Abstract
Background
Comparison of experimentally determined mesophilic and thermophilic homologous protein structures is an important tool for understanding the mechanisms that contribute to thermal stability. Of particular interest are pairs of homologous structures that are structurally very similar, but differ significantly in thermal stability.
Results
We report the X-ray crystal structure of a Thermoanaerobacter tengcongensis ribose binding protein (tteRBP) determined to 1.9 Å resolution. We find that tteRBP is significantly more stable (appTm value ~102°C) than the mesophilic Escherichia coli ribose binding protein (ecRBP) (appTm value ~56°C). The tteRBP has essentially the identical backbone conformation (0.41 Å RMSD of 235/271 Cα positions and 0.65 Å RMSD of 270/271 Cα positions) as ecRBP. Classification of the amino acid substitutions as a function of structure therefore allows the identification of amino acids which potentially contribute to the observed thermal stability of tteRBP in the absence of large structural heterogeneities.
Conclusion
The near identity of backbone structures of this pair of proteins entails that the significant differences in their thermal stabilities are encoded exclusively by the identity of the amino acid side-chains. Furthermore, the degree of sequence divergence is strongly correlated with structure; with a high degree of conservation in the core progressing to increased diversity in the boundary and surface regions. Different factors that may possibly contribute to thermal stability appear to be differentially encoded in each of these regions of the protein. The tteRBP/ecRBP pair therefore offers an opportunity to dissect contributions to thermal stability by side-chains alone in the absence of large structural differences.
Background
The mechanisms that contribute to protein thermal stability are varied, subtle, and complex [1-5]. Various contributing factors to thermal stability have been proposed by comparative analysis of thermophilic and mesophilic proteins [4,6]. Proposed mechanisms can be categorized [5] generally as contributions by the main-chain structure (new folds [7], loop shortening [8]), or by side-chain interactions (increased packing in core [9] or surface [10], alteration of amino acid composition [11-13]), post-translational modifications [14] or co-factor binding [4,15]). Usually increased stability arises from a combination of sequence- and structure-based adaptations resulting in a collection of improvements in the thermophilic protein compared to its mesophilic counterpart [4,6,16,17]. Consequently, the determination of rules for thermal adaptations are difficult to dissect [6]. Of particular interest, therefore, are pairs of naturally evolved proteins that are structurally very similar but differ substantially in thermal stability. Such pairs allow for the dissection of contributions by amino acid diversity to thermal stability in the absence of structural heterogeneity [17-20]. The structure of the Thermoanaerobacter tengcongensis ribose-binding (tteRBP) presented here reveals that this protein and its counterpart in the mesophilic Escherichia coli (ecRBP) form such a pair.
The ribose-binding proteins are members of the periplasmic binding protein (PBP) superfamily whose members play roles in prokaryotic ABC transport [21], chemotaxis [22,23], and intercellular communication [24] systems. The PBP fold consists of two domains each of which adopts a three-layered α/β/α sandwich motif [25]. The two domains are linked by two or three β-strands that form a flexible hinge which permits the domains of the protein to bend towards each other in response to ligand binding at the interface between the two domains [26-28].
Here we report the high-resolution X-ray crystallographic structure of a ribose binding protein (tteRBP) from the hyperthermophilic bacterium T. tengcongensis (optimal growth temp ~80°C) [29]. We find that tteRBP has high sequence and structural similarity to the mesophilic E. coli RBP (ecRBP), although they differ markedly in their thermal stability. The near identity of backbone structure offers an opportunity to address local encoding of thermal stability by amino acid substitutions.
Results and Discussion
Thermal Stability and Ligand Binding
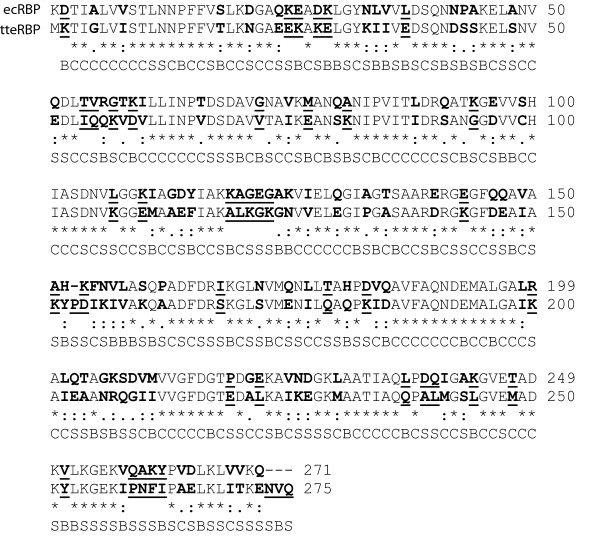
ORF tte0206 in the T. tengcongensis genome sequence [29] was postulated to be a ribose-binding protein homolog (tteRBP) based on its sequence similarity to the known E. coli RBP (57% identity, 76% similarity) (Figure 1) and its position within a putative operon containing ORFs homologous to ABC transporters characteristic of solute transport. The DNA for ORF tte0206, lacking a putative periplasmic signal sequence [30] (residues 1–39), was amplified from T. tengcongensis genomic DNA by the polymerase chain reaction. The resulting DNA fragment was cloned into a pET21a vector in-frame with a C-terminal hexa-histidine tag preceded by a glycine-serine linker. The nucleotide sequence was confirmed by DNA sequencing of the resulting vector. Over-expression of tte0206 produced ~30 mg of pure protein per liter of medium, which was purified by immobilized metal affinity chromatography followed by gel filtration chromatography. tteRBP eluted from the gel filtration column as a broad peak immediately following the void volume of the column (data not shown). For subsequent crystallization and characterization of tteRBP fractions of the broad peak from the gel filtration column, that were consistent with monomeric tteRBP, (fractions with a calculated hydrodynamic radius of 30 kDa ± 15 kDa) were pooled and concentrated to ~15 mg/mL (see Materials and Methods).
Figure 1.
Amino acid sequence comparison of tteRBP and ecRBP. Clustal-W amino acid sequence alignment of tteRBP and ecRBP. Amino acids which are not conserved are in bold type and underlined, amino acids that are conserved but not identical are in bold type (charge inversions are scored as non-conservative here). Core, boundary or surface classification of amino acids is shown below the aligned residues.
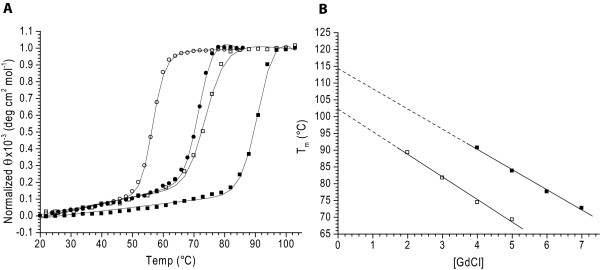
The thermal stability of tteRBP was determined by thermal denaturation using circular dichroism (CD). In the absence of denaturant no significant temperature-dependent change in the CD signal was observed up to 100°C; consequently, heat denaturations were carried out in the presence of varying concentrations of guanidine hydrochloride (GdCl) (Figure 2). Melting curves were found to fit a two-state model [31,32]. The apparent thermal transition midpoint (appTm) of 102°C in the absence of GdCl was determined by linear extrapolation of a series of melting point determinations carried out at different GdCl concentrations [33] (Figure 2). tteRBP is therefore significantly more stable than the mesophilic ecRBP (appTmvalue is 56°C) (Figure 2).
Figure 2.
Thermal denaturation of tteRBP and ecRBP determined by circular dichroism. (A) Thermal denaturation of tteRBP in 4 M GdCl in the absence (open squares) or presence of 1 mM ribose (black squares). Thermal denaturation of ecRBP in the absence (open circle) or presence of 1 mM ribose (black circles). Solid lines in (A) are fit to a two-state model [31, 32] which takes into account the native and denatured baseline slopes. (B) Extrapolated appTm of tteRBP in the absence (open squares) or presence of 1 mM ribose (black squares) obtained from a series of thermal melting curves at different GdCl concentrations. Solid lines represent linear fits to the observations.
Binding of ribose to tteRBP was confirmed by observing ligand-mediated changes in the appTm value in the presence of 4.0 M GdCl. Under these conditions in the absence of ribose, the appTm value is 74°C; and 92°C in the presence of 1 mM ribose (Figure 2). The appTm value of the ribose complex in the absence of GdCl is 114°C; the appTm value for ecRBP under equivalent conditions is 72°C (Figure 2).
Overall Structure of tteRBP
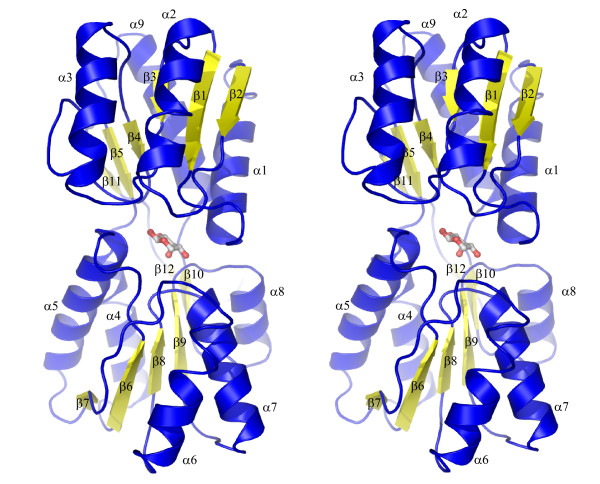
The tteRBP crystal structure was solved to 1.9 Å resolution by molecular replacement [34] using the ribose-bound form of ecRBP as the search model [23]. The tteRBP structure adopts the overall fold and topology that is characteristic of periplasmic ribose-binding proteins (Figure 3). The asymmetric unit contains 346 water molecules and two tteRBP molecules (residues 40–313) in essentially identical conformations (0.12 Å RMSD of backbone atoms) complexed with ribose. Data collection, stereochemistry, and refinement statistics are summarized in Table 1.
Figure 3.
Stereo diagram of the tteRBP structure. Ribose is shown in ball-and-stick representation. The ordering of β-strands (yellow) and α-helices (blue) is indicated.
Table 1.
Data collection and refinement statistics
| tteRBP | |
| Data Collection | |
| Detector Type | Mar225 |
| Wavelength (Å) | 1.0 |
| Resolution (Å) | 15.0–1.9 |
| Measured reflections | 123829 |
| Unique reflections | 16732 |
| Mean I/σ(I)a | 11.6 (4.1) |
| Completeness (%)a | 96.0 (95.1) |
| Rsym (%)a,b | 8.0 (32.4) |
| Redundancya | 3.3 (3.1) |
| Refinement | |
| Resolution (Å) | 15.0–1.9 |
| Num. of Reflections (working set/test set) | 35702/1890 |
| Rcrystc | 19.9 (26.0) |
| Rfreed | 23.4 (30.6) |
| Number of atoms | |
| Protein | 4265 |
| Water | 346 |
| Ligand | 20 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.221 |
| Average B-factor (Å2) | |
| Main Chain | 17.8 |
| Side Chain | 22.7 |
| Solvent | 28.7 |
| Ligand | 10.1 |
| Protein Geometry | |
| Ramachandran outliers (%) | 0.55 |
| Ramachandran favored (%) | 98.7 |
| Rotamer outliers (%) | 0.47 |
aNumber in parentheses represent values in the highest resolution shell.
bRsym = ∑|(I-<I>)|/(∑I), where <I> is the average intensity of multiple measurements.
cRcryst = ∑|Fobs-Fcalc|/(∑|Fobs|)
dRfree is the R-factor based on 5% of the data excluded from refinement.
Structural Diversity of tteRBP and ecRBP
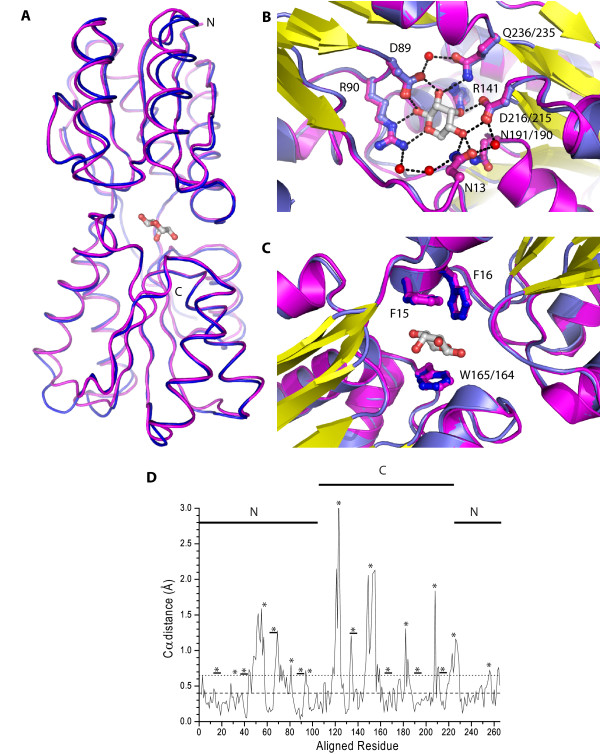
Analysis of main-chain and side-chain geometry of the aligned structures indicates there are few differences in the main-chain geometries of ecRBP and tteRBP (0.4 Å RMSD of 235/271 Cα positions and 0.65 Å RMSD of 270/271 Cα positions and distance between aligned Cα positions range from 0.03–3.1 Å over 270 Cα positions). The loops and turns in the binding pocket retain near-identical conformations. Modest backbone conformational heterogeneity is observed in loops and turns that connect alternating β-strands and α-helices in tteRBP and ecRBP (RMSD of Cα positions for residues 55–61 is 0.9 Å, 117–126 is 1.6 Å and 149–156 is 3.02 Å) (Figure 4). Proline 153 in tteRBP corresponds to a single-residue insertion relative to ecRBP; small structural perturbations associated with this insertion are contained within five amino acids preceding and following this residue (3.1 Å RMSD of Cα positions). tteRBP also contains an additional three amino acids at the C-terminus that are not present in ecRBP.
Figure 4.
Similarity between ecRBP and tteRBP. (A) Backbone atom alignment of tteRBP (blue) and ecRBP (magenta). Loops which have high RMSD are indicated (1/residues 55–61, 2/residues 117–126, 3/residues 149–156). (B) Close-up view of the polar binding pocket residues in tteRBP (blue) and ecRBP (magenta). Ribose is shown in gray. Critical residues involved in ribose binding are indicated (where the tteRBP and ecRBP numbering are different, the former is given first). (C) Close-up view of the non-polar binding pocket amino acids of tteRBP (blue) and ecRBP (magenta). (D) Structural differences in the Cα positions of the aligned models of ecRBP and tteRBP generated by LSQMAN [60]. Dashed and dotted lines indicate the RMSD of 235/271 and 270/271 of the Cα atoms respectively of the aligned structures. The N- and C- terminal residues are indicated with a solid line. Loops and turns are indicated (asterisk), or loops (underlined asterisk) in the binding pocket region.
The amino acid side-chain conformations are also remarkably well conserved (Table 2). Only 24 residues show a significant change in the χ1 torsion, resulting in the adoption of a different side-chain rotamer. Of these, 19 correspond to substitutions, including non-conservative changes; there is therefore a significant bias for non-conservative mutations in the population of residues that exhibit rotameric changes. 17 of the rotamer changes occur in the surface, three in the boundary, and four in the core (Table 2). The non-conservative changes occur mostly on the surface (seven residues). Two of the surface rotamer changes (Q24/E24, D182/D183) involve charged amino acids which results in the formation of two salt bridges, two involve the loss of a salt bridge (K110/E110, K243/L244), one involves the loss of a hydrogen-bond (T178/Q179) and three positions are involved gain of five additional hydrogen-bonds (D52, Q80/S80, R139) relative to ecRBP. The four core changes are conservative substitutions and involve β-branched amino acids, altering packing of the core (V8/I8, I60/V60, T66/V66, V183/I184), and in one case (T66/V66) increasing the hydrophobicity and removing an unsatisfied core hydrogen-bond.
Table 2.
Changes in the χ1 values of ecRBP and tteRBP. Non-conservative substitutions (as defined in [36]; charge inversions are scored as non-conservative here) are underlined.
| ecRBP Residue | tteRBP Residue | ΔX1 | Classification (C/B/S) |
| VAL 8 | ILE 8 | 137 | C |
| GLN 24 | GLU 24 | -97 | S |
| LYS 25 | GLU 25 | -100 | S |
| ASN 33 | LYS 33 | -106 | S |
| ASP 52 | ASP 52 | -283 | S |
| ILE 60 | VAL 60 | -115 | C |
| THR 66 | VAL 66 | -114 | C |
| GLN 80 | SER 80 | 102 | S |
| GLN 91 | SER 91 | -193 | S |
| VAL 98 | VAL 98 | -120 | B |
| LYS 110 | GLU 110 | 244 | B |
| LYS 118 | LYS 118 | 236 | S |
| ARG 139 | ARG 139 | -99 | S |
| GLN 147 | GLU 147 | -240 | S |
| ASN 155 | LYS 156 | -135 | S |
| THR 178 | GLN 179 | -93 | S |
| ASP 182 | LYS 183 | -125 | S |
| VAL 183 | ILE 184 | 125 | C |
| ARG 199 | LYS 200 | 254 | S |
| GLN 202 | GLU 203 | 246 | S |
| SER 207 | GLN 208 | 110 | S |
| GLN 239 | LEU 240 | -246 | S |
| LYS 243 | LEU 244 | -92 | B |
| GLU 246 | GLU 247 | -103 | S |
Polar amino acids, non-polar amino acids, waters, and the hydrogen-bonding interactions are identical in both tteRBP and ecRBP sugar-binding pockets (Figure 4). The total number of hydrogen-bonding interactions [35] is also well conserved among tteRBP and ecRBP (Table 3). Overall, tteRBP has a total of 264 hydrogen-bonds, ecRBP has 257. The hydrogen-bonding pattern outside of the binding pocket varies slightly among tteRBP and ecRBP. tteRBP has an additional three side-chain/main-chain and nine main-chain/main-chain hydrogen-bonds, but has lost five side-chain/side-chain hydrogen-bonds relative to ecRBP (Table 3). Five of the additional seven hydrogen-bonds observed in tteRBP (two main-chain/side-chain, three main-chain/main-chain) are accounted for by the four-residue insertion in tteRBP. There is therefore a net gain of two hydrogen-bonds in tteRBP, which arise from the slight differences in the hydrogen bonding pattern of the side-chain/side-chain and main-chain/main-chain residues. It is also observed that tteRBP has lost two salt bridges relative to ecRBP.
Table 3.
Hydrogen bonding interactions in tteRBP and ecRBP
| Class | tteRBP | ecRBP |
| Side chain/Side chain | 28 | 33 |
| Salt Bridges | 8 | 10 |
| Side chain/Main chain | 44 | 41 |
| Main chain/Main chain | 192 | 183 |
Amino Acid Diversity Among tteRBP and ecRBP
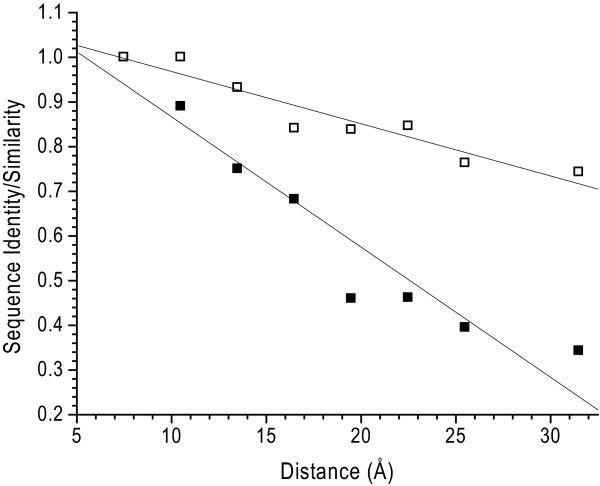
The two RBPs share 57% amino acid identity and 76% similarity (as defined in [36]; in our study charge inversions are scored as non-conservative) (Figure 1). The structures can be divided into core (C), boundary (B), and surface (S) regions using an objective, structure-based classification scheme [37] (Figure 1 and Table 4). The conditional probability of a substitution occurring in a particular region (R) of the protein, p(M|R), is strongly biased (g(M|R) = p(M|R)|p(R)), with g(M|C) = 0.53, g(M|B) = 2.7 and g(M|S) = 1.6 for all interactions and 0.21, 1.36, and 1.74 respectively for non-conservative mutations (g<1, anticorrelated; g = 1, uncorrelated; g>1, positively correlated). The pattern of sequence divergence is also correlated with the distance from the ribose-binding site, as measured by the sequence identity and similarity, in a series of concentric shells centered on the bound ribose (Figure 5). Not surprisingly, the residues in the shell forming the ribose contacts are identical. With increasing distance, there is an approximately monotonic decrease in sequence identity and similarity, with the farthest shell having 73% and 34% similarity and identity respectively (Figure 5).
Table 4.
Divergence patterns in tteRBP and ecRBP. Classification of amino acids into core, boundary, or surface [37] allows identification of the regions which are conserved among tteRBP and ecRBP.
| Mutations | |||
| Region | Residues | All | Non-conservative* |
| Core | 118 | 28 | 5 |
| Boundary | 58 | 33 | 16 |
| Surface | 99 | 58 | 35 |
| Sum | 275 | 119 | 56 |
*As defined in [36]; charge inversions are scored as non-conservative here.
Figure 5.
Structure based sequence comparison of ecRBP and tteRBP. Comparison of sequence identities (black squares) and similarities (open squares) of tteRBP and ecRBP by scoring the number of identical residues in 3 Å concentric shells centered on the mid-point of the bound-ribose. The residues in the last two bins were combined due to the few members in the largest bin. In the primary complementary surface (first shell, 9 Å), the two sequences are identical; at the furthest distance the two sequences are 34% identical and 74% similar. Solid lines are a linear fit of the data to the observations.
Analysis of the amino acid diversity among the core, boundary and surface of ecRBP and tteRBP allows identification of possible determinants of thermal stability in tteRBP (Table 5). A bias is observed for a gain of polar and charged amino acids on the surface of tteRBP (net of twelve charged and three polar substitutions), while the opposite is observed for the tteRBP core, where there is a bias for the loss of polar amino acids (seven net substitutions). There are also a significant number of substitutions of non-β-branched amino acids for β-branched amino acids in the core and boundary of tteRBP (five net substitutions) and a loss of β-branched amino acids in the surface of tteRBP (four net substitutions). Interestingly a large number of β-branched amino acids are conserved in the core and boundary of tteRBP and ecRBP, there is however a bias for the substitution of valine for isoleucine in the thermophile (eight net substitutions).
Table 5.
Amino acid sequence divergence as a function of core, boundary or surface in tteRBP and ecRBP. Differences are classified as substitutions which are found in tteRBP relative to ecRBP.
| Classification | Lose Charge | Lose Polar | Gain Charge | Gain Polar | Non-Branched to Branched | Branched to Non-Branched | V to I | I to V |
| Core | 1 | 7 | 1 | 0 | 5 | 4 | 7 | 2 |
| Boundary | 4 | 3 | 4 | 5 | 6 | 2 | 3 | 0 |
| Surface | 6 | 9 | 18 | 12 | 0 | 4 | 0 | 0 |
Conclusion
We have cloned, expressed, purified, and characterized the structure and stability of the ribose binding protein from the extremophilic bacterium T. tengcongensis. tteRBP is considerably more stable than ecRBP (46°C difference in appTm values of the apo proteins). The amino acid backbone structure of these two proteins are essentially identical (0.41 Å RMSD of 235/271 Cα positions and 0.65 Å RMSD of 270/271 Cα positions), suggesting that all the interactions contributing to differences in thermal stability are encoded entirely in the identity, location, and conformation of the amino acid side-chains.
Comparison of mesophilic and thermophilic protein structures has identified many structural adaptations which are postulated to confer thermal stability [2,6,11,16-18,38]. Numerous side-chain dependent contributions to thermal stability have been proposed, based on amino acid composition of thermophilic proteins and comparison of mesophilic and thermophilic protein sequences and structures, including; increased number of salt-bridges [8], differences in polar/apolar exposed and buried surface areas [8,12,39], introduction of prolines [40], introduction of disulfide bridges [41,42], aromatic interactions [8], helix dipole stabilization [43], post-translational modification [14], alteration of amino acid packing [9,10,44] and secondary structure propensity of amino acids [8,45].
The high structural similarity of the tteRBP/ecRBP pair allows for the dissection of amino acid diversity contributions to thermal stability in the absence of structural heterogeneity. The comparative analysis presented here shows that the substitutions responsible for conferring thermal stability on tteRBP are encoded in side-chain identity and location (core, boundary or surface) which serves to alter surface polarity/charge, removal of unsatisfied core hydrogen bonds and increase in core/boundary side-chain hydrophobicity. In the core of tteRBP there is a bias for the loss of polar amino acids and for the introduction of valine to isoleucine mutations which possibly lower the entropic contribution to the free energy of folding and limits burying core amino acids whose hydrogen bonding potential may remain unsatisfied [38,46]. The large number of valine to isoleucine substitutions in the tteRBP core and boundary leads to an increase in side-chain hydrophobicity and increased packing [44,47]. It is additionally observed in the boundary the substitution of non-β-branched amino acids for β-branched residues which has also been postulated to be important in increasing the packing [48]. Additionally, in a trend that is also observed in other thermophilic proteins, the surface of tteRBP is generally more polar and charged with the introduction of an additional three polar residues and eleven charged residues.
The acquisition of thermal stability in tteRBP arises from contributions by side-chain mediated effects alone. This pair of proteins therefore provides a good test case to examine such contributions experimentally and address some long-standing questions in the acquisition of protein stability [1,5,49]: where in sequence and structure is stability encoded; how many mutations are needed; are mutations punctuated (single mutants cause large changes) or gradual, independent or correlated? Recent advances in protein fabrication automation [50] will assist in addressing these questions by enabling rapid construction of the many sequence variants needed.
Methods
Cloning Over-expression and Purification
The tte0206 gene was amplified from T. tengcongensis genomic DNA by the sticky-end PCR method using the following primers: PO4-TATGA AAACTATAGG ATTAGTGATATCTACTCTTAACAATCC, and TATGAAAACTATAGG ATTAGTGATATCTACTCTTAACAATCC for the 5' end of the gene; PO4- AATTCTAATGGTGATGGTGATGGTGTGATCCCTGTACATTTTCTTTTGTTATGAGTTTAAGTTCTGC, and CTAATGGTGATGGTGATGGTGTGATCCCTGTACATTTTCTTTTGTTATGAGTTTAAGTTCTGC for the 3' end of the gene [51]. The resulting fragment was cloned into the NdeI/EcoRI sites of a pET21a (Novagen) plasmid for over-expression in E. coli. This ORF lacks the putative periplasmic signal sequence [30]. The coding sequence starting at lysine 40 was cloned in-frame with an ATG start codon. A hexahistidine affinity tag and a glycine-serine linker was fused in-frame at the carboxy terminus to facilitate purification by immobilized metal affinity chromatography (IMAC). Protein concentration was determined spectrophotometrically (ε280 = 3800 M-1cm-1) [52]. The resulting gene product was expressed and purified by IMAC as described [33]. Pooled IMAC fractions were concentrated to 12 mL and were loaded onto a Superdex 26/60 S75 (Amersham) gel filtration column that was previously calibrated with blue dextran, bovine serum albumin, chicken serum albumin, chymotrypsin and lysozyme. tteRBP eluted from the column beginning at the void volume and ending at a calculated hydrodynamic radius corresponding to ~20 KDa. For crystallization and characterization, 10 mL fractions corresponding to a calculated hydrodynamic radius corresponding to an apparent molecular weight of 30 KDa ± 15 kDa, were collected and concentrated to 0.5 mM and dialyzed in 10 mM Tris pH7.8, 20 mM NaCl. An average of 30 mg of pure protein produced per liter of medium.
Circular Dichroism
Circular dichroism (CD) measurements were determined on an Aviv Model 202 circular dichroism spectrophotometer. Thermal denaturations were determined by measuring the CD signal at 222 nm (1 cm path length) as a function of temperature, using 1 μM protein (10 mM Tris-HCl pH7.8, 150 mM NaCl), GdCl at various concentrations, in the presence or absence of 1 mM ribose. Protein samples were incubated for 15 minutes prior to collecting data. Each measurement includes a 3-second averaging time for data collection and a 60 second equilibration period at each temperature. Data was fit to a two-state model which accounts for the native and denatured baseline slopes, to determine the apparent Tm values [31,32]. It is not known whether equilibrium was achieved under these conditions; denaturation midpoint temperatures are therefore reported as apparent values (appTm). The appTm values in the absence of denaturant were determined by linear extrapolation [33].
Crystallization and Data Collection
Ribose was added to tteRBP in 3-fold stoichiometric excess prior to crystallization. tteRBP crystals were grown by micro-batch under paraffin oil in drops that contained 2 μl of the protein solution (0.5 mM) mixed with 2 μl of 0.1 M sodium citrate pH 4.0, 50% (w/v) PEG 1000 and 0.1 M potassium phosphate monobasic. The tteRBP crystals diffract to 1.9 Å resolution, belong to the C2 space group (a = 123.18 Å, b = 35.8 Å, c = 118.03 Å, β = 107.02) and typically grew within three weeks at 17°C (Table 1). No stabilizing cryoprotectant was used and crystals were frozen directly in precipitant solution, mounted in a nylon loop and flash frozen in liquid nitrogen. All data were collected at 100 K at the SER-CAT 22 BM beam line at the Advanced Photon Source. The diffraction data were scaled and indexed using SCALA and XDS [53,54].
Structure Determination Methods, Model Building and Refinement
The tteRBP structure was determined by molecular replacement using the ribose-bound form of the ribose binding protein from E. coli [23] as the search model [34]. Rotation, translation, and fitting functions revealed a clear solution yielding higher correlation coefficients and a lower R factor than all the others. Manual model building was carried out in the programs O and COOT and refined using REFMAC5 [55-57]. The final model for the tteRBP complex includes two intact tteRBP monomers (residues 2–275), two ribose molecules, and 346 water molecules. The model exhibits good stereochemistry as determined by PROCHECK and MolProbity; final refinement statistics are listed in Table 1[58,59]. PDB coordinates and structure factors have been deposited in the RCSB Protein Data Bank under the accession code 2IOY.
Authors' contributions
YT constructed the original clone and carried out circular dichroism experiments on purified tteRBP. MJC purified, crystallized and solved the structure of tteRBP, and carried out circular dichroism experiments on ecRBP. MJC, MA and HWH undertook sequence and structural analysis of the tteRBP and ecRBP structures. MJC and HWH wrote the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
This study was funded grant by a grant from HSARPA (W81XWH-05-C-0161) to HWH, a Pioneer Award from the NIH (5 DP1 OD000122-02) to HWH, and a NIH sponsored Biological Chemistry training grant to MJC. The authors would like to acknowledge G. Shirman for protein expression and purification. Data were collected at the Southeast Regional Collaborative Access Team 22-BM at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Contributor Information
Matthew J Cuneo, Email: mjc18@duke.edu.
Yaji Tian, Email: yt11@duke.edu.
Malin Allert, Email: allert@biochem.dueke.du.
Homme W Hellinga, Email: hwh@biochem.duke.edu.
References
- Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tsai CJ, Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–191. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- Jaenicke R, Bohm G. The stability of proteins in extreme environments. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/S0959-440X(98)80094-8. [DOI] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovsky IN, Shakhnovich EI. Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci U S A. 2005;102:12742–12747. doi: 10.1073/pnas.0503890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilagyi A, Zavodszky P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure. 2000;8:493–504. doi: 10.1016/S0969-2126(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC. A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. Embo J. 1996;15:2323–2330. [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S, Varadarajan R. Elucidation of factors responsible for enhanced thermal stability of proteins: a structural genomics based study. Biochemistry. 2002;41:8152–8161. doi: 10.1021/bi025523t. [DOI] [PubMed] [Google Scholar]
- Goldstein RA. Amino-acid interactions in psychrophiles, mesophiles, thermophiles, and hyperthermophiles: insights from the quasi-chemical approximation. Protein Sci. 2007;16:1887–1895. doi: 10.1110/ps.072947007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glyakina AV, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. Different packing of external residues can explain differences in the thermostability of proteins from thermophilic and mesophilic organisms. Bioinformatics. 2007;23:2231–2238. doi: 10.1093/bioinformatics/btm345. [DOI] [PubMed] [Google Scholar]
- Liang HK, Huang CM, Ko MT, Hwang JK. Amino acid coupling patterns in thermophilic proteins. Proteins. 2005;59:58–63. doi: 10.1002/prot.20386. [DOI] [PubMed] [Google Scholar]
- Fukuchi S, Nishikawa K. Protein surface amino acid compositions distinctively differ between thermophilic and mesophilic bacteria. J Mol Biol. 2001;309:835–843. doi: 10.1006/jmbi.2001.4718. [DOI] [PubMed] [Google Scholar]
- Bohm G, Jaenicke R. Relevance of sequence statistics for the properties of extremophilic proteins. Int J Pept Protein Res. 1994;43:97–106. doi: 10.1111/j.1399-3011.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- Jafari-Aghdam J, Khajeh K, Ranjbar B, Nemat-Gorgani M. Deglycosylation of glucoamylase from Aspergillus niger: effects on structure, activity and stability. Biochim Biophys Acta. 2005;1750:61–68. doi: 10.1016/j.bbapap.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Fujii T, Hata Y, Oozeki M, Moriyama H, Wakagi T, Tanaka N, Oshima T. The crystal structure of zinc-containing ferredoxin from the thermoacidophilic archaeon Sulfolobus sp. strain 7. Biochemistry. 1997;36:1505–1513. doi: 10.1021/bi961966j. [DOI] [PubMed] [Google Scholar]
- Yano JK, Poulos TL. New understandings of thermostable and peizostable enzymes. Curr Opin Biotechnol. 2003;14:360–365. doi: 10.1016/S0958-1669(03)00075-2. [DOI] [PubMed] [Google Scholar]
- Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes D, Zeelen JP, Thanki N, Beaucamp N, Alvarez M, Thi MH, Backmann J, Martial JA, Wyns L, Jaenicke R, Wierenga RK. The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparative thermostability structural analysis of ten different TIM structures. Proteins. 1999;37:441–453. doi: 10.1002/(SICI)1097-0134(19991115)37:3<441::AID-PROT11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Kannan N, Vishveshwara S. Aromatic clusters: a determinant of thermal stability of thermophilic proteins. Protein Eng. 2000;13:753–761. doi: 10.1093/protein/13.11.753. [DOI] [PubMed] [Google Scholar]
- Greaves RB, Warwicker J. Mechanisms for stabilisation and the maintenance of solubility in proteins from thermophiles. BMC Struct Biol. 2007;7:18. doi: 10.1186/1472-6807-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Shuman HA, Nikaido H. Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci U S A. 1992;89:2360–2364. doi: 10.1073/pnas.89.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman AJ, Binnie RA, Zhang H, Cole LB, Hermodson MA, Mowbray SL. Probing protein-protein interactions. The ribose-binding protein in bacterial transport and chemotaxis. J Biol Chem. 1994;269:30206–30211. [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- Bjorkman AJ, Mowbray SL. Multiple open forms of ribose-binding protein trace the path of its conformational change. J Mol Biol. 1998;279:651–664. doi: 10.1006/jmbi.1998.1785. [DOI] [PubMed] [Google Scholar]
- Magnusson U, Chaudhuri BN, Ko J, Park C, Jones TA, Mowbray SL. Hinge-bending motion of D-allose-binding protein from Escherichia coli: three open conformations. J Biol Chem. 2002;277:14077–14084. doi: 10.1074/jbc.M200514200. [DOI] [PubMed] [Google Scholar]
- Sharff AJ, Rodseth LE, Spurlino JC, Quiocho FA. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- Bao Q, Tian Y, Li W, Xu Z, Xuan Z, Hu S, Dong W, Yang J, Chen Y, Xue Y, Xu Y, Lai X, Huang L, Dong X, Ma Y, Ling L, Tan H, Chen R, Wang J, Yu J, Yang H. A complete sequence of the T. tengcongensis genome. Genome Res. 2002;12:689–700. doi: 10.1101/gr.219302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/S0968-0004(98)01336-X. [DOI] [PubMed] [Google Scholar]
- Schellman JA. The thermodynamic stability of proteins. Annu Rev Biophys Biophys Chem. 1987;16:115–137. doi: 10.1146/annurev.bb.16.060187.000555. [DOI] [PubMed] [Google Scholar]
- Cohen DS, Pielak GJ. Stability of yeast iso-1-ferricytochrome c as a function of pH and temperature. Protein Sci. 1994;3:1253–1260. doi: 10.1002/pro.5560030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuneo MJ, Changela A, Warren JJ, Beese LS, Hellinga HW. The crystal structure of a thermophilic glucose binding protein reveals adaptations that interconvert mono and di-saccharide binding sites. J Mol Biol. 2006;362:259–270. doi: 10.1016/j.jmb.2006.06.084. [DOI] [PubMed] [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst. 1994;A50:157–163. [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisz MS, Hellinga HW. An empirical model for electrostatic interactions in proteins incorporating multiple geometry-dependent dielectric constants. Proteins. 2003;51:360–377. doi: 10.1002/prot.10332. [DOI] [PubMed] [Google Scholar]
- Weers PM, Abdullahi WE, Cabrera JM, Hsu TC. Role of buried polar residues in helix bundle stability and lipid binding of apolipophorin III: destabilization by threonine 31. Biochemistry. 2005;44:8810–8816. doi: 10.1021/bi050502v. [DOI] [PubMed] [Google Scholar]
- Criswell AR, Bae E, Stec B, Konisky J, Phillips GN., Jr. Structures of thermophilic and mesophilic adenylate kinases from the genus Methanococcus. J Mol Biol. 2003;330:1087–1099. doi: 10.1016/S0022-2836(03)00655-7. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Masuda T, Ohashi H, Mihara H, Suzuki Y. Multiple proline substitutions cumulatively thermostabilize Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Irrefragable proof supporting the proline rule. Eur J Biochem. 1994;226:277–283. doi: 10.1111/j.1432-1033.1994.tb20051.x. [DOI] [PubMed] [Google Scholar]
- Bian Y, Liang X, Fang N, Tang XF, Tang B, Shen P, Peng Z. The roles of surface loop insertions and disulfide bond in the stabilization of thermophilic WF146 protease. FEBS Lett. 2006;580:6007–6014. doi: 10.1016/j.febslet.2006.09.068. [DOI] [PubMed] [Google Scholar]
- Takagi H, Takahashi T, Momose H, Inouye M, Maeda Y, Matsuzawa H, Ohta T. Enhancement of the thermostability of subtilisin E by introduction of a disulfide bond engineered on the basis of structural comparison with a thermophilic serine protease. J Biol Chem. 1990;265:6874–6878. [PubMed] [Google Scholar]
- Hennig M, Darimont B, Sterner R, Kirschner K, Jansonius JN. 2.0 A structure of indole-3-glycerol phosphate synthase from the hyperthermophile Sulfolobus solfataricus: possible determinants of protein stability. Structure. 1995;3:1295–1306. doi: 10.1016/S0969-2126(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Britton KL, Baker PJ, Borges KM, Engel PC, Pasquo A, Rice DW, Robb FT, Scandurra R, Stillman TJ, Yip KS. Insights into thermal stability from a comparison of the glutamate dehydrogenases from Pyrococcus furiosus and Thermococcus litoralis. Eur J Biochem. 1995;229:688–695. doi: 10.1111/j.1432-1033.1995.tb20515.x. [DOI] [PubMed] [Google Scholar]
- Corazza A, Rosano C, Pagano K, Alverdi V, Esposito G, Capanni C, Bemporad F, Plakoutsi G, Stefani M, Chiti F, Zuccotti S, Bolognesi M, Viglino P. Structure, conformational stability, and enzymatic properties of acylphosphatase from the hyperthermophile Sulfolobus solfataricus. Proteins. 2006;62:64–79. doi: 10.1002/prot.20703. [DOI] [PubMed] [Google Scholar]
- Blaber M, Lindstrom JD, Gassner N, Xu J, Heinz DW, Matthews BW. Energetic cost and structural consequences of burying a hydroxyl group within the core of a protein determined from Ala-->Ser and Val-->Thr substitutions in T4 lysozyme. Biochemistry. 1993;32:11363–11373. doi: 10.1021/bi00093a013. [DOI] [PubMed] [Google Scholar]
- Zhu BY, Zhou NE, Kay CM, Hodges RS. Packing and hydrophobicity effects on protein folding and stability: effects of beta-branched amino acids, valine and isoleucine, on the formation and stability of two-stranded alpha-helical coiled coils/leucine zippers. Protein Sci. 1993;2:383–394. doi: 10.1002/pro.5560020310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha MM, Oobatake M, Sarai A. Important amino acid properties for enhanced thermostability from mesophilic to thermophilic proteins. Biophys Chem. 1999;82:51–67. doi: 10.1016/S0301-4622(99)00103-9. [DOI] [PubMed] [Google Scholar]
- Querol E, Perez-Pons JA, Mozo-Villarias A. Analysis of protein conformational characteristics related to thermostability. Protein Eng. 1996;9:265–271. doi: 10.1093/protein/9.3.265. [DOI] [PubMed] [Google Scholar]
- Cox JC, Lape J, Sayed MA, Hellinga HW. Protein fabrication automation. Protein Sci. 2007;16:379–390. doi: 10.1110/ps.062591607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G. Sticky-end PCR: new method for subcloning. Biotechniques. 1998;25:206–208. doi: 10.2144/98252bm05. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. doi: 10.1107/S0021889893005588. [DOI] [Google Scholar]
- Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 ( Pt 2):110–119. doi: 10.1107/S0108767390010224. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Laskowski RA., MacArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]