for yield in both years. There was still large residual heterosis in the F3 generation. The proportion of genotypic variation in the total phenotypic variation ranged from about 50% in tillers per plant to more than 80% in seed weight in the two years (Table 2). Interactions between genotypes and years were statistically significant for three of the four traits (Table 2). However, the interaction effects were small compared with the main effects of genotypes or years (data not shown).
Table 1.
Means and midparent heterosis (% in parentheses) of yield and yield component traits in parents and the progenies
| Trait | Minghui 63
|
Zhenshan 97
|
F1
|
F3
|
||||
|---|---|---|---|---|---|---|---|---|
| 1994 | 1995 | 1994 | 1995 | 1994 | 1995 | 1994 | 1995 | |
| Yield, t/ha | 6.50 | 6.26 | 4.84 | 4.48 | 8.53 (50.4) | 8.91 (66.0) | 7.40 (30.8) | 6.62 (23.3) |
| Tillers/plant | 10.7 | 11.0 | 11.4 | 9.5 | 12.8 (15.8) | 14.3 (39.5) | 11.2 (1.4) | 10.5 (2.4) |
| Grains/panicle | 101.2 | 105.7 | 81.8 | 89.5 | 114.9 (25.6) | 111.9 (14.6) | 112.9 (23.4) | 110.4 (13.1) |
| Grain weight, g/1,000 | 27.7 | 25.6 | 25.1 | 24.8 | 27.9 (5.7) | 26.0 (3.2) | 26.3 (−0.4) | 25.4 (0.8) |
Table 2.
Some summary statistics from two-way analysis of variance of the traits measured in 1994 and 1995
| Trait | SSG in SST,* %
|
G × Y†
|
||
|---|---|---|---|---|
| 1994 | 1995 | F | P | |
| Yield | 51.9 | 74.1 | 1.60 | 0.000 |
| Tillers/plant | 49.0 | 53.2 | 1.14 | 0.098 |
| Grains/panicle | 74.6 | 75.5 | 2.35 | 0.000 |
| Grain weight | 83.0 | 89.0 | 2.06 | 0.000 |
Proportion of genotype sum of squares in total sum of squares.
Interaction between genotypes and years.
Linkage Map.
The survey of 591 molecular markers, including 537 RFLPs and 54 simple sequence repeats, identified 150 loci polymorphic between the parents. Mapmaker analysis at LOD (logarithm of odds) 3.0 placed these marker loci into 14 linkage groups. A map was constructed (not shown) which spanned a total of 1,842 centimorgans in length and well integrated the markers from the two high-density RFLP linkage maps (14, 15).
QTLs for Yield and Yield Component Traits.
QTLs resolved with LOD 2.4 (25) for the four traits are given in Table 3. QTLs with LOD exceeding 2.4 in one year but slightly below this threshold in another year are also included in the list.
Table 3.
Putative QTLs identified for yield and yield component traits in 1994 and 1995
| Trait* | QTL† | Flanking markers | LOD | Var, % | A | D | IL |
|---|---|---|---|---|---|---|---|
| 1994 | |||||||
| Yield | yd1a | R753–C161 | 3.5 | 9.7 | −0.3 | 3.2 | 0 |
| yd1b | RG101–C922 | 2.6 | 5.8 | −1.7 | −0.3 | 3 | |
| yd4 | R514–C2807 | 2.4 | 7.4 | −1.8 | 1.4 | 4 | |
| yd5a | G193–RZ649 | 3.3 | 11.7 | −0.5 | 3.4 | 3 | |
| yd8 | C483–R1629 | 3.2 | 6.6 | 0.8 | 2.4 | 1 | |
| Ti/Pl | tp4 | C820–C56 | 3.1 | 6.1 | −0.4 | −0.1 | 2 |
| tp5 | G1458–G193 | 2.8 | 5.5 | −0.4 | 0.2 | 2 | |
| tp10 | C677–G4003 | 2.7 | 5.3 | −0.2 | −0.5 | 4 | |
| Gr/Pa | gp1a | R753–C161 | 3.1 | 6.1 | −5.2 | 4.4 | 0 |
| gp1b | RG173–RG532 | 4.2 | 10.6 | −7.9 | −3.5 | 4 | |
| gp3 | R1966–G144 | 8.3 | 16.9 | 10.0 | −3.6 | 1 | |
| gp5‖ | G193–RZ649 | 2.4 | 8.9 | −1.3 | 10.3 | 1 | |
| gp6a‖ | RG653–G342 | 2.5 | 4.9 | 4.1 | −5.6 | 4 | |
| Gr wt | gw1‖ | R753–C161 | 3.5 | 12.1 | 0.7 | 1.5 | 1 |
| gw3a‖‖ | R19–RZ403 | 11.4 | 23.6 | −1.8 | −0.4 | 8 | |
| gw5‖ | RG360–C734 | 8.5 | 15.7 | 1.3 | 0.1 | 1 | |
| gw6 | R1952–C226 | 2.6 | 5.2 | 0.7 | 0.5 | 3 | |
| gw7‖‖‖ | RG128–C1023 | 2.3 | 8.6 | −1.0 | 0.2 | 3 | |
| gw9 | R2638–RG570 | 4.1 | 8.9 | 1.2 | −0.3 | 0 | |
| gw11‖ | RM4-RG98 | 2.2 | 5.6 | −0.6 | 0.7 | 1 | |
| 1995 | |||||||
| Yield | yd1a | R753–C161 | 2.6 | 6.2 | −1.1 | 2.1 | 1 |
| yd5b | C624–C246a | 2.7 | 10.2 | −1.4 | 2.9 | 0 | |
| yd5c | RG360–R1674 | 2.5 | 4.8 | 1.5 | 0.2 | 4 | |
| yd6 | R565–R902 | 2.8 | 5.2 | 1.7 | 0.4 | 4 | |
| yd7‖ | RG128–C1023 | 3.5 | 9.4 | −1.8 | 2.3 | 2 | |
| yd11‖ | C950–G389b | 3.2 | 7.0 | −0.5 | 2.7 | 1 | |
| Ti/Pl | tp4 | C820–C56 | 2.6 | 5.5 | −0.3 | −0.3 | 0 |
| tp7 | RZ471–MX2 | 3.2 | 11.3 | 0.5 | −0.5 | 6 | |
| Gr/Pa | gp1a | RM1–R753 | 6.0 | 15.5 | −8.1 | 3.4 | 10 |
| gp1b‖ | RG173–RG532 | 7.1 | 17.8 | −9.1 | −2.8 | 13 | |
| gp3‖ | RZ403–C269 | 9.0 | 16.0 | 8.7 | −0.5 | 4 | |
| gp5 | G193–RZ649 | 2.3 | 8.6 | −0.2 | 8.8 | 3 | |
| gp6b | R1014–G200 | 2.5 | 5.5 | 5.0 | 0.9 | 0 | |
| gp7 | C1023–R1440 | 4.7 | 9.5 | −5.2 | 6.5 | 2 | |
| gp11 | C950–G389b | 3.1 | 5.9 | −0.7 | 7.3 | 0 | |
| Gr wt | gw1 | R753–C161 | 2.6 | 9.7 | 0.5 | 1.3 | 0 |
| gw3b | C944–C746 | 9.5 | 16.8 | −1.4 | 0.2 | 3 | |
| gw3c | R1966–G144 | 11.4 | 22.5 | −1.7 | −0.7 | 9 | |
| gw5 | RG360–R1674 | 12.9 | 24.1 | 1.6 | −0.3 | 0 | |
| gw6 | R565–R902 | 4.7 | 8.6 | 1.0 | 0.3 | 5 | |
| gw7 | RG128–C1023 | 5.2 | 17.7 | −1.4 | −0.4 | 9 | |
| gw8 | C1121–RG333 | 2.7 | 6.7 | 0.7 | −0.7 | 5 | |
| gw9 | R2638–RG570 | 4.0 | 8.5 | 1.1 | −0.4 | 1 | |
| gw11 | RM4–RG98 | 3.5 | 8.6 | −0.9 | 0.6 | 0 | |
Var, variation explained by each QTL; A, additive effect; D, dominance effect; IL, number of loci with which the QTL interacts. Positive values of the additive effect indicate that alleles from Zhenshan 97 are in the direction of increasing the trait scores, and negative values indicate that alleles from Minghui 63 are in the direction of increasing the score. Positive values of the dominance effect indicate that heterozygotes have higher phenotypic values than the respective means of two homozygotes, and negative values indicate that heterozygotes have lower values than the means of the two homozygotes.
Ti/Pl, tillers per plant, Gr/Pa, grains per panicle; Gr wt, grain weight.
Numbers following the two letters represent the chromosomal locations of the QTLs.
Pairs of interacting loci within a trait.
Five and six QTLs for yield were detected in 1994 and 1995, respectively, with one QTL (yd1a) in common between the two years.
For tillers per plant, three QTLs were detected in 1994 and two in 1995. Again, there was one QTL (tp4) in common between the two years.
Five and seven QTLs for grains per panicle were detected in 1994 and 1995; two (gp1b and gp5) were resolved in both years. In addition, it is almost certain that the effects on chromosome 1 revealed by R753-C161 in 1994 and by RM1-R753 in 1995 were due to the same QTL (gp1a), as the 1-LOD support confidence intervals (25) overlapped substantially (map not shown). Similarly, the LOD peaks that appeared in the tightly linked genomic segments (R1966-G144 and RZ403-C269) on chromosome 3 in the two years were also possibly the result of the same QTL (gp3).
Of the seven and nine QTLs for grain weight resolved in 1994 and 1995, four (gw1, gw7, gw9, and gw11) were detected in both years. The LOD peaks in the proximity of RG360 on chromosome 5 resolved in the two years may also be due to the effect of the same QTL (gw5), as was the case for the LOD peaks in the tightly linked blocks R1952–C226 and R565–R902 on chromosome 6.
Taken together, a total of 32 distinct QTLs were identified using the specified criteria. Twelve were observed in both years, and the remaining 20 were detected only in one or the other year. It can also be seen from Table 3 that several QTLs appeared to have pleiotropic effects by simultaneously affecting two or more traits. This is particularly the case for the QTL in the vicinity of the genomic block around R753 on chromosome 1, whereby large effects were simultaneously detected for three of the four traits in both years.
Dominance and Overdominance.
A locus is regarded as exhibiting overdominance if the ratio of the estimated dominance to the absolute value of additive effect is larger than unity. Thus, although the F3 family means usually underestimate dominance by a factor of ½, partial dominance, full dominance, and overdominance were nonetheless still prevalent among the QTLs (Table 3).
For yield, three of the five QTLs recovered in 1994 and four of the six QTLs in 1995 showed overdominance. Another noticeable case is that of grains per panicle, in which overdominance was detected for one of the five QTLs in 1994 and three of the seven QTLs in 1995. A very large overdominance effect was detected in both years for the QTL on chromosome 5 (gp5). For grain weight, one QTL on chromosome 1 displayed overdominance in both years, while the remaining QTLs showed partial to full dominance.
Various levels of negative dominance were observed in both years at several QTLs, including tp4 for tillers per plant, gp1b and gp3 for grains per panicle, and gw9 for grain weight. There were several QTLs at which negative dominance occurred in one year or the other (Table 3). This indicates that heterozygosity was not necessarily always favorable for the expression of the trait even in such a highly heterotic cross.
Relationship Between Marker Heterozygosity and Trait Expression.
Correlations between overall heterozygosity of the F2 individuals and the F3 family means were very small for all four traits (data not shown). They were significant at P ≤ 0.05 only for grain weight (correlation coefficients 0.17 in 1994 and 0.16 in 1995). Thus, the overall heterozygosity made very little contribution to trait expression. This result differed from analysis of marker distance and hybrid performance in diallel crosses (26, 27). A possible explanation for the lack of correlation is cancellation between positive and negative dominance among various loci, as suggested by estimated dominance effects in Table 3.
Digenic Interactions Involving QTLs.
The two-way analyses of variance detected interactions between QTLs in grains per panicle and grain weight in 1994, and in yield and grains per panicle in 1995 (Table 3). The most noteworthy case was that of grain weight in 1994, in which significant interactions were detected in four pairs of QTLs.
Interactions of QTLs with loci in the rest of the genome were also assessed, using markers flanking the QTLs and markers located in the remaining parts of the linkage map. The majority of the QTLs were found to interact with at least one other locus and many of the QTLs interacted with three or more loci (Table 3).
Digenic Interactions Between Loci in the Entire Genome.
Table 4 presents the numbers of two-locus combinations showing significant (P < 0.01) interactions resulting from testing all possible two-locus combinations and also the numbers of various types of interactions determined by orthogonal contrast tests. Only data sets formed of two codominant loci with each marker genotypic class containing 5 or more individuals were included in the calculation, resulting in 7,585 tests for 1994 and 7,681 for 1995. With α = 0.01 for individual tests, the 99% confidence intervals for the expected numbers of spurious interactions (false positive) would be 75.85 ± 20.02 for 1994 and 76.81 ± 20.14 for 1995. Thus, the numbers of interactions detected in both years for all the traits much exceeded expectations based on spurious interactions, indicating that real interactions exist in the genome of this population.
Table 4.
Summary of the significant (P ≤ 0.01) interactions identified in 1994 and 1995 by searching all possible two-locus combinations
| Trait | Interaction | 1994 | 1995 | Common |
|---|---|---|---|---|
| Yield | Positive pairs* | 105 | 165 | 10 |
| AA | 60 | 91 | 8 | |
| AD (DA) | 51 | 73 | 3 | |
| DD | 4 | 18 | 0 | |
| Tillers/plant | Positive pairs* | 105 | 141 | 17 |
| AA | 79 | 105 | 17 | |
| AD (DA) | 28 | 42 | 1 | |
| DD | 10 | 6 | 0 | |
| Grains/panicle | Positive pairs* | 99 | 160 | 15 |
| AA | 52 | 80 | 9 | |
| AD (DA) | 56 | 74 | 10 | |
| DD | 4 | 16 | 0 | |
| Grain weight | Positive pairs* | 125 | 164 | 49 |
| AA | 84 | 102 | 27 | |
| AD (DA) | 47 | 71 | 19 | |
| DD | 15 | 16 | 9 | |
| Number of tests† | 7,585 | 7,681 |
Number of two-locus combinations showing significant interactions (P ≤ 0.01).
Number of possible two-locus combinations tested.
Significant interactions were detected simultaneously in both years for many of the two-locus combinations (Table 4, column 5). If two loci do not interact with each other, the probability for detecting the same interaction at α = 0.01 in both years is less than 0.01%. Thus, in essence, none of the two-locus interactions simultaneously observed in both years should be regarded as a chance event. These interactions, referred to as common interactions for ease of description, can be taken as the minimum of statistically significant interactions.
The proportion of common interactions was the largest for grain weight. They constituted approximately 40% and 30% of the significant interactions detected in 1994 and 1995, respectively. Common interactions ranged from 6% to 17% of the significant interactions for the remaining three traits in the two years (Table 4).
The numbers of three different types of interactions (AA, AD/DA, and DD) determined using the orthogonal contrast test are also given in Table 4. By chance alone, these terms should occur in a proportion of 1 (AA): 2 (AD/DA): 1 (DD), which was obviously not the case. In all four traits, AA interactions were much in excess, and DD and also AD/DA were in deficiency, providing additional evidence that the interactions were not the results of chance events.
Patterns of Interactions.
We used grains per panicle to illustrate the interactions; two-locus combinations showing interactions at P ≤ 0.01 in both years are listed in Table 5. Two observations can be made from this table. First, the types of interactions (i.e., AA, AD, and DD) detected for the majority of the two-locus combinations were consistent in the two years. Second, the majority of the interaction terms individually accounted for less than 5% of the genotypic variation. It is striking that such small interactions could be repeatedly detected under the experimental conditions.
Table 5.
Two-locus interactions for grains per panicle simultaneously detected at P ≤ 0.01 in 1994 and 1995 by two-way analysis of variance based on marker genotypes
| Locus 1* | Locus 2* | 1994
|
1995
|
||||
|---|---|---|---|---|---|---|---|
| Type | P | SSG, %† | Type | P | SSG, %† | ||
| RG532 (1) | RM4 (11) | AA | 0.002 | 4.0 | AA | 0.010 | 2.5 |
| RG173 (1) | RM203 (3) | AA | 0.002 | 4.1 | AA | 0.002 | 3.5 |
| C547x (1) | RG634 (2) | AA | 0.004 | 3.6 | AA | 0.005 | 3.4 |
| RG236 (1) | R1440 (7) | AD | 0.009 | 2.9 | AD | 0.012 | 2.4 |
| DA | 0.022 | 2.2 | DA | 0.002 | 3.5 | ||
| C112 (1) | G389a (11) | AA | 0.006 | 3.3 | AA | 0.019 | 2.3 |
| MX7b (2) | Waxy (6) | DA | 0.006 | 3.3 | DA | 0.000 | 5.4 |
| RM168 (3) | RM18 (7) | AD | 0.001 | 5.0 | AD | 0.000 | 6.9 |
| C1447 (5) | C677 (10) | AA | 0.005 | 3.3 | AA | 0.006 | 3.2 |
| DA | 0.015 | 2.5 | — | — | — | ||
| C1447 (5) | G389a (11) | AA | 0.003 | 3.9 | AA | 0.003 | 3.7 |
| G1458x (5)§ | G342 (6) | AA | 0.004 | 3.6 | AD | 0.002 | 4.1 |
| G193x (5)§ | G342 (6) | AA | 0.004 | 3.7 | AA | 0.018 | 2.3 |
| AD | 0.036 | 1.9 | AD | 0.005 | 3.3 | ||
| RG360 (5) | RG653 (6)§ | AD | 0.003 | 3.7 | AD | 0.000 | 5.3 |
| DA | 0.011 | 2.7 | DA | 0.021 | 2.1 | ||
| RG360 (5) | G342 (6)§ | AD | 0.001 | 5.4 | AD | 0.000 | 9.6 |
| R830 (5) | RZ404 (9) | AA | 0.005 | 3.3 | AA | 0.004 | 3.4 |
| DD | 0.032 | 2.0 | DD | 0.017 | 2.3 | ||
| C1023 (7) | C794 (11) | DA | 0.002 | 4.1 | DA | 0.014 | 2.2 |
Interaction types with P ≤ 0.01 in one year but 0.01 < P < 0.05 in the other year are also listed.
*Numbers in parentheses represent chromosomal locations.
Percentage of genotypic variation explained by the interaction.
Marker loci that are closely linked.
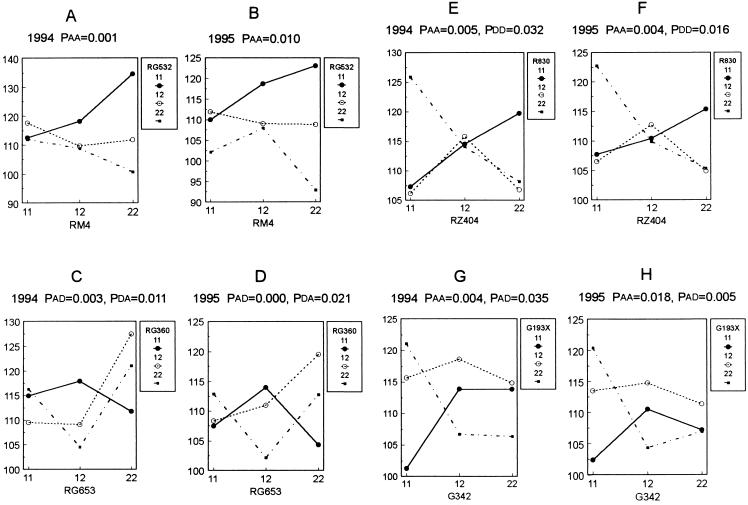
The means of seeds per panicle for the nine genotypic classes for each pair of interacting loci presented in Table 5 are graphed to discern the patterns of interactions. A few examples are given in Fig. 1.
Figure 1.
Patterns of the interactions for grains per panicle displayed by various two-locus combinations in 1994 and 1995. Paa, Pad, Pda, and Pdd are the probabilities for AA, AD, DA, and DD interactions. The vertical axis represents number of grains per panicle. The numbers 11, 12, and 22 represent the three genotypes of each locus: 11, homozygote for Minghui 63 allele; 22, homozygote for Zhenshan 97 allele; and 12, heterozygote.
AA.
The locus pair RG532 and RM4 from chromosomes 1 and 11, respectively, was used to illustrate the AA interaction. As can be seen from Fig. 1 A and B, performance of the two homozygotes of the first locus (marked by RG532) was favored or disfavored differentially by the homozygotes of the second locus (RM4). The two homozygotes of the RG532 locus differed by only 1 and 8 seeds per panicle in 1994 and 1995, respectively, when RM4 was homozygous for the Minghui 63 allele (11). The difference increased to 30 and 31 when RM4 was homozygous for the Zhenshan 97 allele (22), resulting in 11(RG532)/22(RM4) as the favored genotype. Note that a QTL was detected near RG532 (gp1b) in both years, and the interaction clearly contributed to the detection of this QTL.
AD.
The combination of marker loci RG360 (chromosome 5) and RG653 (chromosome 6), showing AD and DA in both years, was chosen as an example to demonstrate AD (Fig. 1 C and D). The critical characteristic of this AD was that the performance of the homozygotes of the first locus depended on the heterozygote of the second locus, or conversely, the phenotype of the heterozygote of the second locus was largely influenced by the homozygotes of the first locus. Apparently, the heterozygote of the second locus (RG653) not only reversed the relative grain numbers of the two homozygotes of the first locus (RG360) but also considerably enlarged the magnitude of the difference between the two homozygotes. The situation was very similar for the DA of the same locus pair.
DD.
A significant DD interaction was detected in both 1994 and 1995 in the R830 (chromosome 5) and RZ404 (chromosome 9) combination (Fig. 1 E and F), which also showed an AA interaction with the pattern differing from that of the RG534 and RM4 combination. This is the only DD detected among the various two-locus combinations listed in Table 5. Heterozygotes of the two loci interacted favorably and the double heterozygote produced more grains per panicle than did single heterozygotes.
Influence of Epistasis on Overdominance.
A large overdominance effect was detected in both years at the QTL gp5 (Table 3), which was found to interact in both years with a locus on chromosome 6. The line graphs of the 9 genotypes (Fig. 1 G and H), based on the flanking markers G193x (chromosome 5) and G342 (chromosome 6), clearly showed that overdominance marked by the G193x locus was dependent on genotypes at the G342 locus. The heterozygote of G193x was superior only when the G342 was heterozygous or homozygous for the Zhenshan 97 allele, and was intermediate when G342 was homozygous for the Minghui 63 allele.
DISCUSSION
The most noticeable finding of the present study based on a highly heterotic cross is the prevalence and importance of epistasis in the rice genome with two pronounced features. First, two-locus analyses resolved much larger numbers of loci contributing to trait expression than single-locus analyses. For example, counting only the interactions simultaneously detected in both years for grain number per panicle, the significant two-locus interactions involved a total of 25 loci located on 9 of the 12 rice chromosomes, compared with 5 and 7 QTLs detected in the two years for this trait. As a second feature, all three types of interactions (AA, AD, and DD) occurred among the various two-locus combinations. It is even more remarkable that multiple interaction terms were found in a considerable proportion of the interacting two-locus combinations in all traits examined (Tables 4 and 5).
Overdominance at the single-locus level was detected at many of the QTLs. More QTLs showed overdominance for yield than for yield component traits, whereby most of the QTLs for yield and a few QTLs for the component traits showed overdominance. This is largely because of the multiplicative relation of yield with the component traits (multiplicative overdominance), as discussed by Zhang et al. (26). However, because QTLs that showed overdominance interacted with at least one other locus, it may not be appropriate to interpret the single-locus marginal effects without specifying the genotypes of the counterpart. The same argument also applies to QTLs that exhibited dominance and/or additive effects, most of which are likely to be embedded in the interlocus interactions involving much of the total genome. Additionally, lack of correlation between heterozygosity and trait expression implies that, collectively, the effects of dominance and/or overdominance made only limited contributions to the heterosis observed in this experimental population.
While the dominant types of interactions (especially DD) may be most relevant to F1 heterosis, the analysis showed that AA seems to be more common than AD and DD in this data set. The deficiency of dominant types of interactions may be partly ascribed to the one-generation lag resulted from collection of marker data from F2 individuals and field data from F3 families; thus both heterozygosity and dominance were reduced by half compared with F2 individuals. Consequently, all the dominant types of effects, including dominance within individual loci and AD and DD between loci, were underestimated in the analysis. Therefore, to determine precisely the extent and significance of dominant types of interactions, it is necessary to have field data collected from F2 populations. Experimental designs are called for in future studies to enable the construction of genetically immortalized F2 populations.
The results also suggest the possibility of higher-order interactions (multilocus epistasis), at least for the most complex trait (yield). There are at least two lines of evidence implying the existence of higher-order interactions. First, fewer QTLs were detected and a smaller proportion of genotypic variation was explained by the QTLs for yield than for two of the yield component traits (Table 3), although the reverse should be expected on the basis of multiplicative relations of yield with the component traits. Also, at the two-locus level, the numbers of interactions detected for yield were not much greater than for its component traits. This suggests the involvement of genetic components that could not be resolved with either single-locus or two-locus analyses. Second, inspection of the significant two-locus interactions revealed a “chain-like” relation among the interacting two-locus combinations such that locus 1 interacted with locus 2, which in turn interacted with locus 3, which interacted with locus 4… Such chains could grow very long to include a large number of loci and even form circles (data not shown), indicating higher-order interactions as in the case of multilocus structure previously observed in an experimental barley population (28) and in natural populations of Avena barbata (9). Unfortunately, the size of the present population did not permit analyses beyond two-locus combinations.
In summary, the results clearly indicate that epistasis plays a significant role in the inheritance of quantitative traits as well as in the genetic basis of heterosis. The relationship between traits and genes in the manifestation of heterosis is much more complex than has commonly been expected on the basis of dominance and overdominance hypotheses.
Acknowledgments
We thank the Cornell Group and the Japanese Rice Genome Project for kindly providing the RFLP probes. This work was supported by a grant from Chinese National Natural Science Foundation and a grant from the Rockefeller Foundation.
ABBREVIATIONS
- QTL
quantitative trait locus
- LOD
logarithm of odds
- RFLP
restriction fragment length polymorphism
- AA
additive by additive interaction
- AD
additive by dominance interaction
- DA
dominance by additive interaction
- DD
dominance by dominance interaction
References
- 1.Davenport C B. Science. 1908;28:454–455. doi: 10.1126/science.28.718.454-b. [DOI] [PubMed] [Google Scholar]
- 2.East E M. Report of the Connecticut Agricultural Experimental Station for Years 1907–1908. 1908. pp. 419–428. [Google Scholar]
- 3.Shull G H. Am Breed Assoc. 1908;4:296–301. [Google Scholar]
- 4.Allard R W. Principles of Plant Breeding. New York: Wiley; 1960. [Google Scholar]
- 5.Stuber C W, Lincoln S, Wolff D W, Helentjaris T, Lander E S. Genetics. 1992;132:823–839. doi: 10.1093/genetics/132.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao J, Li J, Yuan L, Tanksley S D. Genetics. 1995;140:745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright S. Evolution and Genetics of Populations. Vol. 1. Chicago: Univ. Chicago Press; 1968. [Google Scholar]
- 8.Fasoulas A C, Allard R W. Genetics. 1962;47:899–907. doi: 10.1093/genetics/47.7.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allard R W. Euphytica. 1996;92:1–11. [Google Scholar]
- 10.Lark K G, Chase K, Adler F, Mansur L M, Orf J H. Proc Natl Acad Sci USA. 1995;92:4656–4660. doi: 10.1073/pnas.92.10.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu P, Zhang G, Ladha J K, McCouch S R, Huang N. Theor Appl Genet. 1995;91:1177–1183. doi: 10.1007/BF00220926. [DOI] [PubMed] [Google Scholar]
- 12.Fatokun C A, Menancio-Hautea D I, Danesh D, Young N D. Genetics. 1992;132:841–846. doi: 10.1093/genetics/132.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maughan P J, Saghai Maroof M A, Buss G R. Theor Appl Genet. 1996;93:574–579. doi: 10.1007/BF00417950. [DOI] [PubMed] [Google Scholar]
- 14.Causse M A, Fulton T M, Cho Y G, Ahn S N, Chunwongse J, Wu K S, Xiao J H, Yu Z H, Ronald P C, Harrington S E, Second G, McCouch S R, Tanksley S D. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, et al. Nat Genet. 1994;8:365–372. doi: 10.1038/ng1294-365. [DOI] [PubMed] [Google Scholar]
- 16.Murray M G, Thompson W F. Nucleic Acids Res. 1981;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, K. D., Wang, J., Li, H. B., Xu, C. G., Liu, A. M., Li, X. H. & Zhang, Q. (1997) Theor. Appl. Genet., in press.
- 18.Wu K, Tanksley S D. Mol Gen Genet. 1993;241:225–235. doi: 10.1007/BF00280220. [DOI] [PubMed] [Google Scholar]
- 19.Panaud O, Chen X, McCouch S R. Mol Gen Genet. 1996;252:597–607. doi: 10.1007/BF02172406. [DOI] [PubMed] [Google Scholar]
- 20.Lincoln, S., Daly, M. & Lander, E. (1992) Constructing Genetics Maps with Mapmaker/Exp. 3.0, Whitehead Inst. Tech. Rep. (Whitehead Inst., Cambridge, MA).
- 21.Lincoln, S., Daly, M. & Lander, E. (1992b) Mapping Genes Controlling Quantitative Traits with Mapmaker/QTL 1.1, Whitehead Inst. Tech. Rep. (Whitehead Inst., Cambridge, MA).
- 22.Snedecor G W, Cochran W G. Statistical Methods. Ames: Iowa State Univ. Press; 1980. [Google Scholar]
- 23.StatSoft. Statistica. Tulsa, OK: StatSoft; 1991. [Google Scholar]
- 24.Lin S, Min S. Rice Varieties and Their Genealogy in China. Shanghai: Shanghai Science and Technology; 1991. (Chinese). [Google Scholar]
- 25.Lander E S, Botstein D. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Gao Y J, Yang S, Ragab R, Saghai Maroof M A, Li Z B. Theor Appl Genet. 1994;89:185–192. doi: 10.1007/BF00225139. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Gao Y J, Saghai Maroof M A, Yang S H, Li J X. Mol Breed. 1995;1:133–142. [Google Scholar]
- 28.Allard R W, Zhang Q, Saghai Maroof M A, Muona O M. Genetics. 1992;131:957–969. doi: 10.1093/genetics/131.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]