Abstract
Benzodiazepines allosterically modulate γ-aminobutyric acid (GABA) evoked chloride currents of γ-aminobutyric acid type A (GABAA) receptors. Coexpression of either rat γ2 or γ3, in combination with α1 and β2 subunits, results both in receptors displaying high [3H]Ro 15-1788 affinity. However, receptors containing a γ3 subunit display a 178-fold reduced affinity to zolpidem as compared with γ2-containing receptors. Eight chimeras between γ2 and γ3 were constructed followed by nine different point mutations in γ2, each to the homologous amino acid residue found in γ3. Chimeric or mutant γ subunits were coexpressed with α1 and β2 in human embryonic kidney 293 cells to localize amino acid residues responsible for the reduced zolpidem affinity. Substitution of a methionine-to-leucine at position 130 of γ2 (γ2M130L) resulted in a 51-fold reduction in zolpidem affinity whereas the affinity to [3H]Ro 15-1788 remained unchanged. The affinity for diazepam was only decreased by about 2-fold. The same mutation resulted in a 9-fold increase in Cl 218872 affinity. A second mutation (γ2M57I) was found to reduce zolpidem affinity by about 4-fold. Wild-type and γ2M130L-containing receptors were functionally expressed in Xenopus oocytes. Upon mutation allosteric coupling between agonist and modulatory sites is preserved. Dose–response curves for zolpidem and for diazepam showed that the zolpidem but not the diazepam apparent affinity is drastically reduced. The apparent GABA affinity is not significantly affected by the γ2M130L mutation. The identified amino acid residues may define part of the benzodiazepine binding pocket of GABAA receptors. As the modulatory site in the GABAA receptor is homologous to the GABA site, and to all agonist sites of related receptors, γ2M130 may either point to a homologous region important for agonist binding in all receptors or define a new region not underlying this principle.
Neuronal inhibition of the mammalian brain is mainly effected by γ-aminobutyric acid type A (GABAA) receptors. They are ligand gated chloride channels and belong to a larger channel family among nicotinic acetylcholine, glycine, and serotonin-3 receptors. Purification (1) and cDNA cloning (2) of two subunits belonging to the GABAA receptor have initially been reported. Fourteen subunits or subunit isoforms of the probably pentameric receptor have been described (ref. 3; for reviews, see refs. 4–8).
The GABAA channel is functionally modulated by a large number of drugs (for review, see ref. 7). For example, it is subjected to positive allosteric modulation by clinically used drugs acting at the benzodiazepine binding site. Occupation of the binding site by these substances results in sedative, anxiolytic, myotonolytic, and anticonvulsant effects. In addition, substances are known to bind to this site, having no or a negative allosteric effect on the channel function. The location of this binding site on the receptor is of obvious interest.
A major site of photoaffinity labeling by the benzodiazepine [3H]flunitrazepam is a conserved histidine on α subunits (9). Several additional amino acid residues on various α subunit isoforms have been implicated in ligand binding (10–14). Expression of recombinant receptors indicates that a γ subunit is absolutely required for the formation of a benzodiazepine binding site (15, 16). Two point mutations in the γ2 subunit influence the benzodiazepine pharmacology (17–19). Upon deletion of γ2 from mice the [3H]Ro 15-1788 binding site nearly disappears, confirming the essential role of γ subunits (20). Thus, α and γ subunits are both thought to contribute to the benzodiazepine site ligand affinity, and its binding site is assumed to be located at the α/γ subunit interface.
Residues influencing the apparent γ-aminobutyric acid (GABA) affinity have been identified on α and β subunits assuming that the agonist site also is located at subunit interfaces. Residues important for GABA binding are at homologous positions to residues belonging to the agonist binding sites of the nicotinic acetylcholine and glycine receptors (for a review, see ref. 21). It appears that the localization of the neurotransmitter binding sites is conserved between these homologues (21). Remarkably, residues influencing benzodiazepine site ligand affinity at GABAA receptors are located at homologous regions to regions contributing to these agonist sites (14, 18, 19, 21).
Zolpidem, an imidazopyridine, binds with high affinity to α1-containing GABAA receptors and is able to displace the classical benzodiazepines (22). α2 and α3 apparently confer intermediate zolpidem affinity and the α5 subunit very low affinity to triple subunit combinations αxβ2γ2 (23–25). Zolpidem displays lower affinity to γ3 than to γ2 containing receptors (26, 27), pointing to the importance of the γ subunit for zolpidem binding.
By expression of chimeric subunits and radioligand binding we show that the zolpidem affinity is influenced by two distinct regions within the N-terminal domain of the γ2 subunit. Site-directed mutagenesis of residues of the γ2 subunit differing between γ2 and γ3 identified two amino acid residues to be important for benzodiazepine site ligand affinity. Our work represents an important step leading to a rational drug design based on the identification of structural determinants on the receptor protein relevant for ligand binding.
MATERIALS AND METHODS
Construction of Receptor Subunits.
The cDNAs coding for the α1, β2, γ2S, and γ3 subunits of the rat GABAA receptor channel have been described (26, 28–30). All subunits have been cloned in to the polylinker of pBC/CMV (31). This expression vector allows high level expression of a foreign gene under control of the cytomegalovirus (CMV) promoter and also allows in vitro transcription using SP6 RNA polymerase. Chimeras were constructed by PCR amplification of the desired sequences and three fragment ligation using standard molecular biology techniques. Site-directed mutagenesis was done using the QuikChange mutagenesis kit (Stratagene). In vitro-synthesized sequences have been verified by DNA sequencing.
Binding Assays to Membrane Preparations of Transiently Transfected Cells.
Human embryonic kidney 293 cells (HEK 293 cells) (ATCC no. CRL 1573) were transfected with plasmids coding for GABAA receptor subunits by the calcium phosphate precipitation method (32), and membranes were prepared as described (19). Resuspended cell membranes (0.5 ml) were incubated for 90 min on ice in the presence of [3H]Ro 15-1788 (87 Ci/mmol; DuPont/NEN; 1 Ci = 37 GBq) and various concentrations of competing ligands. Membranes (20–50 μg protein/filter) were collected by rapid filtration on GF/C filters presoaked in 0.3% polyethylenimine. After three washing steps with 4 ml of buffer, the filter-retained radioactivity was determined by liquid scintilation counting. Nonspecific binding was determined in the presence of 10 μM Ro 15-1788. Data were fitted by using a nonlinear least-squares method to the equation B(c) = Bmax⋅c/(Kd + c) for binding curves and B(c) = Bmax/(1 + c/IC50) for displacement curves, where c is the concentration of ligand; B, binding; Bmax, maximal binding; and Kd, the dissociation constant. IC50 values obtained form displacement curves were converted to Ki values according to the Cheng–Prusoff equation (33). Protein concentration was determined with the Bio-Rad protein assay kit with BSA as standard. All wild-type, chimeric, and mutant receptor combinations expressed equally well (Bmax 0.5–2 pmol [3H]Ro 15-1788 binding sites/mg of protein).
Functional Expression and Characterization.
Xenopus laevis oocytes were prepared, injected, and defoliculated, and currents were recorded as described (18, 34). Briefly, oocytes were injected with 50 nl of capped, polyadenylated cRNA dissolved in 5 mM K-Hepes (pH 6.8). This solution contained the transcripts coding for the different subunits at concentrations (calculated from the UV absorption) of 13, 13, and 130 nM for α1, β2, and γ2, respectively. Electrophysiological experiments were performed by the two-electrode voltage clamp method at a holding potential of −80 mV. Allosteric potentiation via the benzodiazepine site was measured at a GABA concentration (1–3 μM) eliciting 2–10% of the maximal GABA current amplitude by coapplication of GABA and diazepam or of GABA and zolpidem. Oocytes were only exposed to a single drug in addition to GABA, to avoid contamination, and the perfusion system was cleaned by washing with dimethyl sulfoxide for the same reason. The apparent GABA affinity was measured at a holding potential of −60 mV. Agonist concentrations between 0.03 and 3,000 μM were applied for 20 s, and a washout period of 4–15 min was allowed to ensure full recovery from desensitization. Current responses have been fitted to the Hill equation: I = Imax/(1 + (EC50/[A])n) where I is the peak current at a given concentration of GABA (A), Imax is the maximum current, EC50 is the concentration of agonist eliciting half maximal current, and n is the Hill coefficient.
RESULTS
Binding Properties of Receptors Containing γ2 or γ3.
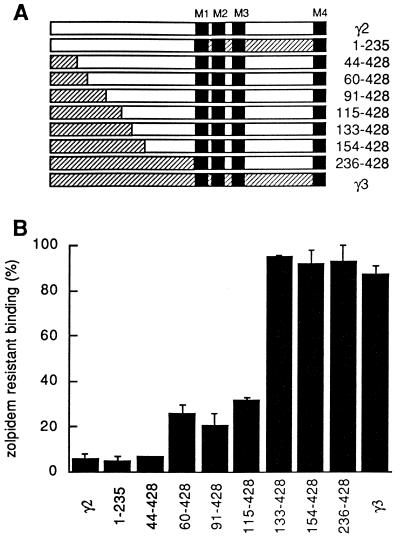
Coexpression of either γ2 or γ3 subunits together with α1 and β2 results in receptors displaying high affinity to the benzodiazepine site antagonist [3H]Ro 15-1788 (Table 1). Receptors containing a γ3 subunit instead of a γ2 subunit differ significantly in their zolpidem binding properties. They exhibit an approximately 178-fold reduced affinity for zolpidem. To locate amino acid residues responsible for reduced zolpidem affinity, eight different γ subunit chimeras were constructed (Fig. 1A).
Table 1.
Binding affinities of benzodiazepine site ligands to wild-type and mutant GABAA receptors
| α1β2γx receptor |
Kd or Ki, nM
|
|||
|---|---|---|---|---|
| [3H]Ro 15-1788 | Diazepam | Zolpidem | Cl 218872 | |
| γ3 | 0.94 ± 0.16 | 180 ± 30 | 2,670 ± 350 | 6.3 ± 0.8 |
| γ2* | 0.61 ± 0.24 | 12 ± 8 | 15 ± 3 | 46 ± 1 |
| γ2M130L | 0.50 ± 0.01 | 30 ± 1 | 770 ± 70 | 5.3 ± 0.6 |
| γ2M57I | 0.77 ± 0.05 | 22 ± 4 | 60 ± 10 | 62 ± 1 |
| γ2T55V | 0.47 ± 0.03 | ND | 24 ± 4 | ND |
Kd values were determined by binding of [3H]Ro 15-1788 to washed membranes of transiently transfected HEK 293 cells. Ki values for each compound were determined by displacement of [3H]Ro 15-1788 binding, and were calculated according to the equation of Cheng and Prusoff (33). IC50 values for each compound were determined by nonlinear least-squares regression assuming a Hill coefficient of 1. Data indicate mean ± SD of two experiments performed in duplicate. ND, not determined.
Data are from Buhr et al. (18).
Figure 1.
Two distinct regions within the amino acid sequence of γ subunits determine zolpidem affinity. (A) Chimeric subunits were constructed by exchanging various parts of the N-terminal extracellular domains between the γ2 (open bars) and γ3 (hatched bars) subunits. Numbers indicate residues of the mature form of the γ2 subunit present in the chimera. Filled boxes denote the four presumed transmembrane segments (M1–M4). (B) Specific binding of 2 nM [3H]Ro 15-1788 in the presence of 1 μM zolpidem to GABAA receptors expressed as triple subunit combinations consisting of γ2, γ3, or chimeric γ subunits each in combination with α1 and β2 subunits. Error bars indicate SD of two independent determinations performed in duplicate.
Binding Properties of Chimeric Receptors.
Chimeric γ subunits were coexpressed with α1 and β2 subunits in cultured HEK 293 cells and assayed for their ability to bind the antagonist [3H]Ro 15-1788 in the presence or absence of zolpidem (1 μM). All wild-type and chimeric receptors displayed comparable high affinity binding of [3H]Ro 15-1788. This was verified by determining specific binding of [3H]Ro 15-1788 at different concentrations. Binding was increased between 10% and 25% upon increasing the radioligand concentration from 2 nM to 6 nM (data not shown), indicating that the Kd for [3H]Ro 15-1788 was between 0.4 nM and 1 nM for all receptors. From these results it was obvious that the antagonist binding site did not undergo remodeling in receptors containing chimeric γ subunits.
Receptors containing a chimeric subunit consisting of the entire N-terminal extracellular domain of the γ2 subunit combined with the transmembrane domain of the γ3 subunit display the same zolpidem sensitivity as wild-type γ2 containing receptors. Furthermore, chimeric receptors containing the entire extracellular domain of the γ3 subunit display identical binding properties as receptors containing a wild-type γ3 subunit. Therefore, it could be concluded that the first 235 amino acids of the mature γ2 subunit determine the difference in zolpidem affinity. A total of 68 amino acid residues are different within the putative N-terminal extracellular domain of both isoforms. Further chimera were constructed and analyzed. Chimeric receptors displayed high, low, and intermediate sensitivity toward zolpidem (Fig. 1B), indicating that two distinct amino acid regions influence zolpidem affinity. One region drastically influences zolpidem sensitivity and is located between amino acid residues 115 and 133 (termed region I) of the γ2 subunit the other region, affecting zolpidem affinity to a lesser extent is located between residues 44 and 59 (termed region II).
Receptors Mutagenized in Region I.
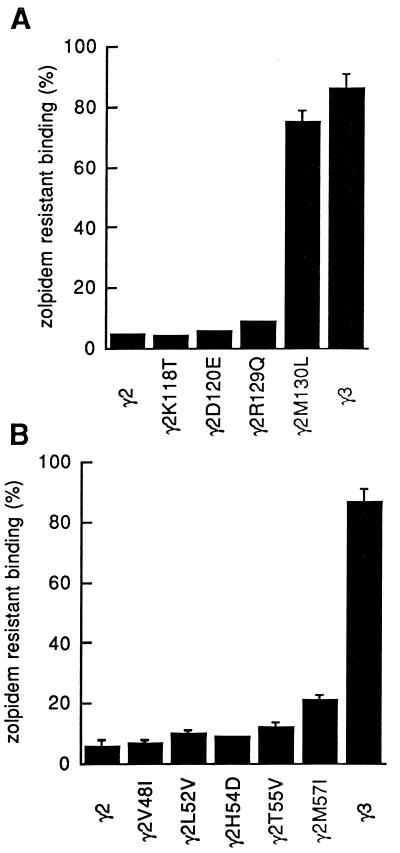
A total of 4 amino acid residues are different between γ2 and γ3 subunits in region I. Each of these four residues of the γ2 subunit has been substituted by the corresponding amino acid residue of the γ3 subunit. Mutants were coexpressed together with the α1 and β2 subunits, and receptors were assayed for [3H]Ro 15-1788 affinity and zolpidem sensitivity. Receptors containing a methionine-to-leucine substitution at position 130 of the γ2 subunit (γ2M130L) displayed a drastically reduced zolpidem sensitivity of [3H]Ro 15-1788 binding (Fig. 2A). Detailed analysis showed that there was little effect on Ro 15-1788 affinity (Fig. 3), whereas the zolpidem affinity was drastically affected (Fig. 4), showing a 51-fold decrease in affinity for zolpidem (Table 1). The diazepam affinity is decreased by 2.5-fold in receptors containing γ2M130L (Table 1). Interestingly, the same mutation resulted in a 9-fold increase in Cl 218872 affinity (Table 1). Receptors containing the γ3 subunit display almost the same high affinity to this ligand as receptors containing γ2M130L (Table 1).
Figure 2.
γ2M130 and γ2M57 are important for zolpidem affinity. Shown is the specific binding of [3H]Ro 15-1788 in the presence of 1 μM zolpidem. (A) Zolpidem sensitivity of receptors containing point mutations in region I (residues 115–132 of mature γ2). The concentration of [3H]Ro 15-1788 was 1 nM. (B) Zolpidem sensitivity of receptors containing point mutations in region II (residues 44–59 of mature γ2). The concentration of [3H]Ro 15-1788 was 2 nM. Error bars indicate SD of two independent determinations performed in duplicate. In some cases the error is to small to show up.
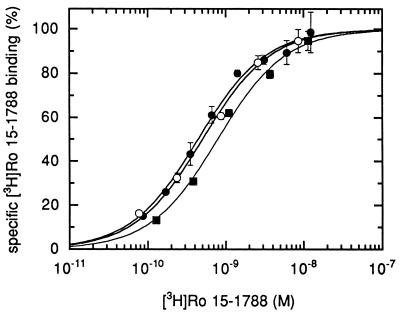
Figure 3.
Receptors containing the mutant γ2M130L subunit display high affinity binding for [3H]Ro 15-1788. Concentration dependence of specific binding of [3H]Ro 15-1788 to membranes prepared from HEK 293 cells transiently transfected with α1β2γ2 (•), α1β2γ2M130L (○), and α1β2γ3 (▪). Each point represents the mean ± SD of one determination performed in duplicate. A second experiment gave similar results.
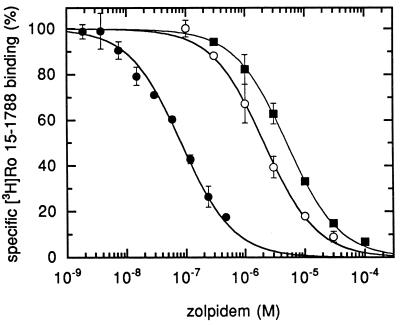
Figure 4.
The γ2M130L mutation drastically reduces zolpidem affinity. Membranes prepared from HEK 293 cells transiently transfected with α1β2γ2 (•), α1β2γ2M130L (○), or α1β2γ3 (▪) were incubated with fixed concentrations of [3H]Ro 15-1788 and various concentrations of competing zolpidem. For receptors containing the γ2 subunit, 2 nM of [3H]Ro 15-1788 was used. For receptors containing γ2M130L or the γ3 subunit, 1 nM of [3H]Ro 15-1788 was used. Assay of γ2 containing receptors with 1 nM [3H]Ro 15-1788 would have resulted in a curve displaced by a factor of 1.6 to the left. Each point represents the mean ± SD of one determination performed in duplicate. A second experiment gave similar results.
Receptors Mutagenized in Region II.
The zolpidem affinity of receptors containing γ2M130L was still about 3.5-fold higher than the affinity of receptors containing the γ3 subunit. Results obtained by chimeric receptors also indicated that at least one additional residue contributes to the zolpidem affinity and that it must be located in region II of the γ2 subunit. Within this region a total of five residues are different between γ2 and γ3. Each of these residues was individually mutagenized to the residue present in the γ3 subunit. Mutant γ2 subunits were coexpressed together with the α1 and β2 subunits. The γ2M57I (methionine-to-isoleucine substitution at position 57 of γ2) containing receptors displayed a significantly reduced zolpidem sensitivity compared with the four other mutant receptors (Fig. 2B). About 79% of specifically bound [3H]Ro 15-1788 (2 nM) was displaced by 1 μM zolpidem. In receptors containing the wild-type γ2 subunit 94% of radioligand was displaced under these conditions. Receptors containing the γ2M57I mutation were directly compared with mutant receptors that displayed a higher sensitivity to zolpidem (γ2T55V, 88% displacement). There was almost no effect on [3H]Ro 15-1788 and zolpidem affinity for the γ2T55V mutation. However, for receptors containing the γ2M57I mutation the affinity for both ligands was reduced (Table 1). The reduction in zolpidem affinity is about 4-fold compared with wild-type receptors. Notably, the difference in zolpidem affinity between γ3- and γ2M130L-containing receptors was similar by about 3.5-fold to the reduction caused by the γ2M57I mutation. Diazepam and Cl 218872 affinity are affected by this mutation to a lesser extent (Table 1).
Functional Effects of γ2M130L.
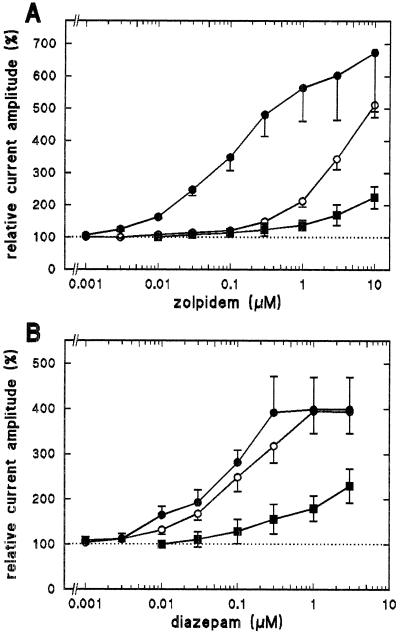
Wild-type and mutant γ subunits were coexpressed with α1 and β2 subunits in Xenopus oocytes to study functional consequences of the γ2M130L mutation. Mutant ion channels expressed GABA-activated chloride currents of comparable maximal current amplitude as wild-type channels (3–4 days after injection, measured at −60 mV; α1β2γ2, 10.4 ± 0.9 μA, n = 3; α1β2γ2M130L, 12.1 ± 1.7 μA, n = 3; α1β2γ3, 3.6 ± 0.8 μA, n = 3). The apparent GABA affinities were 9.6 ± 6.9, 12.5 ± 0.8, and 9.7 ± 1.6 μM (mean ± SD, n = 3) for channels containing γ2, γ2M130L, or γ3, respectively. Therefore, expression and agonist affinity are not affected by the mutation. Allosteric stimulation by zolpidem and diazepam of GABA-activated currents was measured in cumulative dose–response curves (Fig. 5). Stimulation of mutant channels requires much higher concentrations of zolpidem as compared with wild-type γ2-containing channels (Fig. 5A). In contrast to zolpidem, diazepam stimulation is similar in wild-type and mutant γ2 containing channels (Fig. 5B). These results indicate that mutant channels retain allosteric coupling to the agonist site and confirm the importance of γ2M130 for zolpidem binding.
Figure 5.
The γ2M130L mutation drastically affects zolpidem stimulation of GABA-activated currents. Shown are cumulative dose–response curves for zolpidem (A) and for diazepam (B). α1β2γx channels were expressed in Xenopus oocytes. Stimulation was measured at GABA concentrations (1–3 μM) eliciting about 5% of the maximal current amplitude. The same concentration of GABA was coapplied with increasing amounts of zolpidem or of diazepam. α1β2γ2 (•), α1β2γ2M130L (○), or α1β2γ3 (▪). Each point represents the mean ± SD of at least three oocytes.
DISCUSSION
This study demonstrates that one particular amino acid residue at position 130 of the mature γ2 subunit of α1β2γ2S GABAA receptors drastically affects benzodiazepine site ligand affinities. Both α and γ subunits contribute to the benzodiazepine pharmacology of GABAA receptors. Therefore, it is assumed that the benzodiazepine binding site is located at the α/γ subunit interface. Until now, only one amino acid residue on γ subunits influencing benzodiazepine binding affinity (for functional data, see below) has been identified (18, 19). A γ2 or γ3 subunit is required for high affinity binding of the benzodiazepine site antagonist [3H]Ro 15-1788. Receptors containing a γ3 instead of a γ2 subunit display a 178-fold reduced affinity for the ligand zolpidem. The γ3 subunit shows 64.6% sequence identity to the γ2 subunit. About 150 amino acid residues are different between the mature subunits and therefore could be responsible for reduced zolpidem affinity. Chimeric γ subunits were constructed to identify amino acid regions containing the responsible residues. Receptors were expressed and analyzed for the ability of zolpidem to displace [3H]Ro 15-1788 binding.
Coexpression of the γ2 or the γ3 subunit together with α1 and β2 in cultured HEK 293 cells results in receptors displaying high affinity to [3H]Ro 15-1788. All receptors containing chimeric γ subunits expressed equally well and retained high affinity to [3H]Ro 15-1788. These data rule out that this drastic type of mutagenesis had an influence on the overall structure of the receptor complex or a disruptive effect on the benzodiazepine binding pocket. Two regions important for the interaction with zolpidem were identified. One region had a strong effect (termed region I) and the other region had a intermediate effect (termed region II) on zolpidem sensitivity of [3H]Ro 15-1788 binding. In region I and region II a total of four and five amino acid residues are different between γ2 and γ3. Each of these nine residues of the γ2 subunit was individually mutagenized to the residue present in the γ3 subunit, and mutant receptors were expressed and analyzed.
The methionine-to-leucine substitution at position 130 of the γ2 subunit had a dramatic influence on zolpidem sensitivity of [3H]Ro 15-1788 binding. Whereas the affinity to [3H]Ro 15-1788 is not affected the affinity to zolpidem is drastically reduced amounting to a 51-fold change. The affinity for diazepam is reduced by about 2.5-fold. Interestingly, the affinity to Cl 218872 is increased by about 9-fold. Wild-type and mutant subunit combinations were also expressed in Xenopus oocytes and analyzed. These functional data confirmed the importance of γ2M130 for zolpidem stimulation of GABA-activated currents. The apparent zolpidem affinity is drastically reduced whereas diazepam potentiation is much less affected. GABA dose–response curves indicate that the apparent agonist affinity is not affected by the γ2M130L mutation. These results and also the fact that the binding affinity to one benzodiazepine site ligand is not affected at all, whereas other ligands are affected in different ways strongly suggest that γ2M130 plays an important role in the specificity of the binding pocket and that it is most likely part of it.
The analysis of receptors mutagenized in region II identified methionine at position 57 to be important for zolpidem sensitivity of [3H]Ro 15-1788 binding. Zolpidem affinity was reduced by about 4-fold for receptors containing γ2M57I. That this reduction in ligand affinity was significant was concluded from results obtained from γ2T55V containing receptors analyzed in parallel. For receptors containing γ2M57I the affinities to diazepam and to Cl 218872 were reduced to a smaller extent as compared with zolpidem. Methionine as well as isoleucine both have relatively large and hydrophobic side chains. Therefore, this substitution is relative conservative in nature. It is possible that both side chains interact with the hydrophobic benzodiazepine site ligands in a similar way. The relatively modest reduction in ligand affinities could be caused by the bulkiness of isoleucine leading to small distortions of the binding pocket. Other less conservative substitutions of γ2M57 might have more pronounced consequences on benzodiazepine site ligand affinities. It remains to be shown if γ2M57 is really part of the benzodiazepine binding site or if substitution of this residue has more indirect effects on benzodiazepine site ligand affinities.
Additional amino acid residues have been shown to be important for the benzodiazepine pharmacology. The benzodiazepine site antagonist Ro 15-1788 acts agonistic on receptors containing the γ2T142 to serine mutation (17). The authors conclude that the mutation of threonine at position 142 influences the pharmacology by a mechanism most likely distinct from binding of benzodiazepines. Previously it was shown that substitution of a phenylalanine at position 77 of the γ2 subunit has dramatic consequences on benzodiazepine stimulation (18) after functional expression of mutant receptors in Xenopus oocytes. Furthermore, binding experiments after expression in HEK 293 cells indicate that, depending on the amino acid side chain present at this position, ligand affinities were differentially affected (19). The γ2F77I mutation resulted in a complete loss of zolpidem, Cl 218872, and methyl-6,7-dimethoxy-4-ethyl-β-carboline-3-carboxylate (DMCM) affinities (up to 5,000-fold reduction) whereas high affinity to flunitrazepam was retained (19). A phenylalanine-to-tyrosine substitution at position 77 increased the affinity to zolpidem and Cl 218872. In contrast to four different γ2F77 substitutions (to tyrosine, isoleucine, leucine, or tryptophan), which have always similar effects on zolpidem and Cl 218872 affinities, the γ2M130L mutation has differential effects for these two ligands.
Several amino acid residues on α subunit isoforms have also been identified to influence benzodiazepine site ligands affinities. If α3E225 is mutated to a glycine (which is the homologous residue to α1G200), there is at least a 10-fold increase in binding affinities for zolpidem and Cl 218872 (10). Replacement of histidine 101 by arginine in the α1 subunit leads to a complete loss in detectable diazepam, zolpidem, and Cl 218872 binding, while the binding affinity for [3H]Ro 15-1788 is reduced by more than 100-fold (11, 12). Similarly, two additional amino acid residues (proline 161 and isoleucine 210) have been identified in an α6-derived mutant that together alter affinities to benzodiazepine site ligands (13). Residues that influence benzodiazepine pharmacology have been identified on the α1 subunit after functional expression in Xenopus oocytes. α1Y161A and α1T206A both increase stimulation by diazepam and by zolpidem (18) and at least α1T206A also affects benzodiazepine ligand binding affinities upon expression in HEK 293 cells (unpublished results). The same study showed that receptors containing the α1Y209A substitution displayed a reduced zolpidem stimulation. Preliminary binding experiments indicate that this residue also plays an important role in benzodiazepine site ligand binding (unpublished results). After submission of this work, Amin et al. (14) published that two tyrosine residues (α1Y159 and α1Y209) are crucial for benzodiazepine binding and allosteric modulation of GABA-activated currents, pointing again at the importance of α1Y209.
It is not clear if all of the above-mentioned amino acid residues that affect binding of benzodiazepines are in direct contact with benzodiazepines or if the substitutions have an more indirect effect on the benzodiazepine binding site. However, observations that make it likely that at least some of these residues are in direct contact with the ligands comes from an independent approach. Using bovine brain membranes and photoaffinity labeling by [3H]flunitrazepam, Duncalfe et al. (9) showed that the conserved histidine (residue no. 102 of bovine α subunits, corresponding to 101 in rat α1) is the major site of photoincorporation. Furthermore, residues C terminal of residue 104 can be photolabeled by [3H]Ro 15-4513 (35). Most of the residues on α subunits that have been identified by site-directed mutagenesis are downstream of the conserved histidine. It is likely that further analysis of [3H]Ro 15-4513 photoincorporation will point to one of these residues. Similarly, photoincorporation of the agonist [3H]muscimol into α1F64 (36) identified a residue that is important for apparent GABA affinity of channel gating as shown by functional expression of mutant channels (37). Therefore, site-directed mutagenesis and the study of properties of mutant receptors are important strategies for the identification of amino acid residues interacting with ligands.
Interestingly, it appears that the amino acid residues known to be important for benzodiazepine site ligand affinity are located in regions homologous to regions involved in neurotransmitter binding of GABAA and related receptors. For example, α1F64, which is involved in GABA binding, is homologous to γ2F77, which is important for benzodiazepine site specificity. A four loop model of neurotransmitter binding has been suggested by Galzi and Changeux (21) for the members of the acetylcholine receptor subfamily (including GABAA, glycine, and 5HT3 receptors). As for the benzodiazepine binding site, peptide loops of two neighboring subunits are important for neurotransmitter affinity indicating that both binding sites are located at subunit boundaries. γ2M130 is not located in a region homologous to any region involved in agonist binding. As the γ2M130L mutation drastically affects ligand affinity and because of the homology of the benzodiazepine and GABA binding sites it is likely that new residues of the GABA binding site can be identified. Presently, we are testing this hypothesis by systematically mutagenizing the homologous region of the α1 subunit and determining the apparent GABA affinity of mutant ion channels. As the location of the neurotransmitter binding site is strongly conserved in different receptors it is likely that the regions homologous to the region were γ2M130 is located are of general importance. Alternatively, we have described here a region important for the modulatory site that has no related region in the agonist site.
We identify here new amino acid residues on the γ subunit differentially affecting binding of ligands of the benzodiazepine binding site. There is good chance that at least one of these residues is part of the binding pocket and directly interacts with some ligands of this site. The analysis of various substitutions at key positions might help to understand how precisely ligands interact with the binding pocket. Ultimately, such an analysis may provide a rational basis for drug design. However, final verification of such structural predictions has to await crystallization and structural resolution of the GABAA receptor.
Acknowledgments
We thank R. Baur for expert help with Xenopus oocyte expression, Prof. V. Niggli for careful reading of the manuscript (expression and electrophysiology), and Prof. H. Moehler for providing cDNA coding for the γ3 subunit. We are grateful to Prof. H. Reuter, in whose institute this work was carried out, for continuous encouragement. This study was supported by Grants 31-37192.93 from the Swiss National Science Foundation, the European Union Grant BIO4-CT96-0585 (BWW 96.0010), and the Foundation for the Promotion of Scientific Research at the University of Bern.
ABBREVIATIONS
- GABA
γ-aminobutyric acid
- HEK 293 cells
human embryonic kidney 293 cells
References
- 1.Sigel E, Stephenson F A, Mamalaki C, Barnard E A. J Biol Chem. 1983;258:6965–6971. [PubMed] [Google Scholar]
- 2.Schofield P R, Darlison M G, Fujita N, Burt D R, Stephenson F A, Rodriguez H, Rhee L M, Ramachandran J, Reale V, Glencorse T A, Seeburg P H, Barnard E A. Nature (London) 1987;328:221–227. doi: 10.1038/328221a0. [DOI] [PubMed] [Google Scholar]
- 3.Davies P A, Hanna M C, Hales T G, Kirkness E F. Nature (London) 1997;385:820–823. doi: 10.1038/385820a0. [DOI] [PubMed] [Google Scholar]
- 4.Burt D R, Kamatchi G L. FASEB J. 1991;5:2916–2923. doi: 10.1096/fasebj.5.14.1661244. [DOI] [PubMed] [Google Scholar]
- 5.Macdonald R L, Olsen R W. Annu Rev Neurosci. 1994;17:569–602. doi: 10.1146/annurev.ne.17.030194.003033. [DOI] [PubMed] [Google Scholar]
- 6.Dunn S M J, Bateson A N, Martin I L. Int Rev Neurobiol. 1994;36:51–96. doi: 10.1016/s0074-7742(08)60303-7. [DOI] [PubMed] [Google Scholar]
- 7.Sieghart W. Pharmacol Rev. 1995;47:181–233. [PubMed] [Google Scholar]
- 8.Rabow L E, Russek S J, Farb D H. Synapse. 1995;21:189–274. doi: 10.1002/syn.890210302. [DOI] [PubMed] [Google Scholar]
- 9.Duncalfe L L, Carpenter M R, Smilie L B, Martin I L, Dunn M S. J Biol Chem. 1996;271:9209–9214. doi: 10.1074/jbc.271.16.9209. [DOI] [PubMed] [Google Scholar]
- 10.Pritchett D B, Seeburg P H. Proc Natl Acad Sci USA. 1991;88:1421–1425. doi: 10.1073/pnas.88.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wieland H A, Lüddens H, Seeburg P H. J Biol Chem. 1992;267:1426–1429. [PubMed] [Google Scholar]
- 12.Korpi E R, Kleingoor C, Kettenmann H, Seeburg P H. Nature (London) 1993;361:356–359. doi: 10.1038/361356a0. [DOI] [PubMed] [Google Scholar]
- 13.Wieland H A, Lüddens H. J Med Chem. 1994;37:4576–4580. doi: 10.1021/jm00052a019. [DOI] [PubMed] [Google Scholar]
- 14.Amin J, Brooks-Kayal A, Weiss D S. Mol Pharmacol. 1997;51:833–841. doi: 10.1124/mol.51.5.833. [DOI] [PubMed] [Google Scholar]
- 15.Pritchett D B, Sontheimer H, Shivers B D, Ymer S, Kettenmann H, Schofield P R, Seeburg P H. Nature (London) 1989;338:582–585. doi: 10.1038/338582a0. [DOI] [PubMed] [Google Scholar]
- 16.Sigel E, Baur R, Trube G, Möhler H, Malherbe P. Neuron. 1990;5:703–711. doi: 10.1016/0896-6273(90)90224-4. [DOI] [PubMed] [Google Scholar]
- 17.Mihic S J, Whiting P J, Klein R L, Wafford K A, Harris R A. J Biol Chem. 1994;269:32768–32773. [PubMed] [Google Scholar]
- 18.Buhr A, Baur R, Malherbe P, Sigel E. Mol Pharmacol. 1996;49:1080–1084. [PubMed] [Google Scholar]
- 19.Buhr A, Baur R, Sigel E. J Biol Chem. 1997;272:11799–11804. doi: 10.1074/jbc.272.18.11799. [DOI] [PubMed] [Google Scholar]
- 20.Günther U, Benson J, Benke D, Fritschy J-M, Reyes G, Knoflach F, Crestani F, Aguzzi A, Arigoni M, Lang Y, Bluethmann H, Mohler H, Lüscher B. Proc Natl Acad Sci USA. 1995;92:7749–7753. doi: 10.1073/pnas.92.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galzi J L, Changeux J P. Curr Opin Struct Biol. 1994;4:554–565. [Google Scholar]
- 22.Pritchett D B, Seeburg P H. J Neurochem. 1990;54:1802–1804. doi: 10.1111/j.1471-4159.1990.tb01237.x. [DOI] [PubMed] [Google Scholar]
- 23.Carter D B, Thomsen D R, Im W B, Lennon D J, Ngo D M, Gale W, Im H K, Seeburg P H, Smith M W. Bio/Technology. 1992;10:679–681. doi: 10.1038/nbt0692-679. [DOI] [PubMed] [Google Scholar]
- 24.Faure-Halley C, Graham D, Arbilla S, Langer S Z. Eur J Pharmacol. 1993;246:283–287. doi: 10.1016/0922-4106(93)90043-9. [DOI] [PubMed] [Google Scholar]
- 25.Hadingham K L, Wingrove P B, Wafford K A, Bain C, Kemp J A, Palmer K J, Wilson A W, Wilcox A S, Sikela J M, Ragan C I, Whiting P J. Mol Pharmacol. 1993;44:1211–1218. [PubMed] [Google Scholar]
- 26.Herb A, Wisden W, Lüddens H, Puia G, Vicini S, Seeburg P H. Proc Natl Acad Sci USA. 1992;89:1433–1437. doi: 10.1073/pnas.89.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lüddens H, Seeburg P H, Korpi E R. Mol Pharmacol. 1994;45:810–814. [PubMed] [Google Scholar]
- 28.Lolait S J, O’Carroll A-M, Kusano K, Muller J-M, Brownstein M J, Mahan L C. FEBS Lett. 1989;246:145–148. doi: 10.1016/0014-5793(89)80271-6. [DOI] [PubMed] [Google Scholar]
- 29.Malherbe P, Draguhn A, Multhaup G, Beyreuther K, Möhler H. Mol Brain Res. 1990;8:199–208. doi: 10.1016/0169-328x(90)90017-8. [DOI] [PubMed] [Google Scholar]
- 30.Malherbe P, Sigel E, Baur R, Persohn E, Richards J G, Möhler H. J Neurosci. 1990;10:2330–2337. doi: 10.1523/JNEUROSCI.10-07-02330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertocci B, Miggiano V, Da Prada M, Dembic Z, Lahm H-W, Malherbe P. Proc Natl Acad Sci USA. 1991;88:1416–1420. doi: 10.1073/pnas.88.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y C, Prusoff W H. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 34.Sigel E. J Physiol (London) 1987;386:73–90. doi: 10.1113/jphysiol.1987.sp016523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duncalfe L L, Dunn S M J. Eur J Pharmacol. 1996;298:313–319. doi: 10.1016/0014-2999(95)00811-x. [DOI] [PubMed] [Google Scholar]
- 36.Smith G B, Olsen R W. J Biol Chem. 1994;269:20380–20387. [PubMed] [Google Scholar]
- 37.Sigel E, Baur R, Kellenberger S, Malherbe P. EMBO J. 1992;11:2017–2023. doi: 10.1002/j.1460-2075.1992.tb05258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]