Abstract
An in vitro system to study the mechanism of site-specific integration of adeno-associated virus (AAV) was developed. This system is based on two substrates, a linear or circular AAV donor and a circular acceptor containing the preintegration locus AAVS1. In the presence of HeLa extract and the His-Tag-purified Rep68 protein, specific covalent junctions between AAV and AAVS1 were formed and detected by PCR. The majority of the junctions were located within the Rep binding site of both the AAV and the AAVS1 substrates, underlining the involvement of the Rep protein. A limited amount of replication and the presence of nuclear factors promoted the efficiency of the reaction. The process was ATP-dependent, indicating that the helicase activity of Rep may be important in the formation of the junctions. According to current models of integration, the formation of the junctions would represent a first step in the process of AAV integration. This step could be crucial for the site specificity of the recombination event that leads to the integration of AAV into human chromosome 19 in vivo.
Under nonpermissive conditions, adeno-associated virus 2 (AAV) infection of human cell lines results in stable integration of the viral DNA into a specific site on chromosome 19 (1–4). AAV is maintained as a latent provirus until superinfection with a helper virus (e.g., adenovirus) induces the lytic phase of the AAV life cycle (5). Subsequently, AAV gene expression is activated and leads to rescue of the provirus from the chromosome, replication of the viral genome, and production of progeny virus. AAV is unique among eukaryotic DNA viruses in its ability to integrate at a specific site within the human chromosome (19q13.3–qter) and for this reason, and because it is nonpathogenic, has become increasingly attractive as a vector for gene delivery. To enhance the utility of AAV as a vector, it is important to fully understand the mechanism of AAV site-specific integration.
The linear single-stranded AAV genome contains two ORFs and inverted terminal repeats (ITRs) of 145 nt at its ends. The ORF in the left half of the genome encodes four regulatory proteins with overlapping primary sequences (the larger products are extended at the amino termini) known as the Rep proteins. Both the ITRs and the larger Rep proteins are required for site-specific integration (6, 7).
The ITRs contain the Rep binding site (RBS) and a specific cleavage site for the bound Rep (i.e., a terminal resolution site, TRS) and constitute the origin for viral replication (8–12). The AAV preintegration site in chromosome 19, termed AAVS1, has been cloned and a region of 4 kb has been sequenced (3). Genetic analysis of the AAVS1 target locus led to identification of a 33-bp region required and sufficient to target AAV integration (13–15). This short sequence also contains the two signals (RBS and TRS) that are present in the ITR of AAV. Thus, at the genetic level RBS and TRS have been identified as the necessary cis-active sequences in both the incoming viral DNA and in the target site. The large Rep proteins have been shown to be the necessary viral trans-active component (6, 16, 17).
In vitro studies have shown that the RBS and TRS signals within the AAVS1 target locus can serve as an origin for Rep-mediated replication (18). In addition, Rep is able to form complexes by binding to an AAV ITR and an AAVS1 oligonucleotide containing the two signals (19). These findings have been summarized in a model that suggests that as a first step of the integration process, a limited amount of Rep-mediated replication generates a junction between AAV and the AAVS1 sequence (15, 20, 21). Rep, as a hexamer, binds to the RBS of a circular AAV genome and directs AAV to the AAVS1 target locus by forming a complex at the RBS and TRS sites of AAVS1. After nicking the AAVS1 substrate at TRS, the helicase activity of Rep unwinds the DNA and thereby initiates replication by the cellular polymerase complex. Strand switching, a characteristic feature of AAV replication in the absence of adenoviral lytic infection, then will lead to the formation of the AAV–AAVS1 junctions.
To facilitate the investigation of AAV recombination, we decided to develop an in vitro system for AAV site-specific integration. We have designed an assay based on PCR that specifically and with high sensitivity detects the formation of the AAV–AAVS1 junction, which is believed to be the first step to occur in AAV site-specific integration. In accordance with the model, the generation of these AAV–AAVS1 junctions depends on the presence of Rep, the ITR of the AAV substrate, and the AAVS1 target locus.
Materials and Methods
Primers and Probes.
The oligonucleotides were synthesized by Gene Link or Genosys. The AAV primers for the right end were BAAV4 (5′-GCCGGATCCGCGTGGAGATCGAGTGGGAGC-3′; 4239–4259) and BAAV8 (5′-CCCGGATCCTTGGCACCAGATACCTGACTCG-3′; 4383–4401) and for the left end Brml3 (5′-GCCGGATCCTCACGTGACCTCTAATACAGG-3′; 193–214). The AAVS1 primers were H4d1 (5′-GGCAAGCTTCCATCCTCTCCGGACATCGCAC-3′, 426–447) and H5d (5′-GCCAAGCTTCGGCGTTGGTGGAGTCCAGCAC-3′; 703–725). The primer AAVD (14) was used as a hybridization probe to specifically detect the ITR region of AAV, and the primer H4d1 was used to detect the AAVS1 sequence.
DNA Substrates.
The linear AAV substrate was derived by BglII digestion of pBAV2. pBAV2 contains the AAV 2 genome cloned into the BglII site that had been inserted into the ScaI site of pBR322 (C. Giraud and K.I.B., unpublished data). For the circular AAV construct, the BglII fragment was religated. For the “head-to-tail” (HT) substrate the equivalent of a full-length AAV genome was isolated by PpuMI digestion of the recombinant R31 retrieved from the cellular recombination assay (14). A small PpuMI fragment (product 189–249) containing the P5 promoter was lost when the PpuMI AAV DNA was recircularized, recut with BamHI, and cloned into the BamHI site of pUC18, resulting in the construct pJZ1. To prepare the HT substrate, pJZ1 was digested with BamHI. “No-end” DNA was prepared as described (22). Linear AAV DNA lacking the ITRs was prepared by digestion of psub201 with XbaI. For all of the substrates, the vector backbone was cut with additional restriction enzymes. The vector fragments then were separated from the DNA substrates by use of a 5–20% sucrose gradient (23). Sucrose gradient purification was done, because agarose isolation of AAV substrates resulted in very poor replication in the in vitro assay (P. Ward, J.D., and K.I.B., unpublished data). The AAVS1 constructs p220.2/AAVS1 (kilobases 0–1.6) and p220.2/AAVS1 (kilobases 0.51–1.6) have been described (13).
Cell Extracts.
Cytoplasmic extracts from HeLa cells, uninfected or infected with adenovirus, were prepared as described (24, 25). For a nuclear extract enriched with DNA binding proteins, HeLa nuclei were resuspended in one packed nuclei volume of low-salt buffer (20 mM Hepes, pH 7.9/1.5 mM MgCl2/20 mM KCl/0.2 mM EDTA/0.2 mM PMSF/0.5 mM DTT]. Two packed nuclei volumes of extraction buffer (20 mM Hepes, pH 7.9/1.5 mM MgCL2/0.2 M KCl/10 mM EDTA/10 mM EGTA/0.2 mM PMSF/0.5 mM DTT] were added dropwise while stirring. The resulting suspension was tilted 1 h and then centrifuged for 30 min at 25,000 × g. The supernatant was dialyzed twice for 4 h against 50 vol of dialysis buffer (20 mM Hepes, pH 7.9/20% glycerol/100 mM KCl/0.2 mM EDTA/0.2 PMSF/0.5 mM DTT]. The dialysate was centrifuged for 20 min at 25,000 × g and then stored at −80°C. Crude high mobility group (HMG) extracts were prepared by an acid-free method (26) and by perchloric acid extraction (27, 28). A mixture of equal amounts of the two extracts was used in the integration assay.
Integration Assay.
Integration reactions were carried out in 15 μl and contained 40 mM Hepes (pH 7.7), 7 mM MgCl2, 4 mM ATP, 200 μM CTP, 200 μM GTP, 200 μM UTP, 100 μM dATP, 100 μM dCTP, 100 μM dGTP, 100 μM dTTP, 2 mM DTT, 40 mM creatine phosphate, 0.5 μg of creatine phosphokinase, 30 μg of HeLa extract protein, 50 ng of AAV substrate, 100 ng of AAVS1 substrate, and 50 ng of His-Tag-purified Rep68 (molar ratio: AAV/AAVS1/ReP = 1:1:50). Reactions were incubated at 34°C for 6–16 h and then stopped by adding 50 μl of a solution of 20 mM Hepes (pH 7.9), 10 mM KCl, 10 mM EDTA, 1% SDS, and 50 mM NaCl. After proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation, the DNA products were dissolved in 20 μl of 2.5 mM Tris⋅HCl (pH 7.5) and 0.25 mM EDTA. PCR amplification was performed with 1–5 μl of the resuspended DNA pellet as described below.
PCR Amplification and Analysis of the AAV–AAVS1 Junctions.
To facilitate amplification of the G+C-rich regions, the PCRs were carried out with the Advantage-GC PCR kit (CLONTECH). The cycling conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 45 sec and 72°C for 3 min. The PCR products were analyzed on 1.5% agarose gels and then transferred bidirectionally (29) to Hybond-N nylon membranes (Amersham Pharmacia). Southern blot hybridization was performed according to the manufacturer’s protocol using the primers AAVD or H4d1 as probes (Boehringer Mannheim). The probes were labeled with the digoxigenin oligonucleotide 3′-End Labeling Kit (Boehringer Mannheim).
Cloning and Sequencing of the AAV–AAVS1 Junctions.
The restriction sites BamHI and HindIII included within the PCR primers were used to clone the AAV–AAVS1 junctions into pBluescript SK+. Sequencing was done with the primers T3HT (5′-AATTAACCCTCACTAAAGGG-3′) or BAAV8 at the BioResource Center at Cornell University in Ithaca.
Results
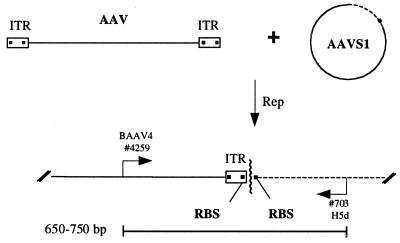
To facilitate the study of AAV site-specific integration, an in vitro system based on the AAV replication assay was developed (Fig. 1). The reaction contained an extract from HeLa nuclei, the His-Tag-purified Rep68, and two substrates for recombination, double-stranded AAV as the donor substrate and a circular plasmid DNA carrying the AAVS1 integration locus as the acceptor substrate. Characterization of provirus configurations in latent cell lines, as well as provirus configurations recovered in Epstein–Barr virus episome integrants (13, 14), has shown that junctions occur preferentially in the vicinity of the RBS and the TRS signals of the two substrates (refs. 6 and 14 and N. Dutheil, personal communication). According to the model (15), the initial events in AAV integration should lead to the formation of intermediates containing a characteristic “AAV–AAVS1” junction as shown in Fig. 1. Therefore, we designed primers that allow specific amplification of the AAV–AAVS1 junction by PCR. As the breakpoints of recombinant junctions analyzed differ, PCR products were expected to vary within a range of 100 bp (650–750 bp).
Figure 1.
Schematic diagram of the assay for AAV site-specific integration. Rep mediates the formation of specific junctions between the linear AAV substrate and the circular substrate containing the preintegration locus AAVS1. The AAV–AAVS1 junctions are detected by PCR with specific primers for the AAV (BAAV4) and the AAVS1 (H5d) sequences. Depending on the position of the crossover points (wavy line), the length of the amplified fragments was expected to vary between 650 and 750 bp.
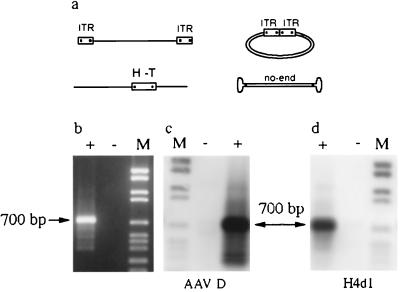
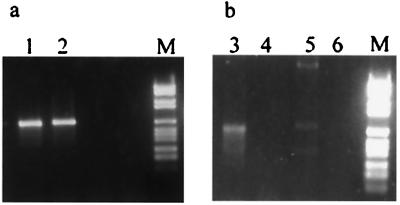
Four different AAV donor substrates were compared in the in vitro integration system (Fig. 2a). Initially, a linear AAV substrate with the ITRs positioned at the ends of the AAV molecule was used. This substrate also was used in its circularized form. The HT AAV substrate was isolated from the recombinant R31 generated in the cellular system (14). Apart from deletion of a short sequence including the P5 promoter, the HT-AAV substrate represents a full AAV genome equivalent containing the fused termini with HT organization at an internal position (positions 257–4555, 1–248, and 249–256 deleted). The appearance of the HT formation of the ITRs in several recombinants was seen as an indication that circularization of the AAV genome might form a precursor to integration (14, 27). Others have suggested that an AAV substrate termed “no-end” DNA represents a precursor for AAV integration (22, 30). No-end AAV DNA is a double-stranded linear AAV genome that has closed hairpin structures at both ends. Surprisingly, all four AAV substrates generated junctions at comparable yields and the PCR products were of a similar size, about 700 bp (Fig. 2 and data not shown). The junctions were detected only if the in vitro reactions were incubated with Rep (Fig. 2b). Bidirectional Southern blot hybridization showed that amplified PCR products were positive for the AAV (an internal oligonucleotide from the AAV ITR) and AAVS1 probes (Fig. 2 c and d).
Figure 2.
PCR detection and analysis of AAV–AAVS1 junctions. (a) Substrates tested in the integration assay are shown. (b) The integration assay was carried out with the HT-AAV substrate in the presence (+) or absence (−) of Rep. The integration products were detected by PCR using the primers BAAV4 and H5d and analyzed by gel electrophoresis. (c) Southern blot hybridization with an AAV probe (AAV D). (d) Southern blot hybridization with an AAVS1 probe (H4d1). Lane M contains the digoxigenin-labeled DNA molecular weight marker VI from Boehringer Mannheim.
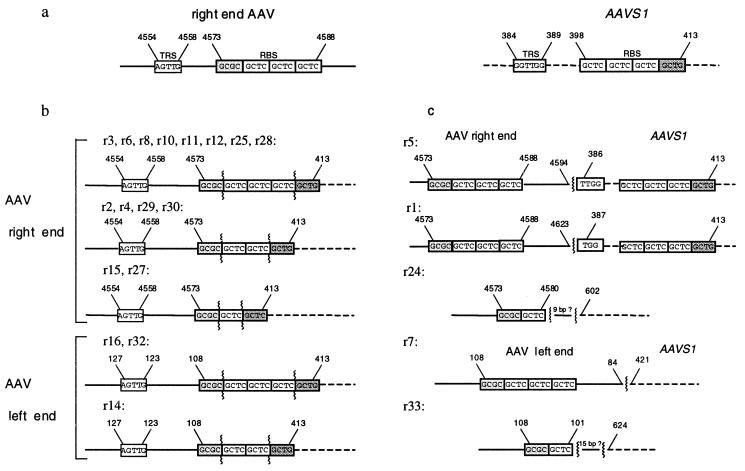
Cloning and sequencing the PCR products revealed the structures for the AAV–AAVS1 junctions (Fig. 3). A summary of the recombinant junctions retrieved from the integration assay is given in Table 1. The junctions were PCR-amplified with an AAV primer, specific for the right (BAAV4, BAAV8) or the left end (Brml3) of the AAV genome, and an AAVS1 primer located close to the RBS (H4d1) or further downstream of the RBS (H5d). Independent of which AAV substrate was used, the majority of the junctions were localized directly at the RBS of AAV and AAVS1, the only difference being the number of the repeats of the RBS (Fig. 3b). Five junctions were located outside of the RBS of AAVS1 and AAV or outside of AAVS1 alone (Fig. 3c).
Figure 3.
Structures of the AAV–AAVS1 junctions generated in the integration assay. (a) The regions of both substrates involved in formation of the AAV–AAVS1 junctions are shown. The elements TRS and RBS are indicated with their respective nucleotide positions. (b) AAV–AAVS1 junctions with the crossover point within the RBS of AAV and AAVS1. (c) AAV–AAVS1 junctions with the crossover point outside of the RBS of AAV and AAVS1 or outside of AAVS1 only.
Table 1.
Summary of the cloned recombinant junctions and the AAV substrates and PCR primers used for generation and amplification
| Recombinant | AAV substrate | AAV primer | AAVS1 primer |
|---|---|---|---|
| r1 | No end DNA | BAAV8 | H4dl |
| r2 | No end DNA | BAAV8 | H4dl |
| r3 | No end DNA | BAAV8 | H4dl |
| r4 | HT-AAV | BAAV8 | H4dl |
| r5 | HT-AAV | BAAV8 | H4dl |
| r6 | HT-AAV | BAAV8 | H4dl |
| r15 | HT-AAV | BAAV4 | H4dl |
| r8 | HT-AAV | BAAV4 | H5d |
| r10 | HT-AAV | BAAV4 | H5d |
| r24 | HT-AAV | BAAV4 | H5d |
| r25 | HT-AAV | BAAV4 | H5d |
| r29 | HT-AAV | BAAV4 | H5d |
| r30 | HT-AAV | BAAV4 | H5d |
| r27 | HT-AAV | BAAV4 | H4d |
| r11 | HT-AAV | BAAV4 | H5d |
| r12 | HT-AAV | BAAV4 | H5d |
| r28 | HT-AAV | BAAV4 | H5d |
| r14 | Linear AAV | Brml 3 | H5d |
| r16 | Linear AAV | Brml 3 | H5d |
| r7 | Circularized AAV | Brml 3 | H5d |
| r32 | Circularized AAV | Brml 3 | H5d |
| r33 | Circularized AAV | Brml 3 | H5d |
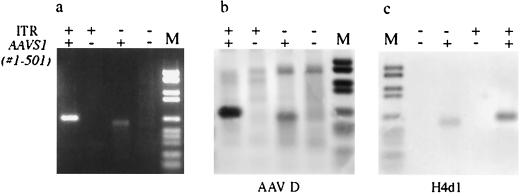
The RBS and TRS signals of the AAVS1 preintegration locus have been shown to be essential to direct integration of AAV into the AAVS1 episome (15). To test the importance of these signals for formation of junctions in our assay, the regions containing the signals were removed from the AAV substrate (deletion of the ITRs) and the AAVS1 insert (deletion of positions 1–501). As expected, the deletion mutants could not generate any junctions, showing that the presence of the RBS and TRS signals are essential (Fig. 4). A weak band was detected with the AAV substrate with the ITRs deleted, indicating that a small amount of nonspecific interaction between the substrates had occurred. However, this signal could be removed completely simply by denaturing the DNA (10 min, 95°C; chill on ice, 5 min) before PCR amplification (Fig. 5, lanes 3 and 4). The weak signal represented a PCR artefact, and it now became important to find out whether the junctions generated with the original substrates were covalent before amplification by PCR. Indeed, PCR products of similar size could be amplified simply by mixing the two substrates and incubating the solution over a longer period (Fig. 5, lane 5). Heat denaturation of the DNA removed the nonspecific signal (Fig. 5, lane 6). In contrast, the PCR signals amplified from the DNA that had been incubated with extract and Rep in the reaction mix were resistant to denaturation, although a decrease of the signal was observed sometimes (Fig. 5, lanes 1 and 2). As a final control, we incubated the AAV and AAVS1 substrates separately in the assay, which should allow only replication of the substrates. The DNA products of the two reactions were mixed at a ratio of 1:1 and amplified by PCR. No signal was detected under these conditions (data not shown), showing that replication of the two substrates did not create intermediates that promote artificial AAV–AAVS1 junctions during PCR. Therefore, it is likely that the majority of the junctions detected by PCR represent covalent AAV–AAVS1 junctions formed during the in vitro integration reaction in the presence of Rep.
Figure 4.
Formation of the AAV–AAVS1 junctions depends on the TRS and RBS signals on both the AAV and the AAVS1 substrates. The integration assay was carried out with AAV and AAVS1 substrates containing (+) or lacking (−) the TRS and RBS signals. (a) Gel analysis of the PCR products. (b) Southern blot hybridization with an AAV probe (AAV D). (c) Southern blot hybridization with an AAVS1 probe (H4d1). Lane M contains the digoxigenin-labeled DNA molecular weight marker VI from Boehringer Mannheim.
Figure 5.
Heat denaturation removes unspecific interactions between the AAV and AAVS1 DNA substrates. The integration assay was carried out with an AAV substrate containing (lanes 1 and 2) or lacking (lanes 3 and 4) the TRS and RBS signals. In lanes 5 and 6, the two DNA substrates alone were incubated in Tris/EDTA buffer at room temperature for 16 h. In lanes 2, 4, and 6, the DNA was heat-denatured (10 min at 95°C and 5 min on ice) before PCR amplification. Lane M contains the DNA molecular weight marker VI from Boehringer Mannheim.
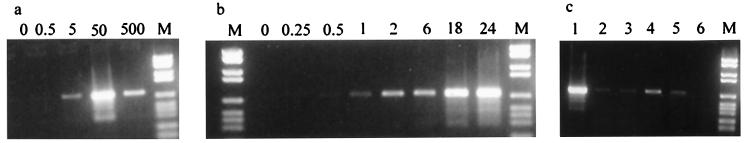
A preliminary characterization of the requirements for the assay was done (Fig. 6). Because the PCRs were carried out without internal controls, only semiquantitative data could be obtained. Titration of the His-Tag Rep68 showed that a molar ratio of 50:1 of the Rep to the set of substrates is optimal (Fig. 6a). A 10-fold increase or decrease of Rep resulted in lower yields of product. The yield of recombinant junctions increased progressively with continued incubation for 24 h (Fig. 6b).
Figure 6.
Characterization of the in vitro integration assay. (a) The integration assay was carried out with increasing ratios of Rep to substrates (0:1, 0.5:1, 5:1, 50:1, and 500:1). (b) The integration reaction was incubated at 34°C for 24 h, aliquots were removed at eight points (0, 0.25, 0.5, 1, 2, 6, 18, and 24 h), and the reaction was stopped by adding SDS. (c) Lane 1 represents the integration assay as described. The integration reaction was carried out in absence of Mg2+ (lane 2), ATP, creatine phosphate, and creatine phosphokinase (lane 3), HeLa extract (lane 4), nucleotides (lane 5), or DNA substrates (lane 6). Lane M contains the DNA molecular weight marker VI from Boehringer Mannheim.
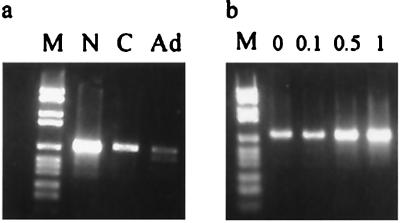
The efficiency of the reaction was reduced drastically when the ATP-regenerating system was not present (Fig. 6c, lanes 1 and 3). This can be easily explained, because Rep itself is an ATPase (11). The yield of product also seems to rely on the presence of the divalent cation Mg2+, which implies the involvement of DNA binding proteins (Fig. 6c, lane 2). The model for AAV site-specific integration suggests that a certain amount of replication is involved in the event and that the AAV–AAVS1 junction is formed by strand switching of the cellular polymerase complex (15). The formation of the junction was reduced significantly when the extract or the nucleotides were removed from the reaction (Fig. 6c, lane 4 and 5). Surprisingly, a small amount of activity was detectable in the absence of the HeLa extract (Fig. 6c, lane 4). Heat treatment did not remove the signal, indicating that the junctions formed were covalent and not created during PCR amplification. Unless the Rep protein was contaminated with replication proteins, one has to conclude that to a certain extent the junctions are formed independent of replication or, alternatively, contamination of the Rep protein with endonuclease and ligase activities might give rise to replication-independent junctions. Comparison of different extracts showed that the integration assay performed best with the nuclear extract that had been prepared in the presence of EDTA to release the DNA binding proteins (Fig. 7a, lane N). The cytoplasmic extracts from uninfected and adenovirus-infected cells did less well (Fig. 7a, lanes C and Ad). The higher yields seen with the nuclear extract indicated that nuclear DNA binding proteins may be involved in promoting the formation of the AAV–AAVS1 junctions. Protein HMG1 is a DNA binding protein that bends DNA and is involved in various recombination processes (31–33). Recently, HMG1 has been suggested to enhance the assembly of the nucleoprotein structure that is involved in AAV site-specific integration (27). HMG1 interacts with Rep and enhances its DNA binding to RBS and nicking activities at TRS. Adding HMG-enriched extract to the cytoplasmic extract did indeed increase the intensity of the signal (Fig. 7b). More detailed studies with purified HMG will be necessary to confirm these initial results.
Figure 7.
Comparison of various HeLa extracts in the integration assay. (a) The reaction was carried out with 30 μg of uninfected nuclear extract (lane N), uninfected cytoplasmic extract (lane C), or adenovirus-infected cytoplasmic extract (lane Ad). (b) Increasing amounts of crude HMG extract (0, 0.1, 0.5, and 1 μg) was added to the integration reaction containing cytoplasmic HeLa extract.
Discussion
We have developed an assay that detects the formation, in an in vitro system, of covalent junctions between AAV DNA and the target sequence for in vivo AAV site-specific integration. In this assay linear AAV is targeted to the AAVS1 insert of a circular substrate when incubated in a cellular extract supplemented with the viral protein Rep. The interaction of the two substrates with each other leads to the formation of a specific AAV–AAVS1 junction that can be detected by PCR. The generation of the AAV–AAVS1 junctions depends on Rep in trans and the presence of the ITR in the AAV donor and the 500-nt sequence known to contain the in vivo target site in the AAVS1 substrate. The products are resistant to heat, extensive treatment with proteinase K, and subsequent phenol and chloroform extraction, suggesting that the products formed contain covalent junctions. Experiments using Escherichia coli transformation did not lead to the detection of circular recombinants (J.D. and K.I.B., unpublished data). We, therefore, think it unlikely that the identified junctions belong to products that represent fully resolved recombinants. However, the AAV–AAVS1 junctions could belong to intermediates of recombination, and their generation would represent the first step of the AAV site-specific recombination event according to proposed models (15, 20, 21).
Seventeen of the 22 junctions were within the RBS. With the model system used to study site-specific integration in vivo (13, 14), only one of the junctions that were sequenced showed joining within the RBS with preservation of the entire sequence (14). All but one, though, were grouped very close to the RBS within AAVS1 (within 15 nt). Thus, the in vitro assay does appear to mimic the in vivo model system. AAV-specific DNA synthesis can be initiated on purified AAV or AAVS1 DNA in vitro (18, 34). The models for AAV integration have proposed that integration begins by a copy choice mechanism in which the elongating strand switches the template being copied (in this case from a viral to a cellular strand or vice versa). The existence of short repeated sequences, such as those found within RBS, may well facilitate the process (35–37). If the in vitro assay is a faithful reflection of integration in vivo, we must suppose that there could be sufficient differences between the in vitro and in vivo mechanisms to explain why the in vitro switch would be so much more specific, although these are unknown. Also, fully half the junctions observed in vivo occur near the RBS, proximal to the leftward most promoter at map position 5. In the in vitro assay, all detected junctions occurred within the ITR. However, this is simply a reflection of the primers used. Junctions near p5 would not have been observed. In any event, the requirements for the observed reaction in vitro and the products seem sufficiently similar to those observed in vivo to warrant further characterization of the reaction.
Our major concern was whether the junctions detected by PCR could have been the result of experimental artifact. Clearly, some of the bands observed were of this nature because they could be eliminated by heating before the PCR. Indeed, these may have represented earlier intermediates held together only by hydrogen bonding, but they did not contain covalent junctions. Formation of apparent covalent junctions resistant to heat denaturation required the ITR in the AAV and the region of AAVS1 carrying the target site for integration. Formation of the covalent junctions also was time-dependent and required an appropriate stoichiometric ratio of AAV Rep protein to DNA substrate. Finally, incubation of the two DNA substrates in separate reactions that were mixed after the reactions were stopped but before the PCR did not yield detectable junctions.
The reaction was enhanced by, but did not seem to totally require, Mg2+, all four deoxynucleotides, or an ATP-generating system. These observations are similar to those of Ward and Berns (34) in their characterization of the in vitro DNA replication assay and in large part are likely to reflect the presence of a large amount of cell extract in the reaction mixture. However, the presence of a detectable band representing a junction in the absence of the cell extract is more puzzling, and we have no good explanation. This is particularly true in the light of the reaction enhancement observed with addition of an extract enriched for HMG proteins to the reaction mixture.
The observation that an extract from uninfected cells was more effective in the in vitro assay for integration than an extract from adenovirus-infected cells was in accord with observations in vivo that adenovirus infection activates AAV replication and rescue from the integrated state (5) but does not promote integration.
The in vitro system for AAV site-specific integration provides a useful tool to define the optimal conditions for AAV integration that will lead to fully resolved recombinants. Testing fractionated cell extracts, chemical inhibitors, or antibodies directed against specific cellular proteins in this assay should allow the identification of the proteins involved in AAV integration and eventually lead to reconstitution of the preintegration complex of AAV.
Apart from providing the information needed for efficient and safe rAAV vectors, the system also could find answers to other interesting questions such as whether AAV actually is able to integrate in a cell that does not have any DNA synthesis at all. This is an important issue for gene therapy for nondividing cells and has until now been difficult to prove in vivo.
Acknowledgments
We thank Catherine Giraud for providing plasmids, Frank Dean for the HeLa nuclei, and Peter Ward for the adenovirus-infected extract. We thank Rob Kotin, Frank Dean, Catherine Giraud, Peter Ward, Patricio Menesis, and Henry Hamilton for helpful discussions and Nenita Cortez for excellent technical support. This work was supported by Grants AI 22251 and GM 50032 from the National Institutes of Health.
Abbreviations
- AAV
adenovirus-associated virus
- ITR
inverted terminal repeat
- TSR
terminal resolution site
- HT
“head-to-tail”
- HMG
high mobility group
References
- 1.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotin R M, Menninger J C, Ward D C, Berns K I. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 3.Kotin R M, Linden R M, Berns K I. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns K I. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balague C, Kalla M, Zhang W-W. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straus S E, Sebring E D, Rose J A. Proc Natl Acad Sci USA. 1976;73:742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauswirth W W, Berns K I. Virology. 1979;93:57–68. doi: 10.1016/0042-6822(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 10.Im D S, Muzyczka N. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Im D S, Muzyczka N. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 12.Im D S, Muzyczka N. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giraud C, Winocour E, Berns K I. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraud C, Winocour E, Berns K I. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linden R M, Winocour E, Berns K I. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelling A N, Smith M G. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 17.Kearns W G, Afione S A, Fulmer S B, Pang M G, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 18.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzman M D, Kyöstiö S R M, Kotin R M, Owens R A. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linden R M, Giraud C, Winocour E, Berns K I. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiorini J A, Wiener S M, Yang L, Smith R H, Safer B, Kilcoin N P, Liu Y, Urcelay E, Kotin R M. Curr Top Microbiol Immunol. 1996;218:25–33. doi: 10.1007/978-3-642-80207-2_2. [DOI] [PubMed] [Google Scholar]
- 22.Snyder R O, Samulski R J, Muzyczka N. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 24.Wobbe C R, Dean F, Weissbach L, Hurwitz J. Proc Natl Acad Sci USA. 1985;81:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward P, Berns K I. J Mol Biol. 1991;218:791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
- 26.Wagner J P, Quill D M, Pettijohn D E. J Biol Chem. 1995;270:7394–7398. doi: 10.1074/jbc.270.13.7394. [DOI] [PubMed] [Google Scholar]
- 27.Costello E, Saudan P, Winocour E, Pizer L, Beard P. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stros M, Nishikawa S, Dixon G H. Eur J Biochem. 1994;225:581–591. doi: 10.1111/j.1432-1033.1994.00581.x. [DOI] [PubMed] [Google Scholar]
- 29.Hudson M C, Reynolds T L. Biotechniques. 1993;14:382. [PubMed] [Google Scholar]
- 30.Yang C C, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bianchi M E, Beltrame M, Paonessa G. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 32.Paull T T, Haykinson M J, Johnson R C. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 33.Segall A M, Goodman S D, Nash H A. EMBO J. 1994;13:4536–4548. doi: 10.1002/j.1460-2075.1994.tb06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward P, Urcelay E, Kotin R, Safer B, Berns K I. J Virol. 1994;68:6029–6037. doi: 10.1128/jvi.68.9.6029-6037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang R S. BioEssays. 1994;16:785–788. doi: 10.1002/bies.950161102. [DOI] [PubMed] [Google Scholar]
- 36.D’Alençon E, Petranovic M, Mechel B, Noiroy P, Aucouturier A, Uzest M, Ehrlich S D. EMBO J. 1994;13:2725–2734. doi: 10.1002/j.1460-2075.1994.tb06563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canceill D, Ehrlich S D. Proc Natl Acad Sci USA. 1996;93:6647–6652. doi: 10.1073/pnas.93.13.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]