Abstract
A M182T substitution was discovered as a second-site suppressor of a missense mutation in TEM-1 β-lactamase. The combination of the M182T substitution with other substitutions in the enzyme indicates the M182T substitution is a global suppressor of missense mutations in β-lactamase. The M182T substitution also is found in natural variants of TEM-1 β-lactamase with altered substrate specificity that have evolved in response to antibiotic therapy. The M182T substitution may have been selected in natural isolates as a suppressor of folding or stability defects resulting from mutations associated with drug resistance. This pathway of protein evolution may occur in other targets of antimicrobial drugs such as the HIV protease.
Keywords: antibiotic resistance, protein evolution
The production of β-lactamase is the principal mechanism of bacterial resistance to β-lactam antibiotics, such as penicillins and cephalosporins. β-lactamase provides resistance by catalyzing the hydrolysis of β-lactam antibiotics to ineffective antimicrobials. TEM-1 β-lactamase is the most prevalent plasmid-mediated β-lactamase in Gram-negative bacteria (1). It is able to efficiently hydrolyze penicillins and most cephalosporins, but not the more recently developed extended-spectrum cephalosporins, such as ceftazidime and cefotaxime (2). In addition, β-lactamase inhibitor compounds, such as clavulanic acid, have been developed. These compounds themselves do not possess antimicrobial activity but instead are used in conjunction with other β-lactams, such as ampicillin (3). Soon after the introduction of extended-spectrum cephalosporins and inhibitors there were reports of transferable resistance to the drugs (4). Cloning and DNA sequencing revealed that much of the resistance was due to TEM β-lactamase enzymes that contained 1–4 amino acid substitutions (4). These substitutions alter the substrate profile of the enzyme such that it can hydrolyze the extended-spectrum cephalosporins or is insensitive to β-lactamase inhibitors (4, 5). Interestingly, most of the substitutions found in inhibitor-resistant enzymes are different from those found in enzymes able to cleave extended-spectrum cephalosporins (5). An exception appears to be the M182T substitution, which has been identified in both inhibitor-resistant enzymes (TEM-32) and extended-spectrum β-lactamases (TEM-43) (refs. 6 and 7; K. Bush, personal communication). In this study, the M182T substitution was identified as a second-site suppressor of an asparagine for leucine substitution at position 76, which is buried in the TEM-1 structure. Further experiments demonstrated the M182T substitution can suppress the effects of deleterious substitutions at other sites in the protein. These findings suggest that the M182T substitution acts as a global suppressor of β-lactamase substitutions that disrupt the folding and/or stability of the enzyme.
MATERIALS AND METHODS
Strains and Plasmids.
Escherichia coli BW313 [Hfr lysA(61–62) dut1 ung1 thi1 recA1 spoT1] was used to propagate plasmid DNA before oligonucleotide-directed mutagenesis (8). Mutagenized DNA was initially introduced into E. coli ES1301 [lacZ53 mutS201::Tn5 thyA36 rha5 metB1 deoCIN(rrnD-rrnE)] (9). E. coli ES1301 also was used as the mutator strain in reversion analysis. E. coli XL1-Blue [recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′::Tn10 (Tetr) proAB ΔlacIq (lacZ)M15]] was used as the host for the assay of antibiotic susceptibility, for immunoblotting, for specific activity measurements, and for the preparation of single-stranded DNA (10). E. coli SB646 [ΔfhuA Δptr ΔdegP ΔompT Δprc::kan] is a protease-deficient strain that was used in immunoblotting and specific activity experiments. This strain was a gift of Steve Bass (Genentech). Mutagenesis was performed on the plasmid pBG66, which was used in previous studies (11). The pBG66 plasmid contains the wild-type blaTEM-1 gene and a cat gene encoding chloramphenicol acetyltransferase. This 4.8-kb plasmid also contains the ColEI and f1 origins of DNA replication.
Construction of Mutants.
The L76N and L76S substitutions were constructed by oligonucleotide-directed mutagenesis using the method of Kunkel et al. (8). The L76 codon was randomized using the following oligonucleotide, where S represents C or G and N represents a mixture of all four nucleotides: L76X 5′-AATACCGCGCCACASNNCAGAACTTTAAAAGTG-3′.
The template for mutagenesis was the pBG66 plasmid containing a SalI linker insertion into codon 76 of the blaTEM-1 gene (12). In addition, a deletion of two nucleotides from codon 76 created a frameshift mutation and rendered this mutant nonfunctional. The L76X oligonucleotide was annealed to a single-stranded DNA template of the SalI insertion mutant, and mutagenesis was performed exactly as described by Huang et al. (12). The L76N and L76S substitutions were identified by DNA sequencing a collection of 40 mutants.
The M182T single mutant was constructed by digesting the pBG66 plasmid containing the L76N:M182T double substitution with HincII and EcoRI which releases a DNA fragment containing the L76 codon, but not the M182 codon. A HincII-EcoRI fragment containing the wild-type L76 codon then was used to replace the released fragment.
The I47Y:E48C mutant was selected from the L46–48 random library of mutants (12). The mutant was selected by transforming the L46–48 plasmid library into E. coli XL1-Blue and spreading the transformed cells on Luria–Bertani (LB) agar supplemented with 12.5 μg/ml chloramphenicol. Individual colonies then were picked and patched onto agar plates containing either 1 mg/ml or 100 μg/ml ampicillin. The I47Y:E48C mutant was picked for DNA sequencing, and further characterization was based on the fact that it grew on plates with 100 μg/ml, but not 1 mg/ml, ampicillin. The I47Y:E48C:M182T mutant was constructed as described above for the M182T mutant.
The M69I and M69I:M182T mutants were constructed by oligonucleotide-directed mutagenesis using the method of Kunkel et al. (8). The following oligonucleotide was used: M69I 5′-CTTTAAAAGTGCTTATCATTGGAAAACG-3′.
The M69I mutant was constructed by annealed the M69I oligonucleotide to a single-stranded DNA template from the pBG66 plasmid containing the wild-type blaTEM-1 gene. The M69I:M182T mutant was constructed by annealing the M69I oligonucleotide to a single-stranded DNA template from the pBG66 plasmid containing the M182T mutation. The mutagenesis protocol was as described by Huang et al. (12).
Selection of Revertants and Immunoblotting.
Revertants of the L76N mutant were isolated by introducing the pBG66 plasmid containing the blaTEM gene with the L76N mutation into E. coli ES1301 by electroporation. A single transformant was picked and grown for 16 hr at 37°C in 10 ml of 2× YT medium supplemented with 12.5 μg/ml chloramphenicol. As a control, the L76N plasmid was introduced into the nonmutator strain, E. coli XL1-Blue, and grown under identical conditions. Plasmid DNA was isolated from each culture by alkaline lysis (13). The plasmid DNA was electroporated into E. coli XL1-Blue, and the transformants were spread on LB agar plates supplemented with 500 μg/ml ampicillin. A portion of the transformation mix also was spread on LB agar supplemented with 12.5 μg/ml chloramphenicol to estimate the total number of cells transformed with plasmid. A total of 11 colonies were recovered from cells transformed with plasmid isolated from the mutator strain ES1301. This represents a mutant frequency of 2 × 10−5. No transformants were obtained with plasmid isolated from the E. coli XL1-Blue control strain. The frequency of mutant isolation from E. coli XL1-Blue was therefore <1.2 × 10−6. Plasmid DNA was isolated from each of the 11 mutants and retransformed into E. coli XL1-Blue. Transformants were spread on LB agar supplemented with 12.5 μg/ml chloramphenicol. Several transformants were picked for each putative mutant and streaked on LB agar supplemented with 500 μg/ml ampicillin to ensure that the high-level ampicillin resistance was due to a plasmid mutation. The DNA sequence of the entire blaTEM gene and 200 bp of the promoter region was determined for 6 of the 11 revertants that were isolated. DNA sequencing was performed by picking isolated single colonies for each revertant and inoculating the colony directly for the PCR to amplify the coding region and the upstream region of blaTEM. The amplified PCR product then was sequenced directly (14).
Steady-state levels of β-lactamase were examined by immunoblotting as described in Palzkill et al. (15).
Determination of Specific Activity of β-Lactamase Mutants.
Cultures of E. coli XL1-Blue containing the mutant β-lactamase to be tested were grown overnight at 37°C in 2 ml of 2× YT medium supplemented with 12.5 μg/ml chloramphenicol (13). Fifty microliters of the overnight culture was used to inoculate 10 ml of 2× YT with 12.5 μg/ml chloramphenicol. This culture was grown to log phase (OD600 = 0.5). The periplasmic contents of the cells were isolated by an osmotic shock procedure (16). The total protein concentration of isolated periplasmic protein preparation was determined by the method of Bradford (17). The β-lactamase activity was determined in 0.5 ml of phosphate buffer (pH 7.0). Ampicillin was present at a concentration of 300 μM in the reactions. The hydrolysis of ampicillin was monitored as the loss of absorbance at 235 nm in a Beckman DU-640 spectrophotometer. The specific activity was calculated as mM ampicillin hydrolyzed per min divided by the total protein concentration added to the reaction. The extinction coefficients used for ampicillin were ɛ = 2,160 M−1·cm−1, Δɛ = 900 M−1·cm−1. For the specific activity measurements at increasing temperatures the reaction mixtures, without ampicillin, were incubated at the temperature to be assayed for 5 min. Then, to initiate the reaction, preheated ampicillin was added to the reaction mixture, and ampicillin hydrolysis was monitored spectrophotometrically as described above. The reactions were monitored in the spectrophotometer at the same temperature as the preincubation temperature by using a water-jacketed, temperature-controlled cuvette.
Minimal Inhibitory Concentrations (MIC) of Ampicillin.
The MIC of ampicillin for the mutants was determined using E. coli XL1-Blue containing the pBG66 plasmid encoding either the wild-type or mutant derivatives of blaTEM. The strains were inoculated into LB medium supplemented with 12.5 μg/ml chloramphenicol and grown for 16 hr to stationary phase. Each culture was then diluted 1:106, and 0.1 ml of each culture was spread on an LB agar plate supplemented with the following concentrations of ampicillin: 0, 25, 50, 75, 100, 150, 250, 500, 750, 1,000, 1,500, 2,000, and 3,000 μg/ml. The LB plates containing 0 μg/ml ampicillin consistently grew 100–200 colonies. The MIC was recorded as the lowest concentration of ampicillin on which colonies did not grow. In practice, this was an all or nothing phenomenon, either 100–200 colonies grew or no colonies grew. Therefore, the MIC determination was not subjective.
RESULTS
In a previous study, we performed saturation mutagenesis in which each of the 263 codons of the gene for TEM-1 β-lactamase were randomized by oligonucleotide-directed mutagenesis (12). Functional random mutants were selected based on their ability to confer ampicillin resistance to E. coli. The DNA sequence of several functional mutants was determined for each set of random mutants. It was found that 43 of the 263 residues tested do not tolerate amino acid substitutions and therefore are critical for the structure and function of the enzyme. The essential residues are largely located in the active site pocket or at buried positions in the protein (12).
One of the critical residues that is located at a buried position in the enzyme is Leu-76. The fact that Leu-76 is essential can be rationalized based on the fact that buried residues, in general, are less tolerant of amino acid substitutions than surface residues (18). However, buried residues are often tolerant of substitutions of other hydrophobic residues and nearly all of the buried residues in TEM-1 β-lactamase can be substituted by other hydrophobic residues (12). Therefore, the compositional constraints on Leu-76 are exceptional. To determine what residues in the protein may be imposing the compositional constraints on Leu-76, a genetic reversion analysis of Leu-76 mutants was performed.
To create a mutation to study at position 76, an asparagine was substituted for leucine by site-directed mutagenesis (8). The substitution reduced the level of ampicillin resistance of E. coli containing a plasmid with the blaTEM gene from 3,000 μg/ml to 100 μg/ml ampicillin (Table 1). In addition, the specific activity of the L76N enzyme was reduced to 2.0% of the wild-type level (Table 1). Therefore, the L76N substitution is clearly deleterious for β-lactamase function.
Table 1.
MIC and enzyme specific activity measurements for TEM β-lactamase variants
| Enzyme | MIC XL1-Blue* | Specific activity XL1-Blue† | Relative activity XL1-Blue, % | Specific activity SB646† | Relative activity SB646, % |
|---|---|---|---|---|---|
| TEM-1 | 3,000 | 45.9 | 100 | 16.6 | 100 |
| L76N | 100 | 0.9 | 2.0 | 1.3 | 7.8 |
| L76N:M182T | 2,000 | 19.1 | 41.6 | 7.9 | 47.6 |
| L76S | 250 | 2.2 | 4.8 | 4.8 | 28.9 |
| L76S:M182T | 1,500 | 30.0 | 65.4 | 10.2 | 61.4 |
| M182T | 2,000 | 32.3 | 70.4 | 8.0 | 48.2 |
| M69I | 750 | 7.3 | 15.9 | 1.5 | 9.0 |
| M69I:M182T | 750 | 8.8 | 19.2 | 1.4 | 8.4 |
| I47Y:E48C | 500 | 4.9 | 10.7 | 2.6 | 15.7 |
| I47Y:E48C:M182T | 750 | 11.0 | 24.0 | 3.3 | 19.9 |
MIC measured in μg/ml of ampicillin.
Specific activity is recorded as mM ampicillin hydrolyzed per mg of protein per min at 37°C.
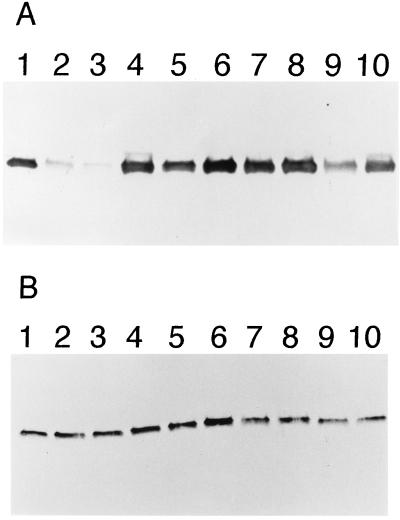
Immunoblotting of the wild-type and L76N enzymes with an anti-β-lactamase polyclonal antibody indicated that the L76N enzyme is poorly expressed with respect to the wild-type enzyme (Fig. 1A, lanes 1–2). The poor expression of the L76N enzyme appears to be due to degradation by proteases after it is secreted to the periplasm because expression levels of L76N return to wild-type levels in E. coli strain SB646, which contains chromosomal deletions of the genes for four periplasmic proteases (S. Bass, personal communication; Fig. 1B, lanes 1–2). Because the position of the L76N substitution is completely buried in the structure of the enzyme, it is unlikely that a new protease site is introduced by the substitution. Rather, it appears the L76N substitution introduces a structural change that alters the folding or stability of the enzyme. This is consistent with the fact that unstable proteins are known to be rapidly proteolyzed in E. coli (19). It also should be noted that proteolysis alone does not account for the entire difference in specific activity between wild-type and L76N. Even in the protease-deficient strain, the L76N enzyme only exhibits 8% of the wild-type specific activity (Table 1). This result is also consistent with a structural change in the enzyme.
Figure 1.
Steady-state levels of wild-type and mutant β-lactamases evaluated by immunoblotting. (A) Immunoblot of β-lactamase mutants in E. coli XL1-Blue. Positions are: 1, wild-type TEM-1 β-lactamase; 2, L76N; 3, L76S; 4, M182T; 5, L76N:M182T; 6, L76S:M182T; 7, M69I; 8, M69I:M182T; 9, I47Y:E48C; 10, I47Y:E48C:M182T. (B) Immunoblot of β-lactamase mutants in E. coli SB646. The order of mutants is as in A.
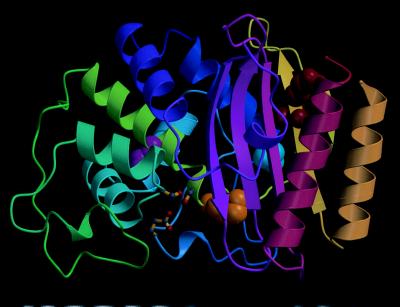
Reversion analysis was performed to identify substitutions that could compensate for the L76N defect. Revertants were identified by introducing the plasmid containing the gene for the L76N β-lactamase into a mutS mismatch repair deficient strain of E. coli (9). The strain was grown to saturation, and the plasmid DNA was isolated and used to transform E. coli XL1-Blue (10). The transformants were spread on agar plates containing 500 μg/ml ampicillin. This concentration is 10-fold higher than the concentration on which E. coli containing the L76N β-lactamase can grow. Transformants were obtained at a frequency of 2 × 10−5. The DNA sequence of the blaTEM gene of six transformants was determined. One mutant contained a 2-bp change that reverted codon 76 to leucine, a second mutant contained a single bp change that mutated codon 76 to isoleucine, and four mutants had a single bp change that converted codon 182 from methionine to threonine. Finding M182T as a suppressor was surprising in that position 182 is located 17 Å from residue 76 in the structure of the enzyme (Fig. 2). Therefore, the M182T substitution does not suppress the L76N defect via a direct interaction with the Asn-76 side chain.
Figure 2.
Diagram of the TEM-1 β-lactamase structure (20) showing the positions of residues I47 and E48 (red), M69 (yellow), L76 (magenta), and M182 (cyan). The position of the active site is indicated by the ball-and-stick representation of the catalytic residues S70, S130, and E166. Figure was made using molscript (21).
Several lines of evidence indicate the M182T substitution restores function to the L76N enzyme. First, the ampicillin-resistance level conferred to E. coli by the plasmid containing the L76N:M182T mutant is 2,000 μg/ml compared with 100 μg/ml for the L76N mutant (Table 1). Second, the specific activity of the L76N:M182T enzyme is 42% of the wild-type specific activity whereas the L76N enzyme is only 2% of wild-type activity (Table 1). This result also shows that the M182T substitution does not restore the function of the L76N enzyme completely to wild-type levels. Finally, immunoblotting with anti-β-lactamase polyclonal antibody indicates the addition of the M182T substitution to the L76N enzyme prevents in vivo proteolysis of the enzyme (Fig. 1A, lanes 2 and 5). Note that in the protease-deficient strain SB646 there is not a large difference in expression levels of L76N versus L76N:M182T (Fig. 1B, lanes 2 and 5). However, the specific activity of L76N:M182T from E. coli SB646 is increased to 48% of the wild-type level compared with 8% for L76N. This suggests the L76N enzyme is only marginally stable, and that in the periplasm of E. coli XL1-Blue cells a significant proportion of enzyme is not properly folded and therefore rapidly proteolyzed. In the periplasm of the protease-deficient strain a proportion of L76N enzyme also is incorrectly folded, but because it is not proteolyzed, it is detected in the immunoblot. However, the improperly folded enzyme is not functional, so the specific activity of the L76N enzyme from the protease-deficient strain remains low.
The M182T substitution next was introduced into the wild-type TEM-1 enzyme to assess its effect. The level of ampicillin resistance of the M182T mutant was slightly lower than that of wild-type TEM-1 (Table 1). Similarly, the specific activity of the M182T enzyme was 70% that of wild-type from E. coli XL-Blue and 49% of wild-type in E. coli SB646 (Table 1). In addition, expression levels of the M182T enzyme were similar to wild-type in both E. coli XL1-Blue and SB646 (Fig. 1). These results suggest that, in isolation, the M182T substitution may be slightly detrimental to β-lactamase function.
We then wished to examine whether M182T could partially restore activity to other amino acid substitutions. First, another substitution at position 76, L76S, was constructed alone and in combination with the M182T substitution. The L76S enzyme was found to behave similarly to the L76N enzyme in terms of antibiotic resistance (Table 1), specific activity (Table 1), and expression levels (Fig. 1A, lane 3). The addition of the M182T substitution to the L76S enzyme resulted in increased ampicillin resistance from an MIC of 250 to 1,500 μg/ml (Table 1). The specific activity increased from 5% to 65% of wild-type activity (Table 1), and the expression levels were raised to levels similar to wild type (Fig. 1A, lane 6). Therefore, the M182T substitution can restore activity to enzymes containing other amino acid substitutions at position 76.
To assess whether M182T can restore activity to enzymes containing substitutions at positions other than residue 76, an enzyme containing a double amino acid substitution, I47Y:E48C, was examined. This mutant was isolated from a library of random substitutions encompassing residues 46–48 (12). The residue 46–48 region of the enzyme was chosen to identify a mutant because it is distant in the structure from both the active site and residue 76. The collection of random mutants were screened for those with low levels of ampicillin resistance and thus reduced levels of activity. Table 1 shows that the I47Y:E48C mutant has an MIC of 500 μg/ml ampicillin. In addition, the I47Y:E48C enzyme retains only 11% of wild-type specific activity in E. coli XL1-Blue. Immunoblotting indicated that the I47Y:E48C enzyme is expressed at lower levels than the wild-type enzyme in E. coli XL1-Blue and at equal levels in the protease deficient strain (Fig. 1 A and B, lane 9).
Addition of the M182T substitution to the I47Y:E48C enzyme resulted in increased enzyme activity. For example, the ampicillin resistance level increased from an MIC of 500 μg/ml for I47Y:E48C to 750 μg/ml for I47Y:E48C:M182T (Table 1). In addition, the specific activity increased from 11% to 24% of wild-type levels (Table 1). Finally, expression levels were increased upon addition of the M182T substitution to near wild-type levels in E. coli XL1-Blue (Fig. 1A, lanes 9 and 10). These results indicate that the M182T substitution can restore function to enzymes with substitutions at multiple positions in the protein.
As stated above, the M182T substitution has been found associated with TEM β-lactamase enzymes that contain other substitutions that result in resistance to β-lactamase inhibitors (TEM-32) or hydrolysis of extended-spectrum cephalosporins (TEM-43)( ref. 6; K. Bush, personal communication). The finding that M182T can suppress mutations found at other positions suggests that this may be its function in the TEM-32 and TEM-43 enzymes. Specifically, the enzymes may acquire substitutions near the active site such as the M69I (TEM-32) and R164H (TEM-43). Residue 69 is on an α-helix immediately adjacent to the catalytic residue serine 70, which serves as the nucleophile for attack on the lactam ring (Fig. 2). Residue 164 forms a salt bridge with aspartate 179. The salt bridge stabilizes an omega loop that makes up the base of the active site pocket (Fig. 2). Substitutions at these positions change the catalytic properties of the enzyme but at the same time may slightly destabilize the enzyme or alter its folding. The M182T substitution then may be selected for its ability to suppress a folding or stability defect. This would explain why the M182T substitution is found in both extended-spectrum and inhibitor-resistant enzymes. However, it should be noted that the L76N and I47Y:E48C substitutions, that are suppressed by M182T as described above, are the products of saturation mutagenesis and have not been found in natural isolates. Therefore, it was important to test whether M182T alters the properties of enzymes with substitutions that have been identified in natural isolates.
This was tested by constructing a single M69I mutant and a M69I:M182T double mutant. The M69I:M182T double mutant is equivalent to the TEM-32 natural mutant. The M69I substitution is known to result in resistance to inhibition by clavulanic acid (6, 7). In addition, it is known that the addition of the M182T substitution to the M69I substitution does not alter the IC-50 for inhibition by clavulanate (6). Therefore, if the M182T substitution provides a selective advantage to TEM-32 it must be via a mechanism other than altered inhibitor binding. It was found that the M69I mutant had an MIC for ampicillin of 750 μg/ml (Table 1). Surprisingly, the addition of the M182T substitution did not change the MIC measurement. In addition, the specific activity for ampicillin of the M69I enzyme is 16% of the wild-type level in E. coli XL1-Blue, and the addition of the M182T substitution increases the level to only 19% of wild-type activity (Table 1). Finally, the expression levels of the M69I and M69I:M182T enzymes are both similar to the wild-type enzyme (Fig. 1A, lanes 7 and 8). Therefore, within the sensitivity of these assays, the M182T substitution does not alter the activity of the M69I enzyme.
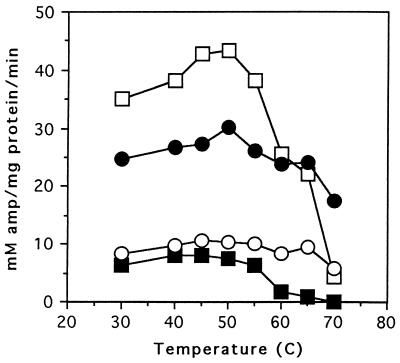
In an effort to increase the sensitivity of the β-lactamase functional assays, the specific activity of enzyme extracts for ampicillin were determined at increasing temperatures (Fig. 3). These experiments were done by preincubating the crude extracts for 5 min at the temperature to be assayed, and then the reaction was performed at this same temperature. The results demonstrate that ampicillin hydrolysis by the M69I enzyme decreases steadily from 45°C to 65°C and is completely absent at 70°C. In contrast, the M69I:M182T enzyme does not begin to lose activity until the temperature is 60°C and retains greater than 50% of its maximal activity at 70°C. In addition, it was found that the M182T enzyme is more heat-resistant than the wild-type β-lactamase. These results indicate that the addition of the M182T substitution to the M69I enzyme does increase the stability of the M69I enzyme. Therefore, the hypothesis that the M182T substitution is selected in natural populations as a suppressor of folding or stability defects resulting from substitutions near the active-site is plausible.
Figure 3.
Graph of specific activities of β-lactamase mutants at increasing temperatures. The x axis displays the temperature at which reactions were performed while the y axis is the specific activity that was measured as mM ampicillin hydrolyzed per mg of total protein per min. □, wild-type TEM-1 β-lactamase; ▪, M69I; ○, M69I:M182T; •, M182T.
DISCUSSION
Experiments performed here have demonstrated that the M182T substitution can partially restore activity to enzymes with defects due to substitutions at positions 47–48, 69, and 76. These substitutions are distant from both each other and M182T in the structure of the enzyme (Fig. 2). Further evidence that M182T acts as a global suppressor is provided by a recent study of chimeric β-lactamases formed between the Proteus vulgaris and TEM-1 β-lactamases (22). The P. vulgaris β-lactamase is 37% identical to TEM-1 β-lactamase (22). When residues 146–184 of the P. vulgaris enzyme were replaced with the analogous residues from TEM-1 β-lactamase the hybrid enzyme was not functional. The plasmid containing the hybrid enzyme then was passaged through an E. coli mutator strain, and mutants with increased ampicillin resistance were selected. Several revertants contained the M182T substitution in the TEM-1 section of the hybrid. Therefore, although only 39 residues of the hybrid originated from TEM-1, the M182T substitution was able to restore activity to the enzyme. Together with the data presented here, this argues strongly that the M182T substitution is a global suppressor.
The fact that the M182T substitution is found in the inhibitor resistant TEM-32 β-lactamase and the extended spectrum TEM-43 β-lactamase suggests that the substitution may be important for the evolution of these enzymes. One possibility is that the active-site proximal substitutions in the TEM-32 (M69I) and TEM-43 (E104K and R164H) may destabilize the enzyme, similar to the L76N substitution, and the M182T substitution may be acquired later to suppress the stability defect. However, immunoblots suggest the M69I enzyme is expressed at levels similar to wild type, and the M69I:M182T double mutant has the same specific activity as the M69I enzyme. It is possible that the effect of the M182T substitution in the context of the M69I substitution is too subtle to detect with these assays. In those cases where the M182T substitution has a clear effect on function, such as the L76 substitutions, or the Proteus/TEM-1 hybrid protein, the expression and function of the starting proteins is extremely poor. Therefore, it is easy to identify increases in expression or activity of the double mutant with M182T. In contrast, the M69I enzyme is not extensively proteolyzed in the periplasm, therefore it is difficult to identify increases in expression using the assays employed here. However, by performing specific activity measurements at increasing temperature it was possible to show that the M182T substitution does stabilize the M69I enzyme. The fact that the M69I substitution is found associated with the M182T substitution in a natural isolate, even though it does not have an extremely deleterious effect on stability and expression, suggests that the bacteria are under strong selective pressure during antibiotic therapy and even subtle changes in catalysis or stability of β-lactamase can be important for the evolution of resistance (23).
To date, the M182T substitution has been identified twice, in the TEM-32 and TEM-43 enzymes. The ability of the substitution to compensate for the deleterious effects of other substitutions suggests the frequency may increase. It is possible that the presence of M182T allows the enzyme to sample substitutions that otherwise would not be selected because of their deleterious effects. Therefore, the M182T substitution may be found associated with new sets of mutations.
Substitutions within a protein that are able to suppress the defect of multiple other substitutions located at distant sites in a protein has been described for staphylococcal nuclease (24). In the nuclease study, second site revertants of mutants with substitutions at several positions were isolated. Subsequent experiments showed that some of the second-site mutations were able to suppress the defects caused by substitutions at multiple positions throughout the protein. Because of the suppression pattern, the substitutions were labeled “global” suppressors. One of the global suppressors subsequently has been shown to function by stabilizing the wild-type staphylococcal nuclease. It acts as a suppressor of multiple substitutions because the overall protein stability is higher, i.e., it displays simple additivity with the other substitutions (25). Global suppressor mutations also have been identified in the λ repressor protein (26). In contrast to the staphylococcal nuclease global suppressor, these global suppressors did not stabilize the protein but appeared to act by increasing the binding affinity for operator DNA (26). Therefore, global suppressors can act via multiple mechanisms.
The observations outlined above with the M182T substitution suggests that it acts via an effect on protein folding or stability. Based on molecular modelling using the wild-type TEM-1 β-lactamase structure, Farzaneh et al. (7) suggest that the presence of a threonine side chain at position 182 may result in a new hydrogen bond between residue 182 and the main chain carbonyl group of residue 64. β-Lactamase has been described as a two-domain protein with a flexible hinge between domains (20). The residue 182-residue 64 hydrogen bond would provide an additional link between domains that could stabilize the protein (7). However, the exact mechanism by which the M182T substitution suppresses multiple substitutions awaits a more detailed biochemical study.
The fact that the M182T substitution is a polymorphism found in β-lactamases in natural isolates raises the possibility that the global suppressor activity of the substitution has been selected in nature. The inhibitor resistant and extended spectrum β-lactamases often contain multiple amino acid substitutions (5). One reason this may occur is that an initial substitution near the active site of the enzyme changes the catalytic activity of the enzyme but also destabilizes the enzyme to some extent. A second mutation, such as M182T, then may correct the stability defect and lead to increased enzyme expression and thus higher fitness. This scenario may apply to other proteins under strong selective pressure due to antimicrobial treatment. For example, the HIV protease in patients under treatment with protease inhibitors has been shown to accumulate amino acid substitutions within the active site and elsewhere that make the enzyme resistant to inhibitor action (27). In addition, high-level resistance to protease inhibitors has been shown to be a multistep process that requires multiple mutations, similar to the acquisition of resistance with β-lactamase. Many of the substitutions in the HIV protease that accumulate early in the selection process also result in reduced viral replicative capacity (27, 28). However, the replicative capacity is partially restored by subsequent mutations that compensate for the viral growth defect. The biochemical basis of the compensatory mutations is not known, but the analogy of this system to the β-lactamase system suggests the compensatory mutations may act as suppressors of folding or stability defects caused by active site mutations.
Acknowledgments
We thank Dr. Mitch Miller and Dr. Kurt Krause for assistance with Fig. 2 and Dr. Steve Bass for the protease-deficient E. coli strain. This work was supported by National Institutes of Health Grant AI32956.
ABBREVIATIONS
- LB
Luria–Bertani
- MIC
minimal inhibitory concentration
References
- 1.Wiedemann B, Kliebe C, Kresken M. J Antimicrob Chemother. 1991;24:1–24. doi: 10.1093/jac/24.suppl_b.1. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. Antimicrob Agents Chemother. 1989;33:264–270. doi: 10.1128/aac.33.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker R H, Eggleston M. Infect Control. 1987;8:36–40. doi: 10.1017/s0195941700066972. [DOI] [PubMed] [Google Scholar]
- 4.Jacoby G A, Medieros A A. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henquell C, Chanal C, Sirot D, Labia R, Sirot J. Antimicrob Agents Chemother. 1995;39:427–430. doi: 10.1128/aac.39.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blazquez J, Baquero M R, Canton R, Alos I, Baquero F. Antimicrob Agents Chemother. 1993;37:2059–2063. doi: 10.1128/aac.37.10.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farzaneh S, Bachir Chaibi E, Peduzzi J, Barthelemy M, Labia R, Blazquez J, Baquero F. Antimicrob Agents Chemother. 1996;40:2434–2436. doi: 10.1128/aac.40.10.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 9.Siegel E, Wain S L, Meltzer S F, Binion M L, Steinberg J L. Mutation Res. 1982;93:25–33. doi: 10.1016/0027-5107(82)90122-1. [DOI] [PubMed] [Google Scholar]
- 10.Bullock W O, Fernandez J M, Short J M. BioTechniques. 1987;5:376–379. [Google Scholar]
- 11.Palzkill T, Botstein D. Proteins. 1992;14:29–44. doi: 10.1002/prot.340140106. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, Petrosino J, Hirsch M, Shenkin P S, Palzkill T. J Mol Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Hanke V, Wink M. BioTechniques. 1994;17:858–860. [PubMed] [Google Scholar]
- 15.Palzkill T, Le Q-Q, Venkatachalam K V, LaRocco M, Ocera H. Mol Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 16.Neu H C, Heppel L A. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 17.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Bowie J U, Reidhaar-Olson J F, Lim W A, Sauer R T. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 19.Parsell D A, Sauer R T. J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 20.Jelsch C, Mourey L, Masson J-M, Samama J-P. Proteins. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]
- 21.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 22.Hosseini-Mazinani S M, Nakajima E, Ihara Y, Kameyama K-Z, Sugimoto K. Antimicrob Agents Chemother. 1996;40:2152–2159. doi: 10.1128/aac.40.9.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean A M. Genetics. 1989;123:441–454. doi: 10.1093/genetics/123.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shortle D, Lin B. Genetics. 1985;110:539–555. doi: 10.1093/genetics/110.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shortle D. J Cell Biochem. 1986;30:281–289. doi: 10.1002/jcb.240300402. [DOI] [PubMed] [Google Scholar]
- 26.Hecht M H, Sauer R T. J Mol Biol. 1985;186:53–63. doi: 10.1016/0022-2836(85)90256-6. [DOI] [PubMed] [Google Scholar]
- 27.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. Nature (London) 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 28.Borman A M, Paulous S, Clavel F. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]