Abstract
Previous studies have shown that the chloride channel gene Clc4 is X-linked and subject to X inactivation in Mus spretus, but that the same gene is autosomal in laboratory strains of mice. This exception to the conservation of linkage of the X chromosome in one of two interfertile mouse species was exploited to compare expression of Clc4 from the X chromosome to that from the autosome. Clc4 was found to be highly expressed in brain tissues of both mouse species. Quantitative analyses of species-specific expression of Clc4 in brain tissues from mice resulting from M. spretus × laboratory strain crosses, demonstrate that each autosomal locus has half the level of Clc4 expression as compared with the single active X-linked locus. In contrast expression of another chloride channel gene, Clc3, which is autosomal in both mouse species is equal between alleles in F1 animals. There is no evidence of imprinting of the Clc4 autosomal locus. These results are consistent with Ohno’s hypothesis of an evolutionary requirement for a higher expression of genes on the single active X chromosome to maintain balance with autosomal gene expression [Ohno, S. (1967) Sex Chromosomes and Sex-Linked Genes (Springer, Berlin)].
The recent finding that two closely related mouse species, Mus spretus and laboratory strains (a mixture of M. musculus musculus and M. musculus domesticus) differ in the chromosomal location of the chloride channel gene, Clc4 (1, 2) presents a unique opportunity for studying the function of the Clc4 gene product and for investigating the relationships of dosage compensation, X chromosome inactivation, and the evolution of the mammalian X chromosome. Ohno’s hypothesis (3) predicted that the mammalian X chromosome as a linkage group would be preserved, Clc4 being one of the few exceptions (4). Ohno also hypothesized that during the evolutionary transition to dosage compensation X-linked genes would be up-regulated on the single active X chromosome of males and females to maintain balance with autosomal genes that are expressed from both alleles (3).
Clc4 is a member of the class of voltage-gated ion channels that all share a predicted 12-transmembrane domain structure based on hydrophobicity analysis (5, 6). As expected from the DNA sequence diversity of the chloride channel encoding genes, different family members now have been shown to differ widely in terms of tissue-specific expression and association with disease phenotypes. For example, CLC1 mutations are associated with autosomal dominant or recessive myotonia (7, 8); whereas, mutations of CLC5, a putative renal chloride channel gene, have been found in different kidney disorders (9). To determine the repertoire of tissue expression for the mouse Clc4 gene, Northern and reverse transcriptase–PCR (RT-PCR) analyses of mRNA were carried out in both M. spretus and laboratory strain C57BL/6Ros. In situ hybridization was done to determine the location of Clc4 expression in embryos and adult mice.
To test directly the possibility of up-regulation of X-linked transcription as predicted by Ohno, we generated backcross mice resulting from matings between the interfertile species, M. spretus and a laboratory strain C57BL/6Ros. Expression of Clc4 from the X-linked and from the autosomal loci was measured within individual backcross animals. DNA sequence variants, frequently found between M. spretus and laboratory strains (10), were exploited to quantify expression from each locus using single nucleotide extension (SNuPE) assays (11). Our results are consistent with the doubling of expression from the X-linked locus as compared with the autosomal locus.
MATERIALS AND METHODS
Mice.
Mice with one or two autosomal copies of Clc4 of BL/6 origin and one or two X-linked copies of Clc4 of M. spretus origin were identified in the progeny of two backcrosses, (BL/6 × M. spretus)F1 × M. spretus and (BL/6 × M. spretus)F1 × BL/6. Mice were genotyped using DNA markers previously mapped close to Clc4, including DXMit30, DXMit160, DXMit183, D7Mit74, D7Mit21, and D7Mit56 (2) [Mouse Genome Database, Mouse Genome Informatics, The Jackson Laboratory; World Wide Web (URL: http://www.informatics.jax.org/) September 9, 1996]. DNA was prepared from the tails followed by PCR amplification using primers specific for each marker (SSLP Genetic Map of the Mouse, Whitehead Institute, Massachusetts Institute of Technology Center for Genome Research, release June 1996). Gel electrophoresis was used to discriminate size differences between species-specific product of amplification from the alleles. Genotyping of 133 male and 128 female progeny from the (BL/6 × M. spretus)F1 × M. spretus backcross revealed that the expected four genotypes were present in approximately equal numbers for each sex. Weight recorded for the male mice of the different genotypes as a function of age showed no significant differences between the genotypes (data not shown). Null males completely missing a Clc4 locus showed no apparent abnormal phenotype. When exposed to cold temperature, the Clc4 null mice did not exhibit any cold-induced myotonia, and no behavioral differences from litter mates were observed. Gross examination as well as histological examination of tissues of null mice compared with age-matched control parental mice failed to reveal any overt pathology, except for testes hypoplasia, an expected finding in interspecific F1 males and predicted from Haldane’s rule (12).
RNA Preparation and Northern Analysis.
Total RNA was prepared from flash-frozen tissues using phenol/guanidium reagent Ultraspec (Biotecx Laboratories, Houston). Poly(A)+ RNA was prepared either from total RNA using biotinylated-oligo(dT) and avidin-bound magnetic beads (Promega) or directly from flash-frozen tissues using a kit (Invitrogen). Poly(A)+ RNA electrophoresed in a denaturing agarose gel containing 50% formamide was blotted onto nylon membranes (Hybond, Bio-Rad) and hybridized to 32P-labeled Clc4 cDNA probe (MR9) at 40°C in the presence of 50% formamide. Blots were washed in 1× standard saline citrate/0.5% SDS at 50°C for 1 hr. Blots were rehybridized to a control probe 36B4, which detects transcripts of the ribosomal phosphoprotein gene, PO (13).
RT-PCR.
First-strand cDNA synthesis was carried out on total RNA or poly(A)+ RNA with reverse transcriptase (SuperScript, BRL) in a 20-μl reaction volume with 5 mM MgCl2, 1× PCR buffer, 5.0 pg oligo(dT) (15 mer) at 42°C for 1 hr. After RNA/DNA hybrids were denatured, first-strand cDNA was used as the template in a PCR containing 400 ng each of upstream and downstream Clc4-specific primers (1) and Clc3-specific primers (14) with 30 cycles of 1 min at 94°C, 1 min at 60°C, 1 min at 72°C, and then a final 5-min extension at 72°C.
SNuPE.
Assays based on the method of Singer-Sam et al. (11) were performed on either total RNA or poly(A)+ RNA. After RT-PCR amplification, the 436-bp Clc4-specific and 169-bp Clc3-specific bands were separated on a 2% agarose in 1× TBE (0.089 M Tris borate/0.089 M boric acid/20 mM EDTA, pH 8.3) agarose gel, followed by purification using Centricon-30 spin separators (Amicon) and determination of concentration by fluorometry. Five nanograms of cDNA was used as template in the SNuPE reactions. A variant at sequence position nucleotide 1,637 between M. spretus and BL/6 was identified by sequence analysis of Clc4 cDNA, and a primer (5′-GTGTAAACTCCAACCCCAGC-3′) was designed that stops one nucleotide short of the variant. Similar analysis resulted in the synthesis of a primer specific for Clc3 (5′-CAAACATGGGGTGTCAGCTG-3′). A single round of primer extension was carried out in the presence of either [32P]dCTP or [32P]dTTP nucleotide, using two separate reactions (C or T) for each sample. Reaction products then were run on a 16% polyacrylamide gel at 250 v for 3 hr before exposure to x-ray film for 5 min to 1.5 hr. Quantification was done by densitometric analysis of films and by phosphoimaging of gels.
RNA in Situ Hybridization.
Whole embryos and adult brains were fixed in 4.0% paraformaldehyde and embedded in paraffin, and ≈7-μm sections were cut. Slides were hybridized to riboprobes as described in Rugarli et al. (15). The riboprobes were derived from two nonoverlapping Clc4 cDNA fragments from position nucleotides 1,111–1,900 and nucleotides 2,558–2,698 subcloned in plasmid pBS SK−. Sense and antisense probes were prepared using the Stratagene RNA transcription kit and labeled with 35S or 33P. After hybridization, and autoradiography with 7- to 28-day exposures, slides were developed and counterstained with 2 μg/ml Hoechst 33258 or hematoxylin and eosin before examination with a dark-field condenser for the silver grains and with fluorescence or transmitted light to visualize cell nuclei.
RESULTS
Clc4 Expression in M. spretus and Laboratory Strains.
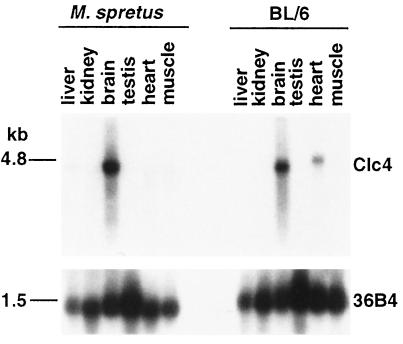
Northern blots were prepared from poly(A)+ RNA extracted from different tissues from two M. spretus and two laboratory strain C57BL/6Ros (hereafter designated as BL/6) mice. Clc4 was highly expressed in brain tissues of both M. spretus and BL/6 mice with a transcript size of about 4.5–5.0 kb (Fig. 1). Lower expression also was seen in BL/6 heart tissues, but not in heart tissues of M. spretus mice (Fig. 1). This pattern was consistent for the different animals examined. A control probe for the ribosomal phosphoprotein gene, PO, hybridized to the same blots, indicated the presence of similar amounts of intact RNA in all lanes (Fig. 1). The tissue-specific expression of Clc4 was confirmed by RT-PCR with high levels of Clc4 amplification in brain tissues of both species and some amplification in heart tissues of BL/6 mice.
Figure 1.
Northern blot analysis of tissue-specific expression of Clc4 in M. spretus and C57BL/6Ros mice. Two micrograms of poly(A)+ RNA from tissues of each species were run on a denaturing agarose gel as indicated above the lanes. The resulting blot was hybridized to a 32P-labeled Clc4 MR9 cDNA probe, resulting in a 4.5- to 5-kb band. The blot was rehybridized to a control ribosomal phosphoprotein cDNA probe, 36B4 (12), producing a 1.5-kb band.
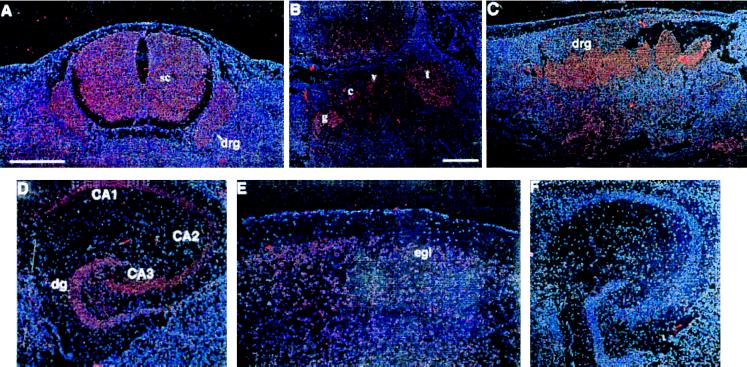
A more detailed analysis of Clc4 expression during mouse development and in the adult brain was done by in situ hybridization to sections of staged embryos and adult tissues. Embryos were from a laboratory strain (ICR) whereas adult tissues were collected both from laboratory strains (IRC and BL/6) and M. spretus mice. Samples were hybridized to either of two nonoverlapping antisense riboprobes. Both probes yielded identical results, and control hybridizations with sense riboprobes gave no signal (Fig. 2F). At 10.5 days Clc4 expression was appreciable but with diffuse staining throughout the embryo. Later in development at 12.5–14.5 days, Clc4 expression was found to be progressively up-regulated in neuroectodermal tissues. At 13.5 days, neurons of the spinal cord and of dorsal root ganglia became positive (Fig. 2A). At 14.5 days, expression was high in cranial sensory ganglia (Fig. 2B) and sympathetic dorsal root ganglia (Fig. 2C). In the adult mouse, low, but detectable, levels of hybridization were found throughout the brain. However, some areas, such as the cerebral cortex, the olfactory bulb, and the hippocampus, displayed a strong hybridization pattern. Within the cerebral cortex, a positive signal was characteristically concentrated in the external granular layer or layer II, and in the olfactory bulb, Clc4 was expressed in the mitral cell layer and in the granular layer (Fig. 2E). The most prominent area of Clc4 expression in the adult mouse brain was the hippocampus with the pyramidal cell layers of CA1 to CA3 fields of Ammons horn densely labeled (Fig. 2D). Granule cells of the dentate gyrus appeared slightly less positive (Fig. 2D). There were no significant differences between hybridization patterns on adult brain sections from laboratory strains (IRC and BL/6) and from M. spretus.
Figure 2.
In situ hybridization of 35S-labeled Clc4 riboprobes to embryos and adult brains form a laboratory strain. (A) A transverse section of a 13.5-day embryo shows expression in the spinal cord (SC) and the dorsal root ganglia (drg). (B and C) Sagittal sections of a 14.5-day embryo show expression in the glossopharyngeal nerve ganglion (g), the cochlear ganglion (c), the vestibular ganglion (v), the trigerminal ganglion (t), and the dorsal root ganglia (drg). (D and E) Horizontal sections of adult brain show expression in the dentate gyrus (dg), the pyramidal cell layers of the CA1, CA2, and CA3 fields of Ammons horn, and in the external granular layer on layer II (eg1). (F) Negative control hybridization using a sense probe. Magnifications are identical for all panels (bar shown in A, 500 μm), except for B (bar, 200 μm).
Clc4 Expression from the X-Linked Locus and the Autosomal Locus in Interspecific Hybrid Mice.
To compare expression of Clc4 from the X-linked locus and the autosomal locus within the same animal, RT-PCR combined with either restriction enzyme or SNuPE analyses were performed on samples from male mice with one X-linked Clc4 locus and one or two chromosome 7 loci, as identified by genotyping with DNA markers flanking the Clc4 loci. Male mice with one X-linked and one autosomal locus derived either maternally or paternally also were compared for evidence of imprinting.
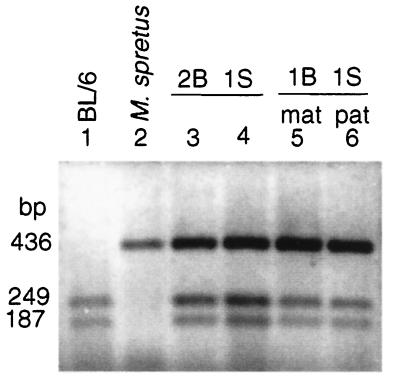
Restriction analyses of RT-PCR products of amplification of Clc4 from brain tissues using EagI to distinguish alleles based on the presence of a restriction site in BL/6 only, indicated that Clc4 expression was higher from the X-linked locus as compared with the autosomal locus (Fig. 3). Quantification of expression by densitometry showed that the observed data were close to the expected results if Clc4 expression from the X-linked locus was twice that from the autosomal locus (Table 1). The parental origin of the BL/6 allele did not alter the lower transcript level of the chromosome 7 locus and thus there was no indication of imprinting of the autosomal locus (Fig. 3 and Table 1).
Figure 3.
EagI digests of Clc4 RT-PCR products amplified from brain tissues of parental mouse species BL/6 and M. spretus and of backcross progeny with different copy numbers of Clc4. Lanes 1 and 2 contain products from parental male controls. Lanes 3 and 4 contain products from two different male mice with one X chromosome locus of M. spretus origin (1S) and two chromosome 7 loci of BL/6 origin (2B). Lane 5 is from a male mouse with one X chromosome locus of M. spretus origin (1S) and one paternally inherited chromosome 7 locus of BL/6 origin (1Bmat). Lane 6 is from a mouse with one X chromosome locus of M. spretus origin (1S) and one maternally inherited chromosome 7 locus of BL/6 origin (1Bpat). Levels of allele-specific transcripts were quantified as described (Table 1).
Table 1.
Quantitative analysis of Clc4 expression from the X-linked locus of M. spretus origin (S) and from the autosomal locus of BL/6 (B) in interspecific hybrid male mice
| Genotype* | Percent of BL/6 locus expression†
|
|||||
|---|---|---|---|---|---|---|
| Expected | Observed | |||||
| Mouse | Ch X | Ch 7 | If B = S no imprinting | If S = 2 × B no imprinting | Restriction analysis‡ | SNuPE analysis‡ |
| 1 | S | BpatS | 50 | 33 | 31.2 ± 5.4 | 33.3 ± 5.9 |
| 2 | S | BmatS | 50 | 33 | 29.0 ± 14.7 | 36.8 ± 5.2 |
| 3 | S | BmatS | 50 | 33 | N.D. | 31.5 |
| 4 | S | BB | 67 | 50 | 46.0 ± 7.0 | 53.9 ± 4.4 |
N.D., not determined.
Genotype at the Clc4 loci on the X chromosome and on chromosome 7 were determined by typing closely linked DNA markers as described. S: M. spretus, B: BL/6, Bmat: BL/6 of maternal origin, Bpat: BL/6 of paternal origin.
The percentage of BL/6 locus expression of Clc4 was calculated as: B product/S + B products × 100.
Average of three measures ± SD.
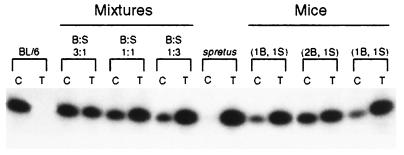
SNuPE assays were used to quantify more precisely the allele-specific expression of Clc4 in mouse tissues. Sequence analysis of a portion of Clc4 (nucleotides 1,515–1,950) amplified from M. spretus and from BL/6 revealed similar DNA sequences with a variant site at nucleotide 1,669. Primers were designed that stopped one nucleotide short of the identified variant, and single nucleotide primer extensions carried out in the presence of either 32P-labeled nucleotide (C or T) and of RT-PCR products of Clc4 amplification from brain tissues of each animal (Fig. 4). Data were quantified by densitometry normalizing values to an artificial 1:1 mixture of the two parental species of cDNA (11). SnuPE analyses on brain tissues from four backcross male mice with one or two copies of the autosomal locus of Clc4 showed that expression from the X-linked locus was double that from the autosomal locus (Fig. 4; Table 1). SNuPE analyses on heart tissues confirmed Clc4 expression in BL/6 heart tissues only (data not shown). Based on the restriction and SNuPE analyses the ratio between the output from the X-linked and the autosomal locus is 2.07 ± 0.07 (mean ± SD). X2 analysis showed that the observed values expressed in % BL/6 output fit the expected values for twice the output from the M. spretus locus at a 0.95 confidence level.
Figure 4.
Examples of SNuPE analyses of Clc4 from brain RNA. (Left) Parental species BL/6 (B) and M. spretus (S) flanking artificial mixtures of the two species cDNAs with B/S ratios of 3:1, 1:1, and 1:3, respectively. (Right) Two backcross mice with one Clc4 locus of BL/6 origin (1B) and one Clc4 locus of M. spretus origin (1S) flanking one mouse with two Clc4 loci of BL/6 origin (2B) and one Clc4 locus of M. spretus origin (1S). For each sample, C indicates the lane with primer extension products obtained in the presence of 32P-labeled dCTP and T in the presence of 32P-labeled dTTP. Amounts of incorporation were quantified as described (Table 1).
Artificial mixtures of parental RNAs from M. spretus and BL/6 brain tissues were combined in different ratios and subjected to RT-PCR and SNuPE (data not shown). These experiments were designed to measure the amount of Clc4 transcript per μg of total cellular RNA. The percent of BL/6 expression observed was again close to that expected if on a total RNA per cell basis the output from the autosomal locus was approximately half that of the X-linked locus. Control mixtures of parental cDNAs with variable amounts of each species Clc4 cDNA were done to show that there was a linear relationship between ratios based on SNuPE measurements and input cDNA ratios. Background levels of misincorporation of T in BL6 and C in M. spretus were below detectibility in control parental SNuPE reactions.
Additional control SNuPE analyses were done to quantitate expression of Clc3, a chloride channel gene with high sequence homology to Clc4 but which is autosomal in both species of mice (14). SNuPE assays were done on brain tissues from five F1 mice from a C57BL/6 × M. spretus cross and from one backcross animal. Results showed that Clc3 expression was nearly equal from the BL/6 and M. spretus alleles as expected (Table 2). The average ratio between output from the BL/6 and M. spretus loci was 0.93 ± 0.06. X2 analysis showed that the observed values expressed in % BL/6 output fit the expected values for equal expression from the loci of the two mouse species at a 0.95 confidence level.
Table 2.
Quantitative analysis of Clc3 expression from the allelic loci of M. spretus origin (S) and of BL/6 origin (B) in F1 mice
| Mouse* | Genotype | Percent of BL/6 locus expression†
|
|
|---|---|---|---|
| Expected | Observed‡ | ||
| 1 | BS | 50 | 48 ± 4.4 |
| 2 | BS | 50 | 48 ± 0.5 |
| 3 | BS | 50 | 51 ± 3.0 |
| 4 | BS | 50 | 47 ± 2.8 |
| 5 | BS | 50 | 47 ± 3.4 |
| 6 | BS | 50 | 48 ± 0.4 |
Mice 1 to 5 were F1 from a Bl/6 × M. spretus cross. Mouse 6 resulted from a [BL/6 × M. spretus] F1 × M. spretus backcross.
The percentage of BL/6 locus expression of Clc3 was calculated as: B product/S + B products × 100.
Average of two measurements ± SD.
DISCUSSION
The finding of a different chromosomal position for Clc4 in two interfertile mouse species is unusual especially because in one of the species (M. spretus) the gene is X-linked and regulated by X inactivation (1, 2). CLC4 is also X-linked in human (6), and thus it is likely that Clc4 ancestral location was on the X chromosome and that the gene subsequently was translocated to an autosome in a subset of Mus species (1, 2). The translocation event that occurred during the divergence of the Mus species must be reconciled with existing theories of the evolution of the sex chromosomes and in particular Ohno’s hypothesis, which predicts the conservation of the X chromosome as a linkage group among mammals (3). The doubling of transcriptional output from X-linked loci, Ohno speculated, would be the first evolutionary step in the accommodation of X-linked genes to their hemizygous existence in males. Consistent with this prediction are the present quantitative measurements of Clc4 expression, which reveal a 2-fold higher expression from the X-linked locus compared with each autosomal locus. The net result is equal Clc4 expression in brain tissues of both sexes of M. spretus (each with a single active X-linked locus) and of laboratory strains of mice (each with two active autosomal loci). The measurements reported here were based mainly on the analysis of allele-specific expression within the same backcross animals to avoid the problem of possible fluctuation of expression between different animals. However, mixing experiments using RNAs from parental mice also confirmed up-regulation of the X-linked Clc4 locus of M. spretus. A control gene Clc3, which is highly homologous to Clc4 and shows a similar pattern of expression in brain (14), was found to be expressed equally from each species-specific allele in F1 mice.
The observed doubling of expression from the X-linked Clc4 locus may reflect either a generalized up-regulation of the X chromosome that is not specific to this locus or up-regulation limited to the Clc4 locus. Another possibility is that differences in Clc4 transcript level could be due to differences in message turnover in the two mouse species or to a different proportion of cells expressing Clc4 in brain tissues of the two mouse species. However, in situ hybridization results indicate a similar distribution of expressing cells in M. spretus and laboratory strains. Because Clc4 is unique in terms of being X-linked and regulated by X inactivation in one mouse species, but autosomal in another interfertile mouse species, extension of our findings to other genes will await the identification of other such genes or the construction of transgenic mice.
It is of interest that in other species, such as Drosophila, dosage compensation is achieved by a generalized doubling of expression from the single X chromosome in males, a process regulated by the X/autosome ratio and mediated by the binding of specific proteins (16). If there is generalized up-regulation of the single active X chromosome in mice, the observed down-regulation of the autosomal Clc4 loci in laboratory strains may result from this gene no longer being within the X chromosome environment, or possibly from autosomal dosage compensation. Autosomal dosage compensation has been observed in Drosophila (17), but there is no evidence of such dosage compensation in mammals (18, 19). Differences in transcript levels related to a gene being in a different chromosomal location also could be the result of what is known as a position effect (20, 21), which would selectively modify Clc4 expression. The underlying mechanisms for the differences in transcript levels and tissue-specific expression for the two chromosomal locations of the mouse Clc4 gene may involve gene-specific promoter and enhancer elements acting in cis, possibly from a distance, in the X-linked or autosomal chromatin context.
Despite the observed doubling of expression from the X-linked locus as compared with the autosomal locus, genotyping of backcross animals reported here and elsewhere (2) suggests that the dosage of Clc4 may be of little or no consequence to the survival of laboratory mice, because mice with increased or decreased expression dosage of Clc4, including Clc4-null mice, are viable. Histological examination of various tissues obtained from Clc4-null mice failed to reveal any observable effect of the loss of the Clc4 gene. Of the gene knockouts catalogued in tbase (The Transgenic/Targeted Mutation Database, Johns Hopkins University, Baltimore, 1996), several exhibit a “normal” phenotype in homozygous mice. Redundancy of gene function often is invoked as an explanation for the lack of phenotype of a null mutant. In the case of Clc4, redundancy of function may be mediated by the highly homologous Clc3 gene, which shows a remarkably similar pattern of expression in neuroectodermal tissues of embryos and adult mice (14). However, phenotypic effects of increased or decreased levels of Clc4 may be subtle, possibly limited to a behavioral response to particular stimuli, or else may be related to changes only observable later in life and thus have escaped detection. Functions associated with specific chloride channels include muscle excitability (22, 23) and cell volume regulation (24), but the functions of CLC4 and CLC3 that are expressed in brain, skeletal muscle, and heart in human are yet to be defined (6, 14). Mutations in other chloride channel genes have been associated with a variety of phenotypes, including human and mouse myotonia (7, 23) and several forms of kidney disease (9). Further analyses, such as electrophysiology, protein localization, and behavioral studies, may be necessary to reveal the phenotype associated with altered dosage or absence of Clc4 in mouse.
In light of these findings, the translocation of the Clc4 locus in murine evolution may have been possible either because Clc4 was immediately down-regulated on the autosome for the reasons discussed above, or because the potential immediate doubling of Clc4 expression due to the translocation resulted in viable mice. The presently observed halving in Clc4 expression from the autosomal locus then may have occurred progressively during evolution as a selective advantage perhaps not perceptible in laboratory mice, which are not subjected to the stringent selective pressures of a wild environment.
The alterations of Clc4 expression associated with the different chromosomal locations of this gene in two interfertile mouse species provides a system for the investigation of models of transcriptional regulation of the X chromosome at the chromatin domain and chromosome levels. Isolation of genomic regions flanking both the X-linked and autosomal Clc4 loci may identify regulatory sequences that are responsible for the differences in Clc4 expression between the two species.
Acknowledgments
We thank Janice Garr for preparation of the manuscript, William Catterall for helpful discussion, and Debbie Tabaczynski and Sushma Thomas for their assistance. This work was supported by National Institutes of Health Grants GM46883 (C.M.D., D.A.A., K.T., and P.A.L.) and GM33160 (V.M.C. and R.W.E.) and March of Dimes Birth Defect Foundation Grant 1013 (C.M.D.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RT-PCR, reverse transcriptase–PCR; SNuPE, single nucleotide extension.
References
- 1.Rugarli E I, Adler D A, Borsani G, Tsuchiya K, Franco B, Hauge X, Disteche C, Chapman V, Ballabio A. Nat Genet. 1995;10:466–471. doi: 10.1038/ng0895-466. [DOI] [PubMed] [Google Scholar]
- 2.Palmer S, Perry J, Ashworth A. Nat Genet. 1995;10:472–476. doi: 10.1038/ng0895-472. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. Sex Chromosomes and Sex-Linked Genes. Berlin: Springer; 1967. pp. 1–140. [Google Scholar]
- 4.Ellis N A. Nat Genet. 1995;10:373–375. doi: 10.1038/ng0895-373. [DOI] [PubMed] [Google Scholar]
- 5.Jentsch T J, Gunther W, Pusch M, Schwappach B. J Physiol Lond. 1995;482:S19–S25. doi: 10.1113/jphysiol.1995.sp020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Sleghtenhorst M A, Bassi M T, Borsani G, Wapenaar M C, Ferrero G B, de Conciliis L, Rugarli E I, Gillo A, Franco B, Zoghbi H Y, Ballabio Hum Mol Genet. 1994;3:547–552. doi: 10.1093/hmg/3.4.547. [DOI] [PubMed] [Google Scholar]
- 7.Koch M C, Steinmeyer K, Lorenz C, Ricker K, Wolf F, Otto M, Zoll B, Lehmann Horn F, Grzeschik K H, Jentsch T J. Science. 1992;257:797–800. doi: 10.1126/science.1379744. [DOI] [PubMed] [Google Scholar]
- 8.George A L, Jr, Crackower M A, Abdalla J A, Hudson A J, Ebers G C. Nat Genet. 1993;3:305–310. doi: 10.1038/ng0493-305. [DOI] [PubMed] [Google Scholar]
- 9.Lloyd S E, Pearce S H, Fisher S E, Steinmeyer K, Schwappach B, Scheinman S J, Harding B, Bolino A, Devoto M, Goodyer P, Rigden S P, Wrong O, Jentsch T J, Craig I W, Thakker R V. Nature (London) 1996;379:445–449. doi: 10.1038/379445a0. [DOI] [PubMed] [Google Scholar]
- 10.Mullins L J, Grant S G, Stephenson D A, Chapman V M. Genomics. 1988;3:187–194. doi: 10.1016/0888-7543(88)90078-x. [DOI] [PubMed] [Google Scholar]
- 11.Singer-Sam J, LeBon J M, Dai A, Riggs A D. PCR Methods Appl. 1992;1:160–163. doi: 10.1101/gr.1.3.160. [DOI] [PubMed] [Google Scholar]
- 12.Haldane J B S. J Genet. 1922;12:101–109. [Google Scholar]
- 13.Laborda J. Nucleic Acids Res. 1991;19:3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borsani G, Rugarli E I, Taglialatela M, Wong C, Ballabio A. Genomics. 1995;27:131–141. doi: 10.1006/geno.1995.1015. [DOI] [PubMed] [Google Scholar]
- 15.Rugarli E I, Lutz B, Kuratani S C, Wawersik S, Borsani G, Ballabio A, Eichele G. Nat Genet. 1993;4:19–26. doi: 10.1038/ng0593-19. [DOI] [PubMed] [Google Scholar]
- 16.Bashaw G J, Baker B S. Curr Opin Genet Dev. 1996;6:496–501. doi: 10.1016/s0959-437x(96)80073-6. [DOI] [PubMed] [Google Scholar]
- 17.Birchler J A, Hiebert J C, Paigen K. Genetics. 1990;124:677–686. [PMC free article] [PubMed] [Google Scholar]
- 18.Eicher E M, Coleman D L. Genetics. 1977;85:647–658. doi: 10.1093/genetics/85.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurnit D M. Proc Natl Acad Sci USA. 1979;76:2372–2375. doi: 10.1073/pnas.76.5.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sturtevant A H. Genetics. 1925;10:117–147. doi: 10.1093/genetics/10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hennikoff S. Trends Genet. 1990;6:422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- 22.Steinmeyer K, Ortland C, Jentsch T J. Nature (London) 1991;354:301–304. doi: 10.1038/354301a0. [DOI] [PubMed] [Google Scholar]
- 23.Steinmeyer K, Klocke R, Ortland C, Gronemeier M, Jockusch H, Gründer S, Jentsch T J. Nature (London) 1991;354:304–308. doi: 10.1038/354304a0. [DOI] [PubMed] [Google Scholar]
- 24.Gründer S, Thiemann A, Pusch M, Jentsch T J. Nature (London) 1992;360:759–762. doi: 10.1038/360759a0. [DOI] [PubMed] [Google Scholar]