Abstract
This study provides insights into the role of nuclear lamins in DNA replication. Our data demonstrate that the Ig-fold motif located in the lamin C terminus binds directly to proliferating cell nuclear antigen (PCNA), the processivity factor necessary for the chain elongation phase of DNA replication. We find that the introduction of a mutation in the Ig-fold, which alters its structure and causes human muscular dystrophy, inhibits PCNA binding. Studies of nuclear assembly and DNA replication show that lamins, PCNA, and chromatin are closely associated in situ. Exposure of replicating nuclei to an excess of the lamin domain containing the Ig-fold inhibits DNA replication in a concentration-dependent fashion. This inhibitory effect is significantly diminished in nuclei exposed to the same domain bearing the Ig-fold mutation. Using the crystal structures of the lamin Ig-fold and PCNA, molecular docking simulations suggest probable interaction sites. These findings also provide insights into the mechanisms underlying the numerous disease-causing mutations located within the lamin Ig-fold.
Introduction
Nuclear lamins are type V intermediate filament (IF) proteins. Like all IF proteins, lamins have a tripartite structure consisting of non–α-helical N-terminal head and C-terminal tail domains flanking an α-helical central rod domain (Goldman et al., 2002; Gruenbaum et al., 2005). However, unique among IF proteins, lamins contain a C-terminal tail motif, the Ig-fold, known to be involved in protein–protein interactions (Dechat et al., 2000; Zastrow et al., 2006). Although present throughout the nucleus, the nuclear lamins are concentrated in the lamina located at the nucleoplasmic surface of the inner nuclear envelope membrane (Shumaker et al., 2003; Gruenbaum et al., 2005). The lamins play important roles in determining the shape, size, and mechanical properties of the nucleus (Goldman et al., 2004). They are also involved in the disassembly (Dessev et al., 1989) and assembly (Nigg, 1992) of the nucleus during cell division, the assembly of the mitotic spindle matrix (Tsai et al., 2006), transcription, and DNA replication (Spann et al., 1997, 2002; Moir et al., 2000a).
Evidence supporting a role for lamins in DNA replication comes from the finding that lamin B1 (LB1) colocalizes with proliferating cell nuclear antigen (PCNA) at sites of DNA synthesis in late S phase (Moir et al., 1994), and that lamin A (LA)/lamin C (LC) is associated with replication sites in early S phase (Kennedy et al., 2000; Johnson et al., 2004). Furthermore, fibroblasts derived from LA knockout mouse embryos replicate their DNA at a slower rate relative to controls as assayed by nucleotide (BrdU) incorporation. When these embryonic fibroblasts are transiently transfected to express GFP-LA, the cells incorporate BrdU at wild-type levels (Johnson et al., 2004).
Biochemical evidence in support of a role for nuclear lamins in DNA replication has been derived from nuclei assembled in Xenopus laevis egg interphase extracts (Blow and Laskey, 1986). DNA replication is initiated in these extracts ∼40 min after the introduction of demembranated sperm chromatin, the time when nuclear envelope assembly is completed (Newport et al., 1990; Goldberg et al., 1995). Importantly, the assembly of nuclei from sperm chromatin requires polymerization of lamin B3 (LB3), the major lamin present in X. laevis eggs (Lourim et al., 1996). This polymerization process involves the head-to-tail interaction of lamin dimers that form the higher order structures located in the lamina and nucleoplasm (Heitlinger et al., 1991). Inhibition of LB3 polymerization by an excess of its non–α-helical C terminus (LB3T) prevents both nuclear envelope assembly and DNA replication (Lopez-Soler et al., 2001). Furthermore, deletion analyses of LB3T have demonstrated that the Ig-fold motif within LB3T (LB3T-Ig) is sufficient to block both lamin polymerization and nuclear envelope assembly in these extracts (Shumaker et al., 2005). These results suggest that lamin polymerization, nuclear assembly, and DNA replication are linked in both a temporal and spatial fashion.
Treatment of replicating X. laevis nuclei with either of the dominant-negative mutants, N-terminally deleted human LA or X. laevis LB3, induces a redistribution of LB3 from the lamina into nucleoplasmic foci. Under these conditions, >90% of DNA synthesis is blocked exclusively at the chain elongation phase of replication (Spann et al., 1997; Moir et al., 2000a). Interestingly, these LB3 foci contain the replication elongation factors PCNA and replication factor C (RFC), which are displaced from their normal association with chromatin (Moir et al., 2000a). In contrast, DNA replication initiation factors such as MCM3 and ORC2 remain associated with chromatin, and DNA polymerase α appears to function normally (Spann et al., 1997; Moir et al., 2000a). These results suggest that there may be a specific interaction between PCNA and/or RFC and the nuclear lamins.
PCNA increases the processivity of DNA polymerase δ during the chain elongation phase of DNA replication by >100-fold. In the absence of PCNA, DNA polymerase δ is readily released from DNA and has a reduced affinity for nucleotides during replication (Lee and Hurwitz, 1990). PCNA forms a homotrimeric (Kong et al., 1992) sliding clamp ring structure, which is loaded onto DNA through its interactions with the AAA+ pentameric clamp loading protein RFC (Yao et al., 2003). The ATP-dependent release of RFC from PCNA on DNA allows DNA polymerase δ to associate with the same face of PCNA formerly occupied by RFC. This face contains the interdomain connecting loop, a sequence of ∼12 amino acids (L121–E132) that is the main binding site for proteins that have been shown to interact with PCNA (Tsurimoto, 1999). Two other regions on PCNA are also involved in protein–protein interactions: the central loop (D41–H44) and the C-terminal tail (K254–E256; Tsurimoto, 1999). These sites are situated close to each other and are localized to the same face of PCNA. All known PCNA binding partners have the conserved binding motif, the PCNA-interacting protein (PIP) box (QXX[h]XX[a][a]; where X is any amino acid, [h] is a residue with a moderately hydrophobic sidechain, and [a] is a residue with an aromatic sidechain; Warbrick et al., 1998).
In this study, the role of lamins in DNA replication is more precisely defined. The results demonstrate that the C-terminal non–α-helical domains of all lamins bind directly to PCNA. The binding site lies within the Ig-fold and an excess of either the C-terminal domain containing this motif or the Ig-fold alone inhibits DNA synthesis. Studies of the steps in the assembly of X. laevis nuclei suggest that lamins are involved in the proper positioning of PCNA on chromatin during the early stages of nuclear assembly. Using their known crystal structures, the most likely binding sites between the Ig-fold and PCNA have been determined by molecular docking simulations. Overall, these results reveal that lamins are essential for the steps in assembly and function of the complex required for the chain elongation phase of DNA replication.
Results
The lamin C-terminal domain containing the Ig-fold inhibits DNA replication
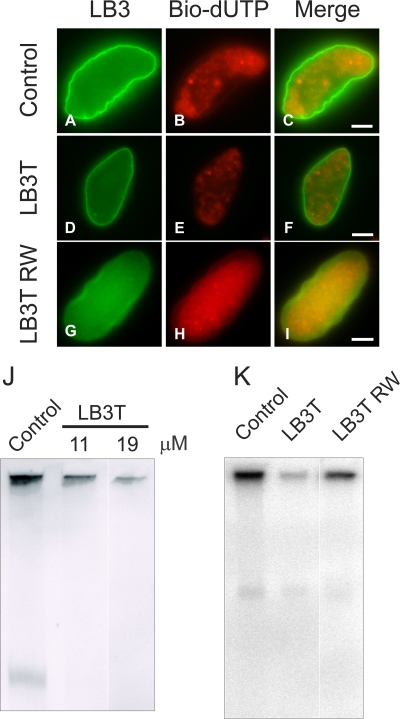
We reasoned that the non–α-helical C-terminal domain of nuclear lamins contained the binding site for PCNA, as previous studies showed that N-terminal–deleted LA and X. laevis LB3 retained their association with PCNA in situ (Spann et al., 1997; Moir et al., 2000a) and that the binding sites for several known lamin-associated proteins are located in this domain (Zastrow et al., 2004). To test this possibility, we added excess LB3T to X. laevis nuclei engaged in DNA replication. Nuclei were assembled for 60 min and LB3T was added for an additional 30 min. Subsequently, biotin-11-2′-deoxyuridine-5′-triphosphate (bio-11-dUTP) was added for 10 min and the nuclei were processed for immunofluorescence. Nuclei treated with ∼19 μM LB3T showed a reduction in nucleotide incorporation as measured by the fluorescence intensity of Texas red streptavidin (Fig. 1, D–F) relative to controls (Fig. 1, A–C). When nuclei were assembled as above but exposed to bio-11-dUTP for 30 min instead of 10 min, there appeared to be a recovery of DNA replication (unpublished data). This recovery of replication is likely caused by the excess of PCNA and LB3 in the X. laevis extract. Nuclei incubated with LB3T containing the mutation R454W (LB3T RW) displayed a bio-11-dUTP pattern similar to controls (Fig. 1, G–I). This mutation in human LA causes Emery-Dreifuss muscular dystrophy (EDMD; Bonne et al., 1999).
Figure 1.
Addition of LB3T to assembled X. laevis nuclei inhibits DNA replication. Nuclei were assembled for 60 min followed by the addition of control buffer (A–C), ∼19 μM LB3T (D–F), or ∼19 μM LB3TRW (G–I) for 30 min followed by 5 μM bio-11-dUTP for 10 min. Nuclei were fixed, pelleted onto poly-l-lysine–coated coverslips, and prepared for immunofluorescence using Hoechst 33342, rhodamine avidin, and anti-LB3. Control nuclei continued to grow after addition of the buffer (A–C) and incorporated bio-11-dUTP (B). Using the same image capture settings, there was less incorporation of bio-11-dUTP after the addition of LB3T (E), and the nuclei appeared to be the same size as those in buffer controls at 60 min. In contrast, LB3T RW had a lesser effect on incorporation of bio-11-dUTP (H). Bars, 5 μm. (J) X. laevis nuclei were assembled for 60 min and different amounts of LB3T were added for 30 min followed by a 10-min incubation in 2 μC [32P]α-dCTP. The reactions were stopped by the addition of replication sample buffer for 10 min. The samples were run on 0.8% agarose gels and dried under vacuum, and radioactivity was measured. Under these conditions, there was a concentration-dependent inhibition of DNA replication attributable to LB3T. When ∼11 μM LB3T was added, there was an ∼50% decrease, and with ∼19 μM LB3T, replication decreased by ∼92%. (K) We also tested the effect of LB3T RW and determined that at a concentration of ∼19 μM, DNA replication was reduced by ∼50%. Each experiment was repeated three times and the results were averaged.
Using a more quantitative assay, we determined the effect of LB3T on DNA synthesis by pulse labeling with [32P]α-dCTP for 10 min under identical conditions (see the previous paragraph). The addition of 11 μM LB3T decreased DNA replication by ∼50 ± 1.0% (n = 3), whereas ∼19 μM LB3T caused a decrease of ∼92 ± 0.5% (n = 3; Fig. 1 J). The LB3T RW mutant was less efficient in inhibiting DNA replication in X. laevis nuclei showing a reduction of ∼50 ± 8.6% relative to controls at a concentration of ∼19 μM (Fig. 1 K).
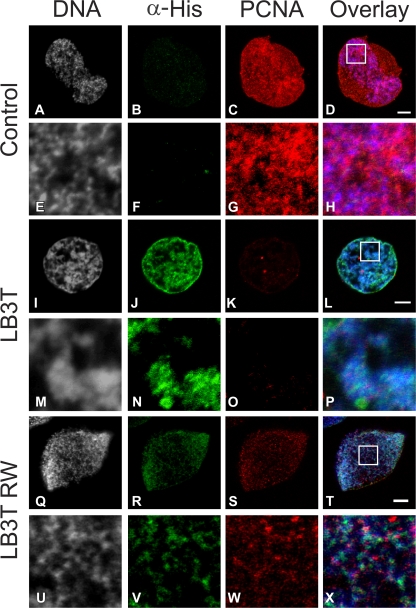
The localization of LB3T and its effect on the normal association of PCNA with chromatin was also examined in X. laevis nuclei 60 min after assembly was initiated. After the addition of His-tagged LB3T for 30 min, the nuclei were prepared for immunofluorescence with antibodies directed against PCNA and His. In controls, most of the PCNA was closely associated with chromatin, which is typical for X. laevis nuclei (Fig. 2, A–H; Spann et al., 1997). When LB3T was added to identical preparations of nuclei, it was incorporated into the lamina region and became associated with chromatin (Fig. 2, I, J, L–N, and P). In these nuclei, there was a substantial decrease in the amount of PCNA associated with chromatin compared with controls (compare Fig. 2, K, L, O, and P with Fig. 2, C, D, G, and H). Overall, the anti-His fluorescence intensity in nuclei treated with LB3T RW appeared to be reduced in comparison with nuclei treated with LB3T (Fig. 2, J, N, R, and V). Furthermore, the nuclei exposed to LB3T RW appeared to have more PCNA associated with chromatin than nuclei treated with LB3T (Fig. 2, K, O, S, and W). These findings suggest that an excess of the lamin C-terminal domain inhibits PCNA binding to chromatin.
Figure 2.
LB3T inhibits the association of PCNA with chromatin. X. laevis nuclei were assembled for 60 min before the addition either in the absence (A–H) or presence (I–P) of ∼19 μM His-tagged LB3T for 30 min. Nuclei were prepared for immunofluorescence using antibodies directed against His6 and PCNA and stained with Hoechst 33342. Control nuclei displayed only background His staining (B, D, F, and H) and a significant amount of PCNA mainly associated with chromatin (C, D, G, and H). In nuclei exposed to LB3T, the His-tagged protein was associated with chromatin (I, J, L–N, and P) and there was a substantial decrease in the amount of PCNA staining (K, L, O, and P). LB3T RW was also associated with chromatin but to a lesser extent than LB3T (Q, R, T–V, and X). Nuclei treated in this manner also had a reduced amount of PCNA associated with chromatin (S, T, W, and X). E–H, M–P, and U–X are 5× views of the boxes in D, L, and T. Bars, 5 μm.
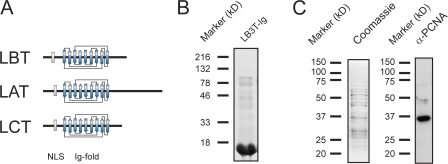
Clues regarding the likely lamin binding site for PCNA come from comparing the lamin C-terminal domains (Fig. 3 A). The lamins contain two highly conserved motifs: a classical nuclear localization sequence (NLS; e.g., residues 413–416 in X. laevis LB3, residues 417–420 in human LA/C, and residues 415–418 in human LB1) and the Ig-fold (residues 437–545 in X. laevis LB3, residues 436–544 in human LA/C, and residues 438–545 in human LB1). Because the Ig-fold motif of all lamins is highly conserved and known to be involved in protein–protein interactions (Dhe-Paganon et al., 2002), we reasoned that this domain might be involved in the binding of lamins to PCNA (Fig. 3 A). To test this possibility, X. laevis egg extracts were used to determine whether X. laevis LB3 Ig-fold (LB3T-Ig) could bind PCNA. Bacterially expressed and purified His-tagged LB3T-Ig (Fig. 3 B) was bound to an Ni-agarose column, and X. laevis high-speed ultracentrifuged supernatant (uS) was passed over the column for 18 h at 4°C. Bound proteins were eluted with a linear gradient from 0.1 to 1.5 M NaCl. PCNA eluted with a peak at 600 mM NaCl and is shown by SDS-PAGE. This peak fraction contains a band of the appropriate size (37 kD) as detected by Coomassie staining. The presence of PCNA was confirmed by immunoblotting with anti-PCNA (Fig. 3 C). These results suggest that the Ig-fold motif could either directly bind to PCNA or may bind to a component of a complex containing PCNA.
Figure 3.
X. laevis LB3T-Ig binds to PCNA in egg extracts. (A) Diagrammatic comparisons of the tail domains of LB3, LA, and LC. Note that the major conserved region among all three tail domains is the Ig-fold motif, which is ∼80% similar between human LA/C and X. laevis LB3. (B) His-tagged LB3T-Ig was bacterially expressed and purified. The purified protein was separated by SDS-PAGE and stained with Coomassie blue. (C) Purified His-tagged LB3T-Ig was bound to a Hi-Trap chelating column, X. laevis high-speed supernatant was passed over the column, and bound proteins were eluted. The fraction shown was eluted with 600 mM NaCl. The eluted proteins were separated by SDS-PAGE and either stained with Coomassie blue or transferred to nitrocellulose and probed with an antibody directed against PCNA. A band of ∼36 kD seen with Coomassie also reacts with the PCNA antibody.
PCNA binds directly to the C-terminal Ig-fold motif of nuclear lamins
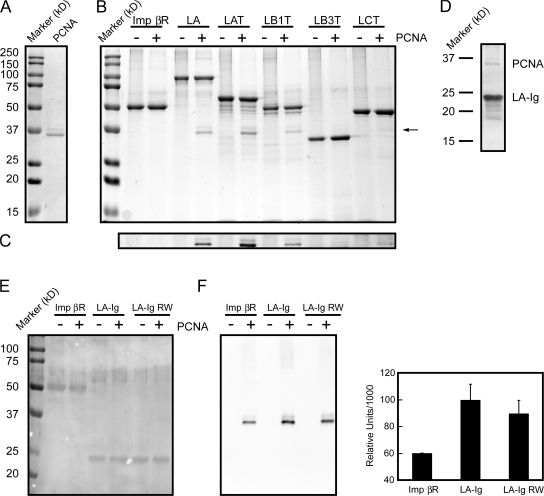
It is obvious from Fig. 3 C that the binding of PCNA to the Ig-fold motif could involve other proteins, as multiple bands are seen on the Coomassie-stained gel. To determine whether the Ig-fold could directly bind to PCNA, we performed a solution binding assay. Purified recombinant lamin C-terminal domains were attached to protein S–agarose beads and incubated with purified recombinant human PCNA (Fig. 4, A and B). The bound proteins were subjected to SDS-PAGE and the gels were stained with Coomassie blue (Fig. 4, A and B). Full-length LA was used as a positive control and the Ran-binding domain of importin β (Imp βR) was used as the negative control for the solution binding assay. The results showed that full-length LA binds directly to PCNA (Fig. 4 B, LA − and +; note the additional band in the + lane indicated by the arrow). Furthermore, the tail domains of LA (Fig. 4 B, LAT − and +), LCT (LCT − and +), LB1 (LB1T − and +), and X. laevis LB3 (LB3T − and +) also bind to PCNA (Fig. 4 B). The low level of human PCNA binding to X. laevis LB3T in this solution binding assay may be caused by species differences. Immunoblotting using the PCNA antibody demonstrates that the light band (Fig. 4, B and C) is PCNA (not depicted). The S-tagged Imp βR did not appear to interact with PCNA under these conditions (Fig. 4 B). We also determined whether the Ig-fold of LA/C (LA-Ig) was sufficient to bind to PCNA. Using the solution binding assay, an interaction between LA-Ig and PCNA could be detected, but this binding was weaker than the interaction between LCT and PCNA. However, the PCNA was readily detected after transfer to nitrocellulose and staining with India ink (Fig. 4 D).
Figure 4.
The lamin Ig-fold motif binds PCNA. (A) Bacterially expressed and purified PCNA was separated by SDS-PAGE and stained with Coomassie blue (PCNA, molecular weight markers on the left). We always observe that the expressed PCNA appeared as one major and one minor band, both of which immunoblotted with anti-PCNA (see E). (B) LA, lamin A tail (LAT), LB1 tail (LB1T), LC tail (LCT), X. laevis LB3 tail (LB3T), and the Ran-binding domain of importin β (Imp βR) were expressed, purified, bound to protein S–agarose beads, and then mixed with purified PCNA. + lanes show experiments where PCNA was added to S-tagged proteins bound to protein S–agarose beads, whereas − lanes show omitted PCNA. The proteins bound to the S–agarose beads were eluted in SDS sample buffer and separated by SDS-PAGE. Gels were stained with Coomassie blue. The results show that LA, LAT, LB1T, LCT, and LB3T bind to PCNA (arrow). (C) The area identified by the arrow (∼37 kD) in Fig. 4 B was enhanced to more easily identify PCNA bound to LB3T. (D) S-tagged LA-Ig was bound to protein S–agarose beads and incubated with PCNA. The bound proteins were separated by SDS-PAGE, transferred to nitrocellulose, and stained with India ink. The location of LA-Ig and PCNA are shown. (E) After immunoblotting, the nitrocellulose blot seen in F (left) was briefly stained with India ink to show that equal amounts of Imp βR (∼50 kD), LA-Ig, and LA Ig-fold R453W (LA-Ig RW; ∼25 kD) were bound to protein S–agarose beads. (F, left) Purified Imp βR, the LA-Ig, and LA-Ig RW were bound to protein S–agarose beads and mixed with purified PCNA. The bound proteins were eluted in SDS sample buffer and separated by SDS-PAGE. The separated proteins were transferred to nitrocellulose and immunoblotted with antibodies directed against PCNA. S-tagged Imp βR was used as a negative control. It was found that the amounts of PCNA bound both to protein S–agarose beads alone (not depicted) and to Imp βR bound to protein S–agarose beads were equivalent (Imp βR +). Therefore, this amount of PCNA binding was considered background. Densitometric analysis showed that after subtracting this background, PCNA binding to LA-Ig RW bound ∼28% less PCNA than wild-type LA-Ig. (F, right) Densitometric analysis of the bound PCNA by immunoblotting seen in F (left). Error bars indicate standard deviation of the mean for six experiments.
As described above (Fig. 1), the EDMD mutation in LB3T (LB3T RW) was less effective in inhibiting DNA replication. This could be attributable to a reduction in the binding of PCNA. Therefore, we determined whether the EDMD mutation (R453W) in the LA Ig-fold (LA-Ig RW; Bonne et al., 1999) had an effect on PCNA binding relative to LA-Ig (Fig. 4, E and F). To quantify the difference in binding, the interactions were assayed by immunoblotting with the Imp βR used as a negative control. It was determined that some PCNA bound to the protein S–agarose beads alone (not depicted) and in equivalent amounts to beads containing S-tagged Imp βR (Fig. 4 F). Under these conditions, PCNA binding to LA-Ig RW was reduced by ∼28% (n = 5) of the amount bound to wild-type LA-Ig (n = 6) as determined densitometrically (Fig. 4 F, right). To make certain that equivalent amounts of Imp βR, LA-Ig, and LA-Ig RW were transferred to nitrocellulose, the proteins were stained with India ink after immunoblotting (Fig. 4 E). This assay demonstrates that LA-Ig binds to PCNA in solution. This interaction is decreased by an EDMD mutation known to destabilize the Ig-fold, which suggests that its structure is important in binding to PCNA.
Because we have shown that the human lamin Ig-fold binds to PCNA, it was of interest to determine the effects of an excess of this domain on DNA replication in human cells. Therefore, HeLa cells were transfected to express a GFP-tagged lamin Ig-fold (GFP-NLS-Ig). After 24 h, BrdU was added to the culture medium for 30 min and the cells were fixed and processed for immunofluorescence. Transfected cells were identified microscopically by GFP fluorescence, which was detected exclusively in the nucleus. Approximately 36% of the cells expressing GFP-NLS-Ig incorporated BrdU (n = 525) compared with ∼51% of those transfected with GFP-LA (n = 352). We observed that the nuclei showing the brightest GFP fluorescence did not incorporate BrdU (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200708155/DC1). These experiments demonstrate that the expression of the lamin Ig-fold domain reduces DNA replication by ∼29% (P = 0.0021) and extend our findings in X. laevis nuclei to mammalian cells.
Lamins are associated with chromatin before PCNA during nuclear assembly
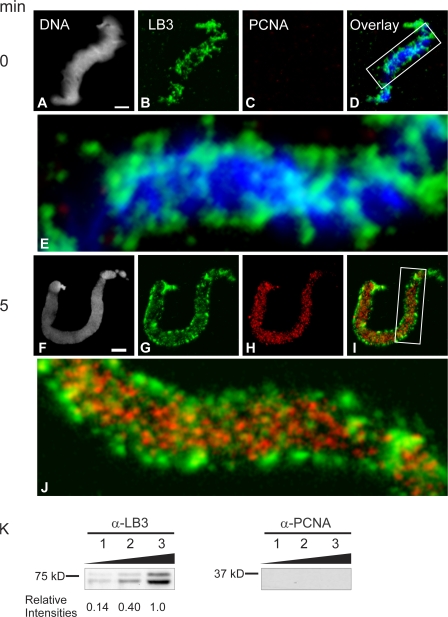
To gain further insights into the spatial and temporal interactions between lamins and PCNA, we examined their organizational states during nuclear assembly and the initiation of replication in X. laevis interphase extracts. Demembranated X. laevis sperm chromatin contains LB3 but not PCNA (Fig. 5, A–D). The LB3 present on sperm head chromatin frequently appears as a filamentous structure at the surface of the condensed chromatin (Fig. 5, B, D, and E). The presence of LB3 and the absence of PCNA in these preparations was confirmed by immunoblotting analyses of different numbers of sperm heads (50,000–200,000; Fig. 5 K).
Figure 5.
LB3 is present on demembranated sperm chromatin and is closely associated with PCNA during nuclear assembly. Demembranated sperm chromatin was exposed to hS, prepared for immunofluorescence with antibodies directed against LB3 and PCNA, and stained with Hoechst 33342 (A–E). Note that PCNA is absent, whereas LB3 is present and frequently appears to be helically wound around the chromatin (D and E). After the addition of X. laevis interphase extracts to sperm head chromatin for 5 min, LB3 is reorganized into patches and foci associated with chromatin (G, I, and J). In addition, PCNA is now present (H) and closely associated with LB3 (I and J). Enlargements of the regions in the boxes in the merged images (D, DNA and LB3; and I, LB3 and PCNA) are shown (E and J, 5.5× view). Bars, 5 μm. (K) The presence of LB3 and the absence of PCNA on demembranated sperm chromatin is also seen by immunoblotting. Demembranated sperm heads in the amount of 50,000 (lane 1), 100,000 (lane 2), or 200,000 (lane 3) were separated by SDS-PAGE, transferred to nitrocellulose, and probed for the presence of LB3 and PCNA. LB3 is seen in all lanes and is quantified relative to lane 3, but no PCNA could be detected.
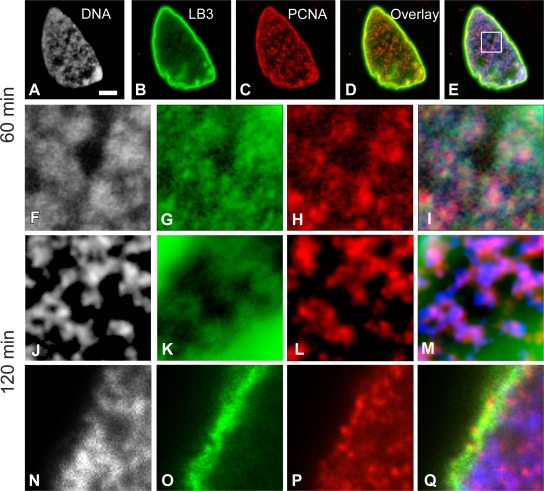
Within 5 min after the initiation of nuclear assembly, the filamentous pattern of LB3 was transformed into small foci and large patches on the surface of decondensing chromatin and throughout the forming nucleoplasm (Fig. 5 G). LB3 was present in the lamina region throughout the ∼40-min process of nuclear envelope assembly and as a relatively diffuse “veil” punctuated with LB3 foci in the nucleoplasm (unpublished data; Moir et al., 2000b). Coincident with these changes in lamin distribution and organization, PCNA became associated with the decondensing chromatin (Fig. 5, H–J). The PCNA pattern consisted primarily of small foci associated with the decondensing chromatin. This pattern is similar to the early S phase patterns seen in mammalian cells (Fig. 5 H; Bravo and Macdonald-Bravo, 1985). The PCNA foci were present throughout the nucleoplasm during nuclear assembly, frequently in close proximity to the lamin foci and patches (Fig. 5, I and J). After the completion of nuclear envelope assembly, LB3 and PCNA retained their close associations with chromatin as replication was initiated and the nucleus increased in size (Fig. 6, A–I). By 120 min after the initiation of nuclear assembly, the chromatin appeared more threadlike (Fig. 6 J), and its association with LB3 and PCNA was retained throughout the nucleoplasm (Fig. 6, J–M) and in the lamina region (Fig. 6, N–Q). These morphological observations show that lamins are closely associated with both chromatin and PCNA, further supporting a role for lamins in the elongation phase of DNA replication.
Figure 6.
LB3 is closely associated with PCNA and chromatin after nuclear envelope assembly. Nuclei were fixed and prepared for immunofluorescence with Hoechst 33342 and antibodies directed against LB3 and PCNA 60 min after assembly was initiated in X. laevis extracts (A–I). There appears to be substantial overlap of DNA, LB3, and PCNA throughout the nucleus and at the nuclear periphery (A–E). The overlay in D shows LB3 and PCNA, whereas E displays DNA, LB3, and PCNA. The area in the box in E was enlarged (6.3×) to show the details of DNA, LB3, PCNA, and the overlay (F–I). We have also examined nuclei after 120 min of assembly both in the nuclear interior (J–M) and at the nuclear periphery (N–Q). These images are approximately the same magnification as in F–I. Frequently, these nuclei have filamentous chromatin (J), which is closely associated with LB3 and PCNA (K–M). The association between these three components is also observed at the nuclear periphery (N–Q). We have used grayscale images of chromatin in A, F, J, and N and blue in the overlays (E, I, M, and Q) for purposes of improved contrast. Bar, 5 μm.
Modeling the PCNA–lamin Ig-fold interaction
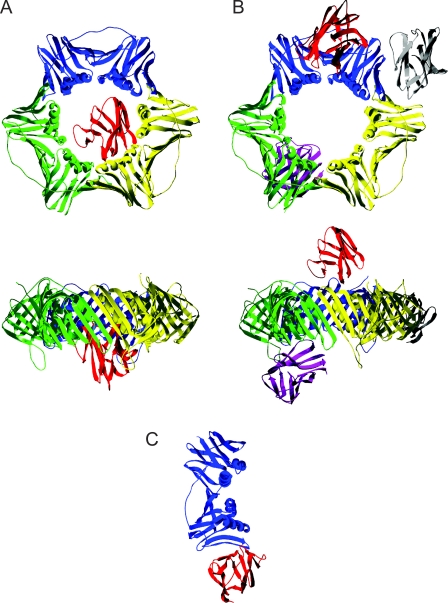
In an attempt to predict the mode of binding between the LA-Ig domain and PCNA in three dimensions, we have performed molecular docking simulations with the known crystal structures of the LA Ig-fold motif (PDB code 1IFR) and the PCNA trimer (PDB code 1VYM) using two different algorithms available through the web-based services GRAMM-X and ClusPro (Comeau et al., 2004; Tovchigrechko and Vakser, 2006). These simulations suggest two potential binding “modes” (Fig. 7). Using the GRAMM-X software, several top-scoring solutions place the Ig-fold inside the “hole” made by the PCNA homotrimer (Fig. 7 A). In each of these solutions, the center of gravity for LA-Ig remains almost the same, even though the orientation of the Ig-fold motif varies. Using the ClusPro software, LA-Ig is docked at the periphery of the homotrimeric PCNA ring (Fig. 7 B). Importantly, the majority of docking solutions suggested by ClusPro show that the binding site involves portions of the PCNA interdomain connecting loop (residues L121–E132), which is known to serve as a binding site for other proteins involved in DNA replication (Tsurimoto, 1999).
Figure 7.
Possible Ig-fold–PCNA binding sites based on molecular docking simulations. (A) Results of the analysis using the GRAMM-X software drawn in two orthogonal views. All of the top 10 solutions placed the Ig-fold (one of them is shown in red) in the center of the PCNA ring (blue, yellow, and green). As clearly seen in the bottom image, the center of gravity for the Ig-fold was somewhat off the plane of PCNA and on the opposite side of the interdomain-connecting loop. (B) The ClusPro docking analysis placed LA-Ig at one of three main locations on PCNA: near the interdomain connecting loop (red), between two subunits of PCNA (gray), or the back side of the PCNA homotrimer (dark purple). As seen in the bottom image, the Ig-fold motifs could be docked on either side of the PCNA ring. (C) The top solution of docking the LA-Ig (red) onto a single PCNA molecule (blue). The docking occurs at the intersubunit interface usually observed within the PCNA trimer.
PCNA's role in DNA replication is thought to begin after RFC binds to and opens the homotrimeric PCNA ring. This complex is then loaded onto primed DNA (Bowman et al., 2004; Yao et al., 2006). The possibility that the PCNA ring is opened is supported by molecular modeling studies (Kazmirski et al., 2005). The “open” ring structure of a sliding clamp appears to have a gap of 4 nm resulting from the opening of one of the three intersubunit interfaces (Millar et al., 2004). This opening should be large enough to accommodate the lamin Ig-fold. Based upon these considerations, we have examined the open ring situation by repeating the docking calculations with PCNA taken as a monomer. In this case, the top docking solution shows the Ig-fold interacting with a side of PCNA that is normally blocked by intersubunit contacts within the homotrimer (Fig. 7 C). This simulation predicts the alignment of two β strands, one derived from the Ig-fold and the other from the PCNA monomer. This results in a continuous β sheet similar to the one present in the intersubunit contact region of the PCNA trimer. This latter model suggests a potential novel regulation of homotrimeric PCNA loading onto primed DNA.
Discussion
In this study, we demonstrate that the DNA replication elongation factor PCNA binds directly to nuclear lamins in their highly conserved Ig-fold motifs. Our results also show that a structurally stable Ig-fold is necessary for PCNA binding. In previous studies, it has been shown that an excess of the Ig-fold inhibits, in a concentration-dependent fashion, lamin polymerization, sperm chromatin decondensation, and nuclear envelope assembly in X. laevis interphase extracts (Lopez-Soler et al., 2001; Shumaker et al., 2005). The inhibition of nuclear assembly by excess Ig-fold could either be attributable to alterations in the lamin–lamin interactions required for their polymerization (Lopez-Soler et al., 2001; Shumaker et al., 2005) or the competitive inhibition of lamin binding partners such as PCNA (Lopez-Soler et al., 2001; Shumaker et al., 2005). Interestingly, it has been shown that the depletion of PCNA from X. laevis extracts results in the formation of small nuclei with abnormal envelopes that are unable to replicate their DNA (Mattock et al., 2001). Other studies have shown that nuclei assembled in extracts “depleted” of LB3 accumulate PCNA (Jenkins et al., 1993) but are also unable to replicate DNA (Newport et al., 1990; Goldberg et al., 1995). These findings and the biochemical and morphological results of this study suggest that the interaction between lamins and PCNA is necessary early in nuclear assembly.
Unlike other binding partners of PCNA, nuclear lamins do not contain the characteristic PIP motif (see Introduction). Interestingly, the amount of PCNA bound is likely to be related to the length of the non–α-helical lamin tail domains. For example, LAT appears to bind more PCNA than either LCT or LA-Ig (Fig. 4). These differences may be attributable to the variability in the regions downstream of the lamin Ig-fold that may be involved in stabilizing their interactions with PCNA. Our docking analysis suggests several models for lamin Ig-fold interactions with PCNA. To better understand which residues on the Ig-fold motif and PCNA interact, mutational analysis and high-resolution structural studies are clearly needed. The models based on the GRAMM-X software suggest that lamins could bind homotrimeric PCNA, thereby facilitating RFC assembly onto a PCNA ring. The ClusPro software indicates that a portion of the PCNA interdomain connecting loop (residues 121–132) could bind LA-Ig, which is unexpected for proteins lacking a PIP domain. The proposed interactions between the lamin Ig-fold and PCNA could spatially and temporally regulate the binding of other proteins to PCNA. Because RFC, in addition to PCNA, localizes to the abnormal nuclear lamin foci after the addition of dominant-negative mutant lamins (Moir et al., 2000a), there is the possibility that lamins also bind RFC. Finally, the crystal structure of the PCNA-RFC complex (PDB code 1SXJ) suggests the possibility that LA-Ig could insert into the cleft between the PCNA ring and RFC (Bowman et al., 2004). If such an interaction indeed takes place, it could have a significant effect on regulating DNA replication.
It is conceivable that the Ig-fold motif could inhibit PCNA activity if it binds to PCNA after RFC opens the PCNA homotrimer. This possibility is supported by evidence from studies of gp45, a homologue of PCNA in the bacteriophage T4. In solution, gp45 has been shown to form an open ring structure when one of the three intersubunit interfaces is separated by ∼4 nm (Millar et al., 2004). This opening is comparable with the size of the Ig fold, which suggests that the Ig-fold could bind to the open interface.
During S phase, PCNA foci form, disassemble, and reassemble in the nucleus. The sequence of the organization of these foci has been well documented and the patterns can be used to distinguish early, middle, and late S phase (Kennedy et al., 2000; Leonhardt et al., 2000). Using fluorescence recovery after photobleaching, GFP-PCNA is stably bound within replication foci, as demonstrated by the slow exchange between the nucleoplasm and these foci (Leonhardt et al., 2000; Sporbert et al., 2002). This indicates that homotrimeric PCNA functions as a stable core within a replication complex and is subsequently disassembled and then reassembled at another replicon (Sporbert et al., 2002). However, the mechanisms regulating the disassembly, reassembly, and repositioning of PCNA remain unknown. We believe that nuclear lamin networks are involved in these dynamic properties of replicons by acting as a scaffold or platform upon which PCNA and other components of the replication–elongation complex are assembled and disassembled in a spatial and temporal sequence during S phase (Shumaker et al., 2003; Gruenbaum et al., 2005).
At the present time, we can only speculate on the precise mechanisms linking the replication machinery to the lamins. The observation that DNA replication is inhibited after the addition of excess lamin tail domain and/or its Ig-fold suggests that these lamin fragments act as competitive inhibitors by preventing the normal interactions between PCNA and chromatin. This idea is supported by our finding that DNA synthesis recovers in in vitro assembled X. laevis nuclei exposed to a known concentration of LB3T for longer periods (∼19 μM LB3T up to 60 rather than 30 min). Under these conditions, it is likely that PCNA and wild-type LB3 continue to become imported into assembled nuclei, thereby diluting the amount of LB3T relative to the LB3 present within each nucleus. In this fashion, the inhibitory effect of LB3T on DNA replication could be reversed. The apparent loss of PCNA in X. laevis nuclei exposed to LB3T and LB3T RW is most likely attributable to its solubility after the fixation and permeablization protocols used for immunofluorescence observations.
It has also been shown that an excess of either LB3T or the Ig-fold inhibits lamin polymerization into the higher-order structures required for normal nuclear assembly (see Fig. 5; Shumaker et al., 2005). When LB3T is added to assembled X. laevis nuclei, it prevents further growth of these nuclei (Shumaker et al., 2005). Here, we show that the addition of LB3T also prevents PCNA from associating with chromatin, thereby inhibiting replication. These findings point to a complex relationship between lamin assembly, nuclear growth, the assembly of PCNA, and the formation of replicons, as well as DNA replication. In addition, we have shown that HeLa cells expressing GFP-NLS-Ig show an ∼29% decrease in the number of cells incorporating BrdU. This result extends the findings regarding a role for nuclear lamins in DNA replication from X. laevis nuclei to mammalian cells. The difference in the level of inhibition of DNA synthesis between X. laevis nuclei exposed to LB3T and mammalian cells expressing GFP-NLS-Ig may be related to the expression levels of GFP-NLS-Ig in individual cells in asynchronous cultures. In support of this, HeLa cells with the highest expression of GFP-NLS-Ig did not incorporate BrdU.
There are >250 allelic variants of the LA gene (LMNA) comprised of ∼170 different mutations causing >15 human diseases known as the “laminopathies” (Gruenbaum et al., 2005; Shumaker et al., 2005). The laminopathies display a broad range of phenotypes including muscular dystrophies, cardiomyopathies, lipodystrophies, and premature aging. Of the ∼170 LA mutations, ∼45 are located in the Ig-fold. Therefore, ∼26% of the mutations are located in only 16% of the protein chain. This suggests that the Ig-fold motif is necessary for lamin function and that alterations in either the structure or stability of the lamin Ig-fold motif can lead to disease. In this study, we have examined a point mutation in the Ig-fold that causes EDMD when present in LA. The addition of LB3T containing this mutation, which is located at a highly conserved site, results in significantly less inhibition of DNA replication. Furthermore, the mutant LA Ig-fold binds ∼29% less PCNA than the wild type, implying that EDMD as well as numerous other laminopathies could be at least in part explained by alterations in the normal process of DNA replication.
In addition to PCNA, nuclear titin has also been shown to bind to the Ig-fold of lamins (Zastrow et al., 2006). Nuclear titin is required for chromosome condensation and separation during mitosis in Drosophila melanogaster (Machado and Andrew, 2000). Because Ig-folds frequently serve as protein–protein interacting motifs (Dhe-Paganon et al., 2002), a logical conclusion is that other many of the proteins known to bind to the C terminus of the lamins (Zastrow et al., 2004) may interact with the Ig-fold.
In conclusion, nuclear lamins most likely regulate the spatial and temporal arrangement of PCNA during DNA replication. Although details of the structure of lamins within the nucleus are yet to be determined, they are known to polymerize into a variety of higher order structures including short and long head-to-tail arrays of dimers and lattice works of 10-nm filaments (Aebi et al., 1986; Sasse et al., 1997). These structures could provide a nuclear scaffold or platform (Goldman et al., 2002) for organizing PCNA and its other binding partners into DNA replication factories. A logical extension of this hypothesis is that lamins are also involved in the regulation of other processes that require PCNA including DNA repair (Toschi and Bravo, 1988), DNA ligation (Mortusewicz et al., 2006), removal of Okazaki fragments (Li et al., 1995), unwinding DNA (Rodriguez-Lopez et al., 2003), DNA methylation (Mortusewicz et al., 2005), cell cycle regulation (Waga et al., 1994), and gene expression (Maga and Hubscher, 2003).
Materials and methods
Nuclear assembly in vitro
X. laevis egg interphase extracts, 200,000 g supernatants (uS), 30,000 g membrane fractions (Sm), and sperm chromatin were prepared essentially as described previously (Newmeyer and Wilson, 1991). Sperm were suspended in 1% vol/vol Triton X-100 in sperm buffer (SB; 80 mM KCl, 15 mM NaCl, 5 mM EDTA, 15 mM Pipes, pH 7.4, 0.2 M sucrose, and 7 mM MgCl2) instead of 0.05% lysolecithin. The chromatin was pelleted at 2,500 g for 10 min through 20% sucrose in SB, resuspended in 1 ml SB containing 1% BSA wt/vol, and pelleted for 10 min at 2,000 g. In some preparations, the chromatin was preswelled with nucleoplasmin for 5 min with heated cytosol (uS boiled for 15 min then centrifuged at 16,000 g for 15 min; Newmeyer et al., 1986).
Cell culture
HeLa cells were maintained in DME high glucose, 10% FBS, and penicillin/streptomycin and grown at 37°C in 5% CO2.
Immunofluorescence and immunoblotting
X. laevis nuclei and sperm chromatin were prepared as previously described (Spann et al., 1997; Shumaker et al., 2005). HeLa cells were also prepared as described previously (Shumaker et al., 2006) and observed at 22°C. Images were taken with either an Axio-Imager Z1 or Axioplan 2 using AxioVision 4.6 (all from Carl Zeiss, Inc.) with multichannel fluorescence and z stack modules. The camera used with the Axio-Imager was the Axiocam MRm r3 (Carl Zeiss, Inc.). For the Axioplan 2, we used the Axiocam MRm r1.1 camera (Carl Zeiss, Inc.). Some images were also taken with a confocal microscope (LSM 510 Meta) using LSM 3.2 SP2 software (both from Carl Zeiss, Inc.). The objectives used were Plan Apochromat 100× NA 1.4 (Carl Zeiss, Inc.). 3D deconvolution was performed using the AxioVision 4.5 deconvolution module (Carl Zeiss, Inc.). Total fluorescence was determined by multiplying the mean nuclear fluorescence intensity by nuclear area. Statistical differences were determined using the Student's t test or χ2 test. Sperm chromatin was suspended in SDS sample buffer (Laemmli, 1970) and immunoblotting was performed as described previously (Shumaker et al., 2006) and quantified by chemiluminescence detection using an ImageStation 440CF and 1D software (Kodak). PCNA binding to LA-Ig and LA-Ig RW was quantified after subtracting the background binding of PCNA to protein S–agarose beads. Figures for plates were prepared in CorelDRAW (Corel Corp.) and image gamma and contrast were adjusted in PhotoPaint (Corel Corp.) and Photoshop (Adobe).
Antibodies
For immunofluorescence, primary antibodies included a mouse mAb directed against X. laevis LB3 (1:1,000, immunofluorescence and immunoblotting; L6-5D5, provided by R. Stick, University of Bremen, Bremen, Germany; Stick, 1988), a rabbit pAb against LB3 (1:1,000; 299), a goat pAb against PCNA (1:50; C-20; Santa Cruz Biotechnology, Inc.), a rat pAb anti-BrdU (1:50; Abcam) and a mouse mAb directed against His6 (1:200; QIAGEN).
For immunoblotting, primary antibodies used were a mouse mAb (1:1,000; PC10; Santa Cruz Biotechnology, Inc.) and a goat pAb (1:1,000; sc-9857; Santa Cruz Biotechnology, Inc.) directed against PCNA and the rabbit anti-LB3 (1:3,000, 299).
Secondary antibodies used for immunofluorescence were goat anti–rabbit IgG–Alexa Fluor 488 (1:400; Invitrogen), donkey anti–mouse IgG–Alexa Fluor 488 (1:400; Invitrogen), donkey anti–goat IgG-Alexa Fluor 568 (1:400; Invitrogen), goat anti–mouse IgG–Alexa Fluor 568 (1:400; Invitrogen), goat anti–rat Alexa Fluor 568 (1:400; Invitrogen), and rhodamine avidin (1:400; GE Healthcare). For immunoblotting, the secondary antibodies were goat anti–mouse IgG, goat anti–rabbit IgG, and donkey anti–goat IgG conjugated to horseradish peroxidase (1:5,000; Invitrogen).
Protein expression and purification
Expression and purification of His-tagged LB3 Ig-fold motif (LB3T-Ig, residues 433–540) has been described previously (Shumaker et al., 2005). The LA Ig-fold motif (LA-Ig, residues 436–544) was prepared by PCR and inserted into the pET30a vector (EMD). The C-terminal tail domains of LA (LAT, residues 243–664), LC (LCT, residues 243–572), LB1 (LB1T, residues 244–586), and LB3 (LB3T, residues 384–583) were prepared by PCR and inserted into pET30a. The Ran-binding domain of importin β (residues 1–380) and the LA Ig-fold motif (residues 440–544) were also cloned by PCR and inserted into the pET-30a vector. The LB3T clone was also mutagenized using the Quick-Change Mutagenesis kit (Stratagene) to convert Arg 454 to Trp (LB3T RW; Shumaker et al., 2005). The LA-Ig clone was also mutagenized to convert Arg 453 to Trp (LA-Ig RW). The LB3T and LB3T RW proteins were expressed and purified as described previously (Spann et al., 1997; Lopez-Soler et al., 2001). The constructs in the pET 30 vectors were grown in LB broth then induced with 1 mM IPTG. The bacteria were pelleted, suspended in lysis buffer (50 mM Tris, pH 7.4, 50 mM NaCl, 5 mM EDTA, 5% glycerol, and 2 mM DTT), and sonicated. The samples were centrifuged at 16,000 g, the expressed proteins were purified on a His-trap column with fast protein liquid chromatography (Äkta), and the fractions containing the protein of interest were further purified on a mono Q column (GE Biosciences) using a linear gradient of 50–750 mM NaCl in 50 mM Tris, pH 8, and 2 mM EDTA.
Affinity chromatography
A HiTrap chelating HP 1-ml column (GE Biosciences) was loaded with NiSO4 according to the manufacturer's instructions. Approximately 1.5 mg of purified His-tagged LB3T, LB3T-Ig protein was added to the column and washed with column buffer (20 mM NaPO4, pH 7.4, and 0.5 M NaCl). 15 mg uS was diluted to 4 ml in 20 mM NaPO4, pH 7.4, and 100 mM NaCl and repeatedly passed over the column for 18 h at 4°C using a peristaltic pump (GE Biosciences). Bound proteins were eluted with a fast protein liquid chromatography column with a gradient of 0.1–1.5 M NaCl in 20 mM NaPO4, pH 7.4.
Solution binding assay
Purified LA, LAT, LB1T, LCT, LB3T, LA-Ig, LA-Ig RW, and Imp βR (5 μg) were bound to protein S–agarose beads (EMD) for 2 h at 22°C in binding buffer (1× PBS, 10% glycerol, 0.1% Tween-20, 50 μg/ml BSA, and 1 mg/ml gelatin) and washed three times with binding buffer followed by the addition of 20 μg of purified PCNA in binding buffer. The samples were rotated for 18 h at 4°C, washed three times in binding buffer, suspended in SDS sample buffer, and separated on an SDS-PAGE gel. The gels were then stained with Coomassie blue G250. PCNA binding to LA-Ig, LA-Ig RW, and Imp βR was performed similarly, except that 10 mM phosphate, pH 7.4, and 50 mM NaCl were used instead of 1× PBS, and 50 μg of purified PCNA was used in the binding assay. Specific binding of PCNA to LA-Ig and LA-Ig RW was determined by densitometry. The amount of PCNA bound to Imp βR was similar to the amount of PCNA bound to protein S–agarose beads alone and was subtracted as background.
DNA replication assay
Nuclei were assembled in X. laevis interphase extracts for 60 min and, in some experiments, lamin protein fragments were added at a concentration of ∼10–20 μM for 30 min followed by the addition of 2 μCi [32P]α-dCTP for 10 min and analyzed as described previously (Spann et al., 1997). The amount of DNA synthesis in each lane was analyzed with a phosphoimager (Cyclone Storage Phosphor System; PerkinElmer) using 1D Image Analysis software (Kodak). Alternatively, 5 μM bio-11-dUTP (Sigma-Aldrich) was substituted for [32P]α-dCTP and analyzed by fluorescence microscopy as described previously (Spann et al., 1997). HeLa cells were transfected with pEGFP-LMNA (Moir et al., 2000b) or pEGFP-NLS-LMNB1 Ig (SV40 NLS and LB1 residues 434–537) vectors in a Gene Pulser II (Bio-Rad Laboratories) at 160 V and 500 μF, and 24 h later, DNA replication was examined as described previously (Solovei et al., 2004).
Molecular docking analysis
Crystal structures of the human LA-Ig (PDB code 1IFR) and human PCNA trimer (1VYM) were downloaded from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) and analyzed using two online servers for molecular docking: GRAMM-X (http://vakser.bioinformatics.ku.edu/resources/gramm/grammx) and ClusPro (http://nrc.bu.edu/cluster/) with the DOT docking algorithm. LA-Ig was docked as a rigid body onto the PCNA trimer using default settings. Resulting molecular models were examined using Deepview (GlaxoSmithKline and the Swiss Institute of Bioinformatics; Guex and Peitsch, 1997) and the images were prepared using POV-Ray 3.6c (Persistence of Vision Raytracer Pty. Ltd.).
Online supplemental material
Fig. S1 shows that HeLa cells expressing GFP-NLS-Ig show a decrease in the incorporation of BrdU. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708155/DC1.
Supplemental Material
Acknowledgments
We thank Reimer Stick for the L6-5D5 antibody. Due to the limitations on the number of references many informative papers could not be referenced.
This work was supported by a grant from the National Cancer Institute (RO1 CA31760) and an Ellison Senior Scholar Award to R.D. Goldman.
D.K. Shumaker and L. Solimando contributed equally to this paper.
A. Grunwald's present address is Albert Einstein College of Medicine, New York, NY 10461.
Abbreviations used in this paper: 11-dUTP, biotin-11-2′-deoxyuridine-5′-triphosphate; EDMD, Emery-Dreifuss muscular dystrophy; IF, intermediate filament; LA, lamin A; LB1, lamin B1; LB3, lamin B3; LC, lamin C; NLS, nuclear localization sequence; PCNA, proliferating cell nuclear antigen; PIP, PCNA-interacting protein; RFC, replication factor C; uS, ultracentrifuged supernatant.
References
- Aebi, U., J. Cohn, L. Buhle, and L. Gerace. 1986. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 323:560–564. [DOI] [PubMed] [Google Scholar]
- Blow, J.J., and R.A. Laskey. 1986. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 47:577–587. [DOI] [PubMed] [Google Scholar]
- Bonne, G., M.R. Di Barletta, S. Varnous, H.M. Becane, E.H. Hammouda, L. Merlini, F. Muntoni, C.R. Greenberg, F. Gary, J.A. Urtizberea, et al. 1999. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 21:285–288. [DOI] [PubMed] [Google Scholar]
- Bowman, G.D., M. O'Donnell, and J. Kuriyan. 2004. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 429:724–730. [DOI] [PubMed] [Google Scholar]
- Bravo, R., and H. Macdonald-Bravo. 1985. Changes in the nuclear distribution of cyclin (PCNA) but not its synthesis depend on DNA replication. EMBO J. 4:655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau, S.R., D.W. Gatchell, S. Vajda, and C.J. Camacho. 2004. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 20:45–50. [DOI] [PubMed] [Google Scholar]
- Dechat, T., B. Korbei, O.A. Vaughan, S. Vlcek, C.J. Hutchison, and R. Foisner. 2000. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J. Cell Sci. 113:3473–3484. [DOI] [PubMed] [Google Scholar]
- Dessev, G., R. Palazzo, L. Rebhun, and R. Goldman. 1989. Disassembly of the nuclear envelope of spisula oocytes in a cell-free system. Dev. Biol. 131:496–504. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon, S., E.D. Werner, Y.I. Chi, and S.E. Shoelson. 2002. Structure of the globular tail of nuclear lamin. J. Biol. Chem. 277:17381–17384. [DOI] [PubMed] [Google Scholar]
- Goldberg, M., H. Jenkins, T. Allen, W.G. Whitfield, and C.J. Hutchison. 1995. Xenopus lamin B3 has a direct role in the assembly of a replication competent nucleus: evidence from cell-free egg extracts. J. Cell Sci. 108:3451–3461. [DOI] [PubMed] [Google Scholar]
- Goldman, R.D., Y. Gruenbaum, R.D. Moir, D.K. Shumaker, and T.P. Spann. 2002. Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16:533–547. [DOI] [PubMed] [Google Scholar]
- Goldman, R.D., D.K. Shumaker, M.R. Erdos, M. Eriksson, A.E. Goldman, L.B. Gordon, Y. Gruenbaum, S. Khuon, M. Mendez, R. Varga, and F.S. Collins. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 101:8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., and M.C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., A. Margalit, R.D. Goldman, D.K. Shumaker, and K.L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21–31. [DOI] [PubMed] [Google Scholar]
- Heitlinger, E., M. Peter, M. Haner, A. Lustig, U. Aebi, and E.A. Nigg. 1991. Expression of chicken lamin B2 in Escherichia coli: characterization of its structure, assembly, and molecular interactions. J. Cell Biol. 113:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, H., T. Holman, C. Lyon, B. Lane, R. Stick, and C. Hutchison. 1993. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J. Cell Sci. 106:275–285. [DOI] [PubMed] [Google Scholar]
- Johnson, B.R., R.T. Nitta, R.L. Frock, L. Mounkes, D.A. Barbie, C.L. Stewart, E. Harlow, and B.K. Kennedy. 2004. A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc. Natl. Acad. Sci. USA. 101:9677–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmirski, S.L., Y. Zhao, G.D. Bowman, M. O'Donnell, and J. Kuriyan. 2005. Out-of-plane motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA. 102:13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, B.K., D.A. Barbie, M. Classon, N. Dyson, and E. Harlow. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14:2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X.P., R. Onrust, M. O'Donnell, and J. Kuriyan. 1992. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 69:425–437. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lee, S.H., and J. Hurwitz. 1990. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc. Natl. Acad. Sci. USA. 87:5672–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt, H., H.P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M.C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., J. Li, J. Harrington, M.R. Lieber, and P.M. Burgers. 1995. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 270:22109–22112. [DOI] [PubMed] [Google Scholar]
- Lopez-Soler, R.I., R.D. Moir, T.P. Spann, R. Stick, and R.D. Goldman. 2001. A role for nuclear lamins in nuclear envelope assembly. J. Cell Biol. 154:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourim, D., A. Kempf, and G. Krohne. 1996. Characterization and quantitation of three B-type lamins in Xenopus oocytes and eggs: increase of lamin LI protein synthesis during meiotic maturation. J. Cell Sci. 109:1775–1785. [DOI] [PubMed] [Google Scholar]
- Machado, C., and D.J. Andrew. 2000. D-Titin: a giant protein with dual roles in chromosomes and muscles. J. Cell Biol. 151:639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga, G., and U. Hubscher. 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116:3051–3060. [DOI] [PubMed] [Google Scholar]
- Mattock, H., P. Jares, D.I. Zheleva, D.P. Lane, E. Warbrick, and J.J. Blow. 2001. Use of peptides from p21 (Waf1/Cip1) to investigate PCNA function in Xenopus egg extracts. Exp. Cell Res. 265:242–251. [DOI] [PubMed] [Google Scholar]
- Millar, D., M.A. Trakselis, and S.J. Benkovic. 2004. On the solution structure of the T4 sliding clamp (gp45). Biochemistry. 43:12723–12727. [DOI] [PubMed] [Google Scholar]
- Moir, R.D., M. Montag-Lowy, and R.D. Goldman. 1994. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J. Cell Biol. 125:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir, R.D., T.P. Spann, H. Herrmann, and R.D. Goldman. 2000. a. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 149:1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir, R.D., M. Yoon, S. Khuon, and R.D. Goldman. 2000. b. Nuclear Lamins A and B1. Different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 151:1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz, O., L. Schermelleh, J. Walter, M.C. Cardoso, and H. Leonhardt. 2005. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. USA. 102:8905–8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz, O., U. Rothbauer, M.C. Cardoso, and H. Leonhardt. 2006. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 34:3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer, D.D., and K.L. Wilson. 1991. Egg extracts for nuclear import and nuclear assembly reactions. Methods Cell Biol. 36:607–634. [DOI] [PubMed] [Google Scholar]
- Newmeyer, D.D., D.R. Finlay, and D.J. Forbes. 1986. In vitro transport of a fluorescent nuclear protein and exclusion of non-nuclear proteins. J. Cell Biol. 103:2091–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport, J.W., K.L. Wilson, and W.G. Dunphy. 1990. A lamin-independent pathway for nuclear envelope assembly. J. Cell Biol. 111:2247–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A. 1992. Assembly-disassembly of the nuclear lamina. Curr. Opin. Cell Biol. 4:105–109. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez, A.M., D.A. Jackson, J.O. Nehlin, F. Iborra, A.V. Warren, and L.S. Cox. 2003. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA. Mech. Ageing Dev. 124:167–174. [DOI] [PubMed] [Google Scholar]
- Sasse, B., A. Lustig, U. Aebi, and N. Stuurman. 1997. In vitro assembly of Drosophila lamin Dm0–lamin polymerization properties are conserved. Eur. J. Biochem. 250:30–38. [DOI] [PubMed] [Google Scholar]
- Shumaker, D.K., E.R. Kuczmarski, and R.D. Goldman. 2003. The Nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr. Opin. Cell Biol. 15:358–366. [DOI] [PubMed] [Google Scholar]
- Shumaker, D.K., R.I. Lopez-Soler, S.A. Adam, H. Herrmann, R.D. Moir, T.P. Spann, and R.D. Goldman. 2005. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc. Natl. Acad. Sci. USA. 102:15494–15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker, D.K., T. Dechat, A. Kohlmaier, S.A. Adam, M.R. Bozovsky, M.R. Erdos, M. Eriksson, A.E. Goldman, S. Khuon, F.S. Collins, et al. 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 103:8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei, I., L. Schermelleh, K. During, A. Engelhardt, S. Stein, C. Cremer, and T. Cremer. 2004. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma. 112:410–423. [DOI] [PubMed] [Google Scholar]
- Spann, T.P., R.D. Moir, A.E. Goldman, R. Stick, and R.D. Goldman. 1997. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 136:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann, T.P., A.E. Goldman, C. Wang, S. Huang, and R.D. Goldman. 2002. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 156:603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporbert, A., A. Gahl, R. Ankerhold, H. Leonhardt, and M.C. Cardoso. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 10:1355–1365. [DOI] [PubMed] [Google Scholar]
- Stick, R. 1988. cDNA cloning of the developmentally regulated lamin LIII of Xenopus laevis. EMBO J. 7:3189–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschi, L., and R. Bravo. 1988. Changes in cyclin/proliferating cell nuclear antigen distribution during DNA repair synthesis. J. Cell Biol. 107:1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovchigrechko, A., and I.A. Vakser. 2006. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34:W310–W314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, M.Y., S. Wang, J.M. Heidinger, D.K. Shumaker, S.A. Adam, R.D. Goldman, and Y. Zheng. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 311:1887–1893. [DOI] [PubMed] [Google Scholar]
- Tsurimoto, T. 1999. PCNA binding proteins. Front. Biosci. 4:D849–D858. [DOI] [PubMed] [Google Scholar]
- Waga, S., G.J. Hannon, D. Beach, and B. Stillman. 1994. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 369:574–578. [DOI] [PubMed] [Google Scholar]
- Warbrick, E., W. Heatherington, D.P. Lane, and D.M. Glover. 1998. PCNA binding proteins in Drosophila melanogaster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res. 26:3925–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, N., L. Coryell, D. Zhang, R.E. Georgescu, J. Finkelstein, M.M. Coman, M.M. Hingorani, and M. O'Donnell. 2003. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J. Biol. Chem. 278:50744–50753. [DOI] [PubMed] [Google Scholar]
- Yao, N.Y., A. Johnson, G.D. Bowman, J. Kuriyan, and M. O'Donnell. 2006. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J. Biol. Chem. 281:17528–17539. [DOI] [PubMed] [Google Scholar]
- Zastrow, M.S., S. Vlcek, and K.L. Wilson. 2004. Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 117:979–987. [DOI] [PubMed] [Google Scholar]
- Zastrow, M.S., D.B. Flaherty, G.M. Benian, and K.L. Wilson. 2006. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J. Cell Sci. 119:239–249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.