Abstract
CTLA-4 plays a critical role in regulating the immune response. It is mainly located in cytoplasmic vesicles and is expressed only transiently on the surface after T cell activation. In this study, we demonstrate that CTLA-4 is associated with AP50, the medium chain of the clathrin-associated coated pit adaptor protein complex AP2. In a yeast two-hybrid screen, three individual cDNA clones that encode mouse AP50 were isolated, all of which can interact specifically with the cytoplasmic domain of mouse CTLA-4, but not with the cytoplasmic domain of mouse CD28. We have shown that CTLA-4 can bind specifically to AP50 when CTLA-4 and AP50 are cotransfected into human 293T cells. A Y201 to F201 mutation in the YVKM intracellular localization motif of the CTLA-4 cytoplasmic domain significantly diminished its binding to AP50. We also found that AP50 bound to a CTLA-4 peptide containing unphosphorylated Y201 but not to a peptide containing phosphorylated Y201. Conversely, the p85 subunit of phosphatidylinositol 3-kinase and, to a lesser extent, protein tyrosine phosphatase SYP (SHP-2) and SHP (SHP-1) bind only to the CTLA-4 peptide containing phosphorylated Y201. Therefore, the phosphorylation status of Y201 in the CTLA-4 cytoplasmic domain determines the binding specificity of CTLA-4. These results suggest that AP50 and the coated pit adaptor complex AP2 may play an important role in regulating the intracellular trafficking and function of CTLA-4.
The T cell surface molecule CD28, upon engagement of its counter-receptors, B7–1 (CD80) and B7–2 (CD86), on the antigen-presenting cell, provides a major costimulatory signal for T cell activation (1–3). Like CD28, CTLA-4 is a member of the immunoglobulin superfamily, and the two molecules share extensive structure and sequence similarity (4, 5). Both molecules bind to the same ligands, B7–1 (CD80) and B7–2 (CD86), although CTLA-4 binds with a much higher affinity (6, 7). An initial study showed that an anti-human CTLA-4 antibody enhanced anti-CD28-mediated costimulation; however, later studies using anti-murine CTLA-4 monoclonal antibodies suggested that CTLA-4 played an inhibitory role during T cell activation (reviewed in ref. 8). The importance of CTLA-4 in down-regulating the immune response was further demonstrated in the observation that CTLA-4-deficient mice die of a rampant T cell lymphoproliferative disorder within 3–4 weeks after birth (9–11).
CTLA-4 is expressed transiently on the cell surface 24–48 hr after T cell activation (12–14). It is mainly localized in intracellular vesicles whose location may overlap the Golgi apparatus (15), transferrin-containing endosomes, and perforin-containing vesicles (16). Surface CTLA-4 is rapidly internalized, and both cell surface and intracellular CTLA-4 molecules accumulate at the sites of contact when T cells are stimulated by antigen-presenting cells or anti-CD3-coated beads (16, 17). Therefore, the intracellular localization of CTLA-4 and its focal release toward the T cell antigen receptor (TCR)/major histocompatibility complex (MHC) contact point may provide a unique mechanism in regulating CTLA-4 function. It was reported that an 11 amino acid residue sequence, TTGVYVKMPPT, including the YVKM motif, in the cytoplasmic domain of CTLA-4 is essential for intracellular localization (15). Interestingly, the YVKM motif has also been suggested to serve as a binding site for both the p85 domain of the phosphatidylinositol (PI) 3-kinase and the protein tyrosine phosphatase SYP (18, 19), indicating that the same YVKM motif may also be important in CTLA-4 signaling.
Clathrin-coated pits and vesicles are involved in the intracellular trafficking of membrane proteins (20). Coated vesicles that bud from plasma membrane contain the adaptor complex AP2, whereas those that bud from the trans Golgi network (TGN) contain adaptor complex AP1. The AP2 complex contains two large chains, α and β adaptin, one medium chain, AP50, and one small chain, AP17. Correspondingly, the AP1 complex contains two large chains, β′ and γ adaptin, one medium chain, AP47, and one small chain, AP19. It has been suggested that sorting motifs such as the dileucine- and tyrosine-based motifs in the cytoplasmic tails of the membrane receptors contain essential signals for the receptors to interact with the adaptor complexes and determine the endocytosis and targeting of these receptors to intracellular compartments (21, 22).
Many of the tyrosine-based motifs possess the consensus sequence YXXΦ, where Φ is a bulky hydrophobic amino acid. Different tyrosine-based motifs can interact specifically with either AP1 or AP2 or both types of adaptor protein complexes (21, 22). It was recently demonstrated in vitro that triplicates of the tyrosine-based motif YQRL in the tail of an integral membrane protein, TGN 38, can interact with AP50 and AP47, the medium chains of the AP2 and AP1 complexes (23). It was further demonstrated that amino acid residues inside or surrounding the YQRL motif may influence the binding specificity of the tyrosine-based motif toward AP50 or AP47 (24, 25). However, it remains to be demonstrated in cellular systems that membrane receptors can interact directly with AP47 and AP50.
In this study, we have sought to investigate the biochemical mechanisms involved in CTLA-4 signaling and intracellular trafficking by identifying proteins that can interact with the CTLA-4 cytoplasmic domain. Three independent clones encoding murine AP50 were isolated, using CTLA-4 as bait in a yeast two-hybrid screen of an activated mouse T cell library. We demonstrate that CTLA-4, but not CD28, can bind specifically with AP50 and that Y201 of the CTLA-4 cytoplasmic domain is required for the binding. We further demonstrate that AP50 binds only to the unphosphorylated CTLA-4 peptide containing the YVKM motif and not to the phosphorylated peptide. These results suggest an important role for AP50 and its associated adaptor protein complex AP2 in the intracellular trafficking and function of CTLA-4.
MATERIALS AND METHODS
Reagents and Antibodies.
Antibodies used in the study include monoclonal anti-murine CTLA-4 antibody 9H10 (14), polyclonal anti-CTLA-4 antibody Q20 (Santa Cruz Biotechnology), monoclonal anti-hemagglutinin (HA) (Babco), polyclonal anti-SHP (SHP-1), SYP (SHP-2) (Santa Cruz Biotechnology), and polyclonal anti-p85 and anti-Grb-2 antibodies (Upstate Biotechnology). Control bait constructs such as pAS2-RB, pAS2-lamin, and pAS2-SNF-1 were kindly provided by S. Elledge (26)
Yeast Two-Hybrid Screening.
The cDNA encoding murine CTLA-4 cytoplasmic domain was PCR-amplified from plasmid F14F4 (4) and inserted into the BamHI and SalI sites of pAS2-CYH (27). Similarly, the cytoplasmic domain of CD28 was also PCR-amplified from a murine CD28 construct (28) and inserted into pAS2-CYH. The pAS2-CTLA-4 bait construct was transfected into yeast strain Y190 along with a mouse peripheral T cell library constructed in the GAL4 activation domain plasmid pACT, and the two-hybrid screen was performed as previously described (26).
Protein Expression.
To express AP50 as an HA-tagged protein, the 1.6-kb XhoI fragment of clone 107 was inserted into pCI-HA, which contains an oligonucleotide encoding the HA peptide inserted into the 5′ end of the multiple cloning sites of the mammalian expression vector pCI (Promega). Plasmid pCI-HA-AP50 was transfected into human 293T cells by using the Lipofectamine reagents (Life Technologies) and following the manufacturer’s instructions. The CTLA-4 cytoplasmic domain was also expressed as a glutathione S-transferase (GST) fusion protein by inserting the PCR product of CTLA-4 cytoplasmic domain into pGEX-1N (Pharmacia Biotech). The resulting plasmid pGEX-CTLA-4 was used to transform bacterial strain DH5a, and GST-CTLA-4 fusion protein was purified by using glutathione-coupled agarose (Sigma) as previously described (29).
Mutagenesis and Expression of CTLA-4 in 293T Cells.
A 1.5-kb fragment of CTLA-4 cDNA containing the entire CTLA-4 ORF was inserted into the HindIII/XhoI sites of the pBluescript SK (+) II vector (Strategene) and used as mutagenesis template. Site-directed mutagenesis was performed using the Muta-Gene T7 kit (Bio-Rad) according to the manufacturer’s instructions. Each mutation was verified by DNA sequencing. The mutated CTLA-4 cDNAs were subcloned into the HindIII/XhoI sites of mammalian expression vector pCDNA-1 (Invitrogen). CTLA-4 cDNAs were transfected into the human 293T cells with or without the AP50 expression vector by using the Lipofectamine reagent, following manufacturer’s protocol.
In Vitro Binding Assays.
293T cells transfected with pCI-HA-AP50 were harvested and lysed in 1% NP40 lysis buffer (1% Nonidet P-40/300 mM NaCl/50 mM Tris⋅HCl, pH 7.5/5 mM EDTA/10 mM iodoacetamide/10 μg/ml aprotinin/10 μg/ml leupeptin/1 mM phenylmethanesulfonyl fluoride). Cell lysates were centrifuged and HA-AP50-containing supernatants were incubated at 4°C with GST fusion proteins immobilized on glutathione-agarose for 3–6 hr. The affinity resin was then washed three times with 1% NP40 lysis buffer and boiled in SDS sample buffer, and the eluted proteins were resolved by SDS/10% PAGE and analyzed by immunoblotting.
CD28 and CTLA-4 peptides were also used for in vitro protein binding assays. The peptide RRNRLLQSDYMNMTPRRPGL, which corresponds to residues 164–183 of the CD28 cytoplasmic domain, and KKRSPLTTGVYVKMPPTEPE, which corresponds to residues 191–210 of the CTLA-4 cytoplasmic domain, were synthesized in both unphosphorylated and phosphorylated forms using solid-phase synthesis (Cancer Research Laboratory, University of California at Berkeley). Briefly, the nonphosphorylated peptides were synthesized using N-Fmoc-protected amino acids and dicyclohexyl carbodiimide (DCC) mediated coupling in dimethyl formamide in the presence of 1-hydroxybenzotriazole (HOBT). The phosphopeptides were synthesized in the same manner except that the DCC-mediated coupling in the presence of HOBT was carried out in 1-methyl-2-pyrrolidinone (NMP). Tyrosine residues were coupled without side-chain protection. After synthesis, the un-O-protected tyrosine residue was phosphorylated, using a 10-fold molar excess of the unhindered di-t-butyl-N,N-diethylphosphoramidite with tetrazole and subsequent oxidation of trivalent phosphorus with a 30-fold molar excess of t-butyl hydroperoxide. Peptides were cleaved, deprotected, and analyzed by HPLC and electrospray ionization mass spectrometry. Peptides were coupled to the Affi-Gel 10 gel (Bio-Rad) according to the manufacturer’s protocol. Immobilized CD28 and CTLA-4 peptides were used in in vitro binding assays with HA-AP50 or Jurkat cell lysates as described above. The peptide dephosphorylations were performed by incubating Affi-Gel-coupled phosphopeptides with calf intestine alkaline phosphatase (Pharmacia) in a concentration of 50 units/ml at 37°C for 2 hr. After the reaction, the coupled peptides were washed in 1% NP40 lysis buffer and used for the binding reaction.
Immunoprecipitation and Immunoblotting.
293T cells transfected with CTLA-4 and HA-AP50 were harvested and lysed overnight at 4°C in 1% NP40 lysis buffer. Cell lysates were clarified by centrifugation and immunoprecipitated using protein G beads coated with anti-CTLA-4 antibody 9H10. After 6–8 hr of incubation at 4°C, the immunoprecipitates were washed using 1% NP40 lysis buffer, and bound proteins were eluted by boiling in SDS sample buffer, separated by SDS/10% PAGE, and transferred onto a poly(vinylidene difluoride) (PVDF) membrane. Membranes were blocked with 4% BSA in PBS and then incubated with anti-HA antibody and subsequently with a horseradish peroxidase-conjugated donkey anti-mouse secondary antibody before visualization using the ECL reagents (Amersham). Membranes were stripped according to the instruction for ECL and reprobed with anti-CTLA-4 polyclonal antibody Q20.
RESULTS
Interaction of AP50 with CTLA-4 in a Yeast Two-Hybrid Screen.
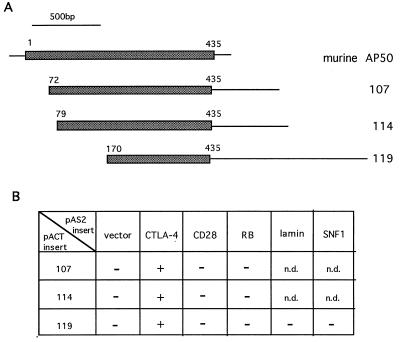
The cytoplasmic tail of CTLA-4 was used as the “bait” to screen a murine T cell library (30). Screening of approximately 5 × 106 colonies yielded 3 independent clones which encoded portions of murine AP50, the medium chain of the clathrin-associated coated pit adaptor protein complex AP2 (Fig. 1A). All three clones, namely 107, 114, and 119, interact specifically with the cytoplasmic domain of CTLA-4, but not the cytoplasmic domain of CD28, the GAL4 DNA-binding domain, or the retinoblastoma protein (Fig. 1B). The plasmid for clone 119 was also recovered from yeast and retransfected into yeast to test its specificity, using bait constructs encoding lamin and SNF1 protein (Fig. 1B). Full-length murine AP50 contains an ORF encoding 435 amino acids (23). These CTLA-4-interacting clones contain ORFs encoding 364, 358, and 267 amino acids (Fig. 1A). The fact that the shortest clone, 119, which lacks the N-terminal 169 amino acid residues, can still interact with CTLA-4, indicates that the N-terminal 169 amino acid residues of AP50 are not essential for CTLA-4 binding.
Figure 1.
Yeast two-hybrid cloning of murine AP50. (A) Schematic diagram of the cDNA clones encoding murine AP50. The cDNA inserts from clones 107, 114, and 119 are 1.6, 1.6, and 2.0 kb, respectively. (B) Yeast specificity test. Plasmids (pAS2-CYH) encoding the GAL4 DNA-binding domain fused to the proteins indicated on the top of each column were transfected into yeast strain Y187 and mated with yeast strain Y190 containing the plasmids (pACT) encoding the GAL4 activation domain fused with AP50 proteins. The yeast diploids which contain both plasmids were picked and tested in the β-galactosidase assay. RB, retinoblastoma protein; n.d., not determined.
Binding of CTLA-4 to AP50 in Vitro.
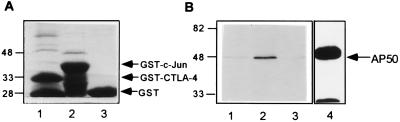
To study the interaction between CTLA-4 and AP50 further, the cytoplasmic domain of CTLA-4 was linked to the GST protein and expressed in Escherichia coli as the GST-fusion protein (Fig. 2A, lane 1). As controls, the GST protein (lane 3) and the GST-c-Jun fusion protein (lane 2) were also expressed in E. coli. AP50 cDNA from clone 107 was expressed in human 293T cells as an HA-tagged protein. The HA-tagged AP50 that was immunoprecipitated from the 293T cell lysates migrated as a single protein band around 48 kDa on SDS/PAGE (Fig. 2B, lane 4).
Figure 2.
Interaction between CTLA-4 and AP50 in vitro. (A) Expression of CTLA-4 cytoplasmic domain as a GST-fusion protein. GST-CTLA-4 (lane 1), GST-c-Jun (lane 2), and GST (lane 3) were separated by SDS/10% PAGE and stained with Coomassie blue. (B) Binding of HA-tagged-AP50 to GST-fusion proteins. Lysates of 293T cells containing HA-AP50 were incubated with immobilized GST protein (lane 1), GST-CTLA-4 (lane 2), or GST-c-Jun (lane 3). Bound HA-AP50 was eluted from glutathione-agarose by boiling in SDS sample buffer, separated by SDS/10% PAGE, and transferred onto PVDF membrane. HA-AP50 was detected by blotting with an anti-HA antibody. HA-AP50 in the 293T cell lysate was also immunoprecipitated using anti-HA-coated protein G agarose (lane 4).
The GST-CTLA-4 fusion protein was immobilized on glutathione-agarose and incubated with 293T cell lysates containing HA-AP50. Bound proteins were eluted with SDS sample buffer and separated by SDS/PAGE, and the presence of HA-AP50 was detected by Western blotting with an anti-HA antibody. As shown in Fig. 2B, lane 2, a significant amount of AP50 protein was associated with the GST-CTLA-4. The specificity of this interaction was demonstrated by the failure of GST itself (Fig. 2B, lane 1) or GST-c-Jun (Fig. 2B, lane 3) to bind to HA-AP50. These results demonstrate that the cytoplasmic domain of CTLA-4 is sufficient to confer specific interaction with AP50.
Structural Requirements for the Binding of CTLA-4 to AP50.
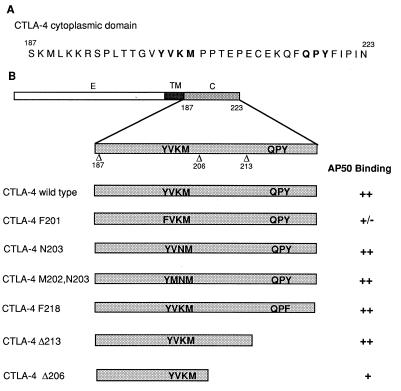
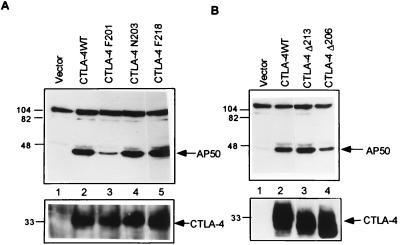
An 11 amino acid residue motif in the cytoplasmic domain of CTLA-4 was reported to be essential for the intracellular localization of CTLA-4 (15). To determine whether this intracellular localization motif is required for AP50 binding and to determine other structural requirements for AP50 binding, we generated a set of CTLA-4 cytoplasmic tail mutants (Fig. 3B). Constructs containing wild-type or mutated CTLA-4 cDNAs were transfected into the human 293T cells along with the pCI-HA-AP50 expression vector. CTLA-4 was immunoprecipitated from cell lysates with monoclonal antibody 9H10 and the coprecipitation of AP50 was revealed by Western blotting with anti-HA. As shown in Fig. 4A, wild-type CTLA-4 associated with abundant AP50 (lane 2), while no AP50 was obtained in the precipitates from cells transfected with empty pCDNA-1 vectors (lane 1). Minimal AP50 coimmunoprecipitated with CTLA-4 F201, indicating that the Y201 is important for AP50 binding (Fig. 4A, lane 3). The mutation of a downstream tyrosine residue, Y218, to F218 had no effect on the interaction between CTLA-4 and AP50 (Fig. 4A, lane 5) suggesting that Y218 is not important for AP50 binding. Similarly, changing of K203 into N203 in the YVKM motif also did not affect AP50 binding (Fig. 4A, lane 4). As shown in Fig. 4A Lower, similar amounts of wild-type and mutated CTLA-4 proteins were present in the immunoprecipitates, confirming that the different amounts of AP50 coprecipitated with CTLA-4 are due to differences in the binding capacity of wild-type and mutated CTLA-4. These results clearly demonstrate that Y201 in the intracellular localization motif of the CTLA-4 tail is critical for the binding of AP50.
Figure 3.
CTLA-4 mutants and their interaction with AP50. (A) Amino acid sequence of the CTLA-4 cytoplasmic domain. Boldface letters indicate the tyrosine-containing motifs that are conserved between CD28 and CTLA-4. (B) List of point mutations and deletion mutations in the cytoplasmic domain of CTLA-4. Domains: E, external; TM, transmembrane; C, cytoplasmic. The binding capacities of wild-type CTLA-4 and the mutants to AP50 are summarized on the basis of representative data shown in Fig. 4 and other results that are not shown.
Figure 4.
Coimmunoprecipitation of AP50 with CTLA-4 mutants from 293T cells. Lysates of 293T cells cotransfected with CTLA-4 and AP50 were immunoprecipitated with anti-CTLA-4 antibody 9H10. The presence of AP50 in the immunoprecipitates was detected by immunoblotting with anti-HA. The membranes in A and B Upper were stripped and reblotted with polyclonal anti-CTLA-4 antibody Q20 (Lower).
To investigate whether other regions of the CTLA-4 cytoplasmic domain are also important for AP50 binding, we examined the interaction of CTLA-4 truncation mutants with AP50. CTLA-4 Δ213 bound AP50 at levels similar to those of the wild-type CTLA-4 (Fig. 4B, lane 3), indicating that the C-terminal 11 amino acid residues, including the QPY218 motif, are not important for AP50 binding. Removal of an additional 7 amino acid residues significantly reduced the binding ability of CTLA-4 to AP50, as the mutant CTLA-4 Δ206 did not precipitate AP50 as effectively as CTLA-4 wild type and CTLA-4 Δ213 (Fig. 4B, lane 4). These results suggest that the 7 amino acid motif PTEPECE is important for AP50 binding.
Phosphorylation Status of Y201 Determines the Binding of CTLA-4 to AP50 or SH2 Domain-Containing Proteins.
The data presented above indicate that Y201 in the YVKM motif of CTLA-4 cytoplasmic domain is important for the binding of AP50, a component of the coated vesicle adaptor complex AP2. Previously, the same YVKM motif was reported to be involved in the binding of protein tyrosine phosphatase SYP and the p85 subunit of PI 3-kinase (18, 19). These results raise an interesting question: how could the same YVKM motif be involved in the binding of several proteins with apparently different functions? Since it was previously demonstrated that both p85 and SYP bound only to the CTLA-4 containing phosphorylated Y201 (18, 19), it suggests that the phosphorylation status of Y201 might determine the binding specificity. Therefore, we examined the effects of phosphorylation of Y201 in the YVKM motif on AP50 binding.
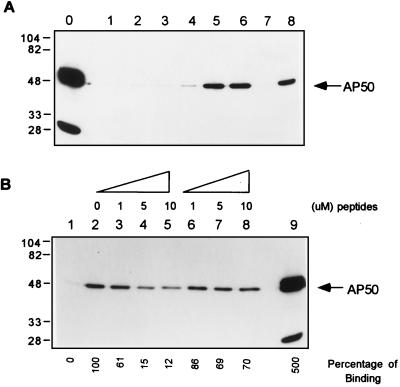
CTLA-4 peptides containing either nonphosphorylated or phosphorylated Y201 were chemically synthesized and coupled to Affi-Gel. In vitro binding assays was performed using Affi-Gel-coupled CTLA-4 peptides and HA-AP50. As shown in Fig. 5A, lane 5, unphosphorylated CTLA-4 peptide readily bound to AP50. The phosphorylated peptide, however, did not bind significant amounts of AP50 (Fig. 5A, lane 7). The ability of phosphorylated peptide to bind AP50 was fully restored if it was first dephosphorylated by digestion with alkaline phosphatase (Fig. 5A, lane 8). Pretreatment of unphosphorylated peptide with alkaline phosphatase had no effect on AP50 binding (Fig. 5A, lane 6). Furthermore, the binding of AP50 to immobilized unphosphorylated CTLA-4 peptide was effectively inhibited by adding excess unphosphorylated CTLA-4 peptide during the binding reaction, while addition of the phosphorylated peptide had no significant effect (Fig. 5B). Together, these results demonstrated that the phosphorylation status of Y201 determines the binding capacity of CTLA-4 for AP50. Our results also indicated that neither phosphorylated nor unphosphorylated peptides of the corresponding region of CD28 bound AP50 effectively (Fig. 5A, lanes 2–4). These data confirm the results from the yeast specificity test that CD28 does not interact with AP50.
Figure 5.
Interaction between CD28 and CTLA-4 peptides and AP50. (A) Binding of phosphorylated or unphosphorylated CD28 and CTLA-4 peptides with AP50. Lysates of HA-AP50 transfected 293T cells were incubated with Affi-Gel (lane 1) or Affi-Gel-coupled unphosphorylated CD28 peptide (lane 2) or phosphorylated CD28 peptide (lane 3) or unphosphorylated CTLA-4 peptide (lane 5) or phosphorylated CTLA-4 peptide (lane 7). The Affi-Gel-coupled phosphorylated CD28 (lane 4) and unphosphorylated CTLA-4 peptide (lane 6) and phosphorylated CTLA-4 peptide (lane 8) were also first digested with alkaline phosphatase before incubation with AP50. (B) Competition of AP50 binding to Affi-Gel-coupled CTLA-4 peptide. Affi-Gel (lane 1) or CTLA-4 peptide-coupled Affi-Gel (lanes 2–8) was incubated with equal amounts of HA-AP50 in the presence of the indicated amounts of unphosphorylated (lanes 3–5) and phosphorylated (lanes 6–8) CTLA-4 peptides. The Affi-Gel-bound AP50 was analyzed as described for Fig. 2. The total amount of AP50 in each binding reaction is shown in A, lane 0, and B, lane 9, by immunoprecipitation with anti-HA antibody.
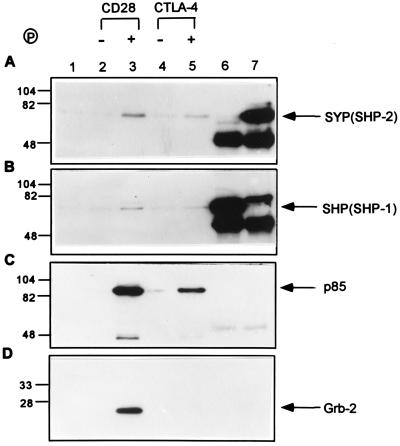
We also examined the binding of phosphorylated and unphosphorylated CTLA-4 and CD28 peptides to protein tyrosine phosphatases SYP (SHP-2) and SHP (SHP-1), the p85 subunit of PI 3-kinase, and the SH2/SH3 adaptor protein Grb-2 (Fig. 6). Lysates of Jurkat cells were used as a source of proteins for the binding assay because Jurkat cells contain large amounts of these proteins (refs. 31 and 32; Fig. 6 A and B, lanes 6 and 7). Our results showed only minimal amounts of SYP and SHP, if any, bound to unphosphorylated CD28 and CTLA-4 peptides (Fig. 6 A and B, lanes 2 and 4). The phosphorylated form of CD28 bound to a small but significant amount of both SYP and SHP (Fig. 6 A and B, lane 3). The tyrosine-phosphorylated CTLA-4 peptide also bound to tyrosine phosphatase SYP and SHP, although at lower levels as compared with the phosphorylated CD28 peptide (Fig. 6 A and B, lanes 3 and 5). In addition, both phosphorylated CD28 and CTLA-4 peptide bound to a significant amount of the p85 subunit of the PI 3-kinase (Fig. 6C, lanes 3 and 5). Under the same conditions, only phosphorylated CD28 bound to the Grb-2 protein, while no binding was detected between phosphorylated CTLA-4 and Grb-2 (Fig. 6D, lanes 3 and 5). Together, these results demonstrated that CD28 and CTLA-4, upon tyrosine phosphorylation, can bind selectively to SH2 domain-containing signaling molecules.
Figure 6.
Interaction between CD28 and CTLA-4 peptides with SH2 domain-containing proteins. (A) Jurkat cell lysates were incubated with Affi-Gel (lane 1), Affi-Gel-coupled unphosphorylated CD28 peptide (lane 2), phosphorylated CD28 peptide (lane 3), unphosphorylated CTLA-4 peptide (lane 4), or phosphorylated CTLA-4 peptide (lane 5). Jurkat cell lysates were also immunoprecipitated with anti-SHP (lane 6) and anti-SYP (lane 7) polyclonal antibodies. Proteins bound to the Affi-Gel were separated by SDS/PAGE and Western blotted with anti-SYP polyclonal antibody. The same PVDF membrane was stripped and blotted again with anti-SHP (B), anti-p85 (C), and anti-Grb-2 (D). A lysate of approximately 3 × 106 Jurkat cells was used for each binding reaction.
DISCUSSION
An intriguing difference exists between the expression patterns of CD28 and CTLA-4. Whereas CD28 is constitutively expressed on the plasma membrane, expression of CTLA-4 is both tightly regulated and highly compartmentalized (15–17). We have demonstrated in both in vitro and in vivo that the cytoplasmic domain of CTLA-4, but not CD28, can associate specifically with AP50, the medium chain of the clathrin-coated pit adaptor complex AP2. The interaction most likely involves other members of the endogenous subunits of the AP complex. Although the mechanisms controlling CTLA-4 trafficking and function are largely unknown, our results suggest that AP50 and clathrin-coated pits may play an important role in restricting CTLA-4 to its intracellular locations and/or for the intracellular trafficking of CTLA-4.
We have further identified two regions in the CTLA-4 cytoplasmic domain that are important for AP50 binding: Y201 in the YVKM motif and a 7 amino acid sequence, PTEPECE. Mutation of Y201 to F201 greatly diminished and removal of the PTEPECE motif significantly reduced the binding of CTLA-4 to AP50. These two regions are either located in or overlap an 11 amino acid sequence previously reported to comprise an intracellular localization motif in the CTLA-4 tail (15). Together, these data provide a strong indication that the association between CTLA-4 and AP50 is important in the intracellular localization of CTLA-4.
We also made additional mutations, including N203 and M202 plus N203, which convert the YVKM motif of CTLA-4 to a sequence more similar to that found in CD28 (Fig. 3B). Since these mutations did not affect the binding of CTLA-4 tail to AP50 (Fig. 3B), we suggest that the four amino acid residue motif YMNM is not sufficient to determine the lack of binding of CD28 to AP50. These results further support the observation that other residues surrounding the YMNM motif may be important for the binding specificity. The differential association of CTLA-4 and CD28 with AP50 are consistent with the previous hypothesis that the binding specificity to the medium chains of the adaptor protein complexes might be influenced by the amino acid residues inside or surrounding the tyrosine-based sorting motifs (24, 25). It remains to be determined exactly which residues surrounding the CD28 and CTLA-4 YXXM motif are important for the differential AP50 binding.
Our results indicate that the phosphorylation status of Y201 is a key in determining binding specificity of CTLA-4 to AP50 and other cellular signaling proteins. Therefore, unphosphorylated CTLA-4 might associate with AP50 and thus be actively internalized from plasma membrane via coated pits and/or retained in the intracellular locations. Upon phosphorylation of Y201, perhaps as a consequence of kinase cascade initiated from TCR or CD28 signaling, CTLA-4 molecules might dissociate from AP50 and consequently be released from vesicles to begin recruiting cellular molecules toward the sites of TCR engagement. Phosphorylated CTLA-4 molecules might also stay on the plasma membrane for a longer period of time without being internalized. Thus, phosphorylation at Y201 and AP50 association might provide a unique mechanism for regulating the intracellular trafficking and signaling functions of CTLA-4.
It has become apparent that CTLA-4 plays an inhibitory role in T cell activation. We previously reported that CTLA-4-mediated inhibition could be reversed by phorbol ester, suggesting that the regulation of tyrosine phosphorylation might be involved in the process (33). However, the CTLA-4 inhibitory pathway seems to be intact in motheaten viable mice, indicating that normal levels of the tyrosine phosphatase SHP are not essential for CTLA-4 function (33). Other studies showed that several tyrosine kinases and kinase substrates are hyperphosphorylated in CTLA-4−/− mice (19). It was further demonstrated that CTLA-4 is associated with the tyrosine phosphatase SYP in activated T cells (19). This led to the suggestion that the inhibitory function of CTLA-4 might be a result of its association with the tyrosine phosphatases and its inhibition of phosphorylation events induced by the TCR and/or CD28. In our studies, we detected an interaction, although very weak, between phosphorylated CTLA-4 peptide and tyrosine phosphatase SYP and SHP. Interestingly, under the same conditions, we found a more pronounced association between the corresponding phosphorylated CD28 peptide and both SYP and SHP. It will be important to determine the functional significance of the binding of CTLA-4 and CD28 to these tyrosine phosphatases.
Our data indicate that both phosphorylated CD28 and CTLA-4 peptides bind to the p85 subunit of PI 3-kinase. Although the p85 subunit and PI 3-kinase activity have been implicated in the CD28 signaling pathway, it remains controversial as to whether they are required for CD28 signaling (34, 35). The function of PI 3-kinase in CTLA-4 signaling is also unclear. PI 3-kinase was previously suggested to play an important role in the ligand-induced endocytosis of the receptor for platelet-derived growth factor (36). It will be of interest to examine whether PI 3-kinase has any function in the trafficking of CTLA-4. Since p85 is a SH2/SH3 adaptor protein itself, it will also be interesting to determine whether p85 could mediate the binding of other cellular signaling proteins to CD28 and CTLA-4. Our results also indicate that phosphorylated CD28, but not CTLA-4, can interact with the Grb-2 adaptor protein. Since Grb-2 was previously suggested to serve as a linkage between CD28 and the Ras signaling pathway (31), the lack of interaction between CTLA-4 and Grb-2 may reflect the different signaling pathways used by CTLA-4 and CD28.
In summary, we have provided evidence that the phosphorylation of a single tyrosine residue in the cytoplasmic domain of CTLA-4 influences both the intracellular localization and the signaling function of CTLA-4 receptor. The linkage between coated pit-mediated trafficking and signaling by cell surface receptors provides an important facet for understanding the function and the regulation of receptor-ligand interaction, particularly for understanding T cell activation.
Acknowledgments
We thank Dr. Steven Elledge for providing the reagents for the yeast two-hybrid screen; Mr. Herb Kasler and Dr. Astar Winoto for providing the GST-c-Jun and pCI-HA constructs; Ms. Chun Tsai, Ms. Nisha Kabra, and Ms. Shirley Liang for generating the DNA constructs and for performing some of the preliminary experiments; and Dr. Cynthia Chambers for critically reading the manuscript. This work is supported by National Institutes of Health Grant CA40041 to J.P.A., and Y.Z. is a fellow of the Leukemia Society of America.
ABBREVIATIONS
- TCR
T cell antigen receptor
- PI
phosphatidylinositol
- HA
hemagglutinin
- GST
glutathione S-transferase
- AP
adaptor protein
- NP40
Nonidet P-40
- PVDF
poly(vinylidene difluoride)
References
- 1.Linsley P S, Ledbetter J A. Annu Rev Immunol. 1993;11:191–212. doi: 10.1146/annurev.iy.11.040193.001203. [DOI] [PubMed] [Google Scholar]
- 2.June C H, Bluestone J A, Nadler L M, Thompson C B. Immunol Today. 1994;15:321–331. doi: 10.1016/0167-5699(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 3.Allison J P. Curr Opin Immunol. 1994;6:414–419. doi: 10.1016/0952-7915(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 4.Brunet J F, Denizot F, Luciani M F, Roux-Dosseto M, Suzan M, Mattei M F, Golstein P. Nature (London) 1987;328:267–270. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 5.Harper K, Balzano C, Rouvier E, Mattei M G, Luciani M F, Golstein P. J Immunol. 1991;147:1037–1044. [PubMed] [Google Scholar]
- 6.Linsley P S, Greene J L, Brady W, Bajorath J, Ledbetter J A, Peach R. Immunity. 1994;1:793–801. doi: 10.1016/s1074-7613(94)80021-9. [DOI] [PubMed] [Google Scholar]
- 7.van der Merwe P A, Bodian D L, Daenke S, Linsley P, Davis S J. J Exp Med. 1997;185:393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers C A, Krummel M F, Boitel B, Hurwitz A A, Sullivan T J, Fournier S, Cassell D, Brunner M, Allison J P. Immunol Rev. 1996;153:27–46. doi: 10.1111/j.1600-065x.1996.tb00919.x. [DOI] [PubMed] [Google Scholar]
- 9.Tivol E A, Borriello F, Schweitzer A N, Lynch W P, Bluestone J A, Sharpe A H. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 10.Waterhouse P, Penninger J M, Timms E, Wakeham A, Shahinian A, Lee K P, Thompson C B, Griesser H, Mak T W. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 11.Chambers C, Cado D, Truong T, Allison J P. Proc Natl Acad Sci USA. 1997;94:9296–9301. doi: 10.1073/pnas.94.17.9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linsley P S, Greene J L, Tan P, Bradshaw J, Ledbetter J A, Anasetti C, Damle N K. J Exp Med. 1992;176:1595–1604. doi: 10.1084/jem.176.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walunas T L, Lenschow D J, Bakker C Y, Linsley P S, Freeman G J, Green J M, Thompson C B, Bluestone J A. Immunity. 1994;1:405–413. doi: 10.1016/1074-7613(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 14.Krummel M F, Allison J P. J Exp Med. 1995;182:459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung H T, Bradshaw J, Cleaveland J S, Linsley P S. J Biol Chem. 1995;270:25107–25114. doi: 10.1074/jbc.270.42.25107. [DOI] [PubMed] [Google Scholar]
- 16.Linsley P S, Bradshaw J, Greene J, Peach R, Bennett K L, Mittler R S. Immunity. 1996;4:535–543. doi: 10.1016/s1074-7613(00)80480-x. [DOI] [PubMed] [Google Scholar]
- 17.Alegre M L, Noel P J, Eisfelder B J, Chuang E, Clark M R, Reiner S L, Thompson C B. J Immunol. 1996;157:4762–4770. [PubMed] [Google Scholar]
- 18.Schneider H, Prasad V S, Shoelson S E, Rudd C E. J Exp Med. 1995;181:351–355. doi: 10.1084/jem.181.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marengere L E M, Waterhouse P, Duncan G S, Mittrucker H W, Feng G S, Mak T W. Science. 1996;272:1170–1173. doi: 10.1126/science.272.5265.1170. [DOI] [PubMed] [Google Scholar]
- 20.Robinson M. Trends Cell Biol. 1992;2:293–297. doi: 10.1016/0962-8924(92)90118-7. [DOI] [PubMed] [Google Scholar]
- 21.Trowbridge I S, Collawn J F, Hopkins C R. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 22.Sandoval I V, Bakke O. Trends Cell Biol. 1994;4:292–297. doi: 10.1016/0962-8924(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 23.Ohno H, Fournier M C, Poy G, Bonifacino J S. J Biol Chem. 1996;271:29009–29015. doi: 10.1074/jbc.271.46.29009. [DOI] [PubMed] [Google Scholar]
- 24.Ohno H, Stewart J, Fournier M C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J S. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 25.Boll W, Ohno H, Songyang Z, Rapoport I, Cantley L C, Bonifacino J S, Kirchhausen T. EMBO J. 1996;15:5789–5795. [PMC free article] [PubMed] [Google Scholar]
- 26.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 27.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 28.Gross J A, St. John T, Allison J P. J Immunol. 1990;144:3201–3210. [PubMed] [Google Scholar]
- 29.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 30.Staudinger J, Perry M, Elledge S J, Olson E N. J Biol Chem. 1993;268:4608–4611. [PubMed] [Google Scholar]
- 31.Schneider H, Cai Y C, Prasad K V S, Shoelson S E, Rudd C. Eur J Immunol. 1995;25:1044–1050. doi: 10.1002/eji.1830250428. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y-L, Phillips C, Bjorndahl J M, Trevillyan J M. Eur J Immunol. 1994;24:2732–2739. doi: 10.1002/eji.1830241124. [DOI] [PubMed] [Google Scholar]
- 33.Chambers C A, Allison J P. Eur J Immunol. 1996;26:3224–3229. doi: 10.1002/eji.1830261257. [DOI] [PubMed] [Google Scholar]
- 34.Rudd C E. Immunity. 1996;4:527–534. doi: 10.1016/s1074-7613(00)80479-3. [DOI] [PubMed] [Google Scholar]
- 35.Hutchcroft J E, Bierer B E. J Immunol. 1996;156:4071–4074. [PubMed] [Google Scholar]
- 36.Joly M, Kazlauskas A, Fay F S, Corvera S. Science. 1994;263:684–687. doi: 10.1126/science.8303278. [DOI] [PubMed] [Google Scholar]