Abstract
The endoplasmic reticulum (ER) protein GT1 (UDP-glucose: glycoprotein glucosyltransferase) is the central enzyme that modifies N-linked carbohydrates based upon the properties of the polypeptide backbone of the maturing substrate. GT1 adds glucose residues to nonglucosylated proteins that fail the quality control test, supporting ER retention through persistent binding to the lectin chaperones calnexin and calreticulin. How GT1 functions in its native environment on a maturing substrate is poorly understood. We analyzed the reglucosylation of a maturing model glycoprotein, influenza hemagglutinin (HA), in the intact mammalian ER. GT1 reglucosylated N-linked glycans in the slow-folding stem domain of HA once the nascent chain was released from the ribosome. Maturation mutants that disrupted the oxidation or oligomerization of HA also supported region-specific reglucosylation by GT1. Therefore, GT1 acts as an ER quality control sensor by posttranslationally reglucosylating glycans on slow-folding or nonnative domains to recruit chaperones specifically to critical aberrant regions.
Introduction
Protein maturation and quality control in the ER involves a network of molecular chaperones, foldases, and sorting factors that are recruited at specific stages to optimize the folding process and monitor the integrity of the protein product (Ellgaard et al., 1999; Hebert and Molinari, 2007). Encoded in the composition of N-linked glycans is information about the foldedness of the modified protein (Helenius and Aebi, 2004; Hebert et al., 2005). They serve as cellular maturation and quality control tags by recruiting, in a carbohydrate composition–dependent manner, a variety of factors that assist and guide the nascent chain through the early secretory pathway. A central player in the quality control decision in the ER is GT1 (UDP-glucose: glycoprotein glucosyltransferase; Trombetta and Parodi, 2003). GT1 is the main ER enzyme that alters the glycan composition of a substrate based on structural features of the modified protein (Sousa et al., 1992; Sousa and Parodi, 1995; Ritter and Helenius, 2000; Trombetta and Helenius, 2000; Caramelo et al., 2003; Taylor et al., 2003). Though little is known about its activity in its native biological environment, in vitro studies support the hypothesis that GT1 is the key quality control decision maker in the ER.
Nascent secretory proteins are targeted to the ER for their cotranslational translocation into the ER lumen. 14-member N-linked glycans (Glc3Man9GlcNAc2) are added en bloc to the elongating polypeptides during translocation by the oligosaccharyltransferase complex (Kornfeld and Kornfeld, 1985). Glycans are then immediately trimmed by glucosidases I and II, creating monoglucosylated glycans that provide platforms for the recruitment of the lectin chaperones calnexin and calreticulin (Ou et al., 1993; Hammond et al., 1994; Hebert et al., 1995). These lectin chaperones transiently interact with nascent glycoproteins and increase their folding efficiency by preventing premature folding and aggregation and recruiting the associated oxidoreductase ERp57 (Hebert et al., 1996; Vassilakos et al., 1996; Oliver et al., 1997; Zapun et al., 1998). Generally, folding is thought to occur upon release of the substrate from the chaperone (Bukau and Horwich, 1998). In the case of the lectin chaperones, this is directed by the removal of the final glucose residue by glucosidase II and the relatively weak affinity (micromolar) of the chaperones for glycans (Hebert et al., 1995, 1996; Kapoor et al., 2003).
Proteins that have not attained their native conformations can be targeted for reentry into the calnexin binding cycle by their reglucosylation via GT1 (Helenius, 1994; Hebert et al., 1995; Cannon and Helenius, 1999; Trombetta and Parodi, 2003). Labriola et al. (1995, 1999) have shown that GT1 reglucosylation counteracts glucosidase II activity and drives lectin chaperone association with substrate glycoproteins in Trypanosoma cruzi, a unicellular parasite which transfers unglucosylated glycans. In vitro studies of GT1 activity using purified proteins have revealed that GT1 preferentially reglucosylates nonnative glycoproteins (Sousa and Parodi, 1995; Trombetta and Helenius, 2000). This selectivity is driven by the exposure of hydrophobic amino acids on the protein surface, a hallmark of aberrantly folded proteins (Sousa and Parodi, 1995; Caramelo et al., 2003; Taylor et al., 2003). GT1 has a high sensitivity for structural imperfections, as it selectively reglucosylates misfolded domains of an otherwise native protein (Ritter and Helenius, 2000). Both an enzymatically active protein with a single point mutation and the removal of a native disulfide bond have been shown to support reglucosylation by purified GT1 (Taylor et al., 2004; Ritter et al., 2005). GT1 reglucosylation of endogenous transferrin limited the formation of disulfide-bonded aggregates and heat-induced misfolding (Wada et al., 1997). In vitro studies have found that GT1 appears to display a preference for molten globule near-native proteins and can reglucosylate natively folded proteins that have impaired oligomerization (Caramelo et al., 2003, 2004; Keith et al., 2005).
Previous in vitro studies have used small soluble well-characterized engineered substrates. Although these studies have provided a foundation for understanding the function of GT1 in ER quality control, there is a lack of information on the role of GT1 in its native environment using a dynamic maturing substrate. Influenza hemagglutinin (HA) is arguably the most thoroughly understood protein in regards to its maturation in the mammalian secretory pathway (Braakman et al. 1991, 1992a; Chen et al., 1995; Hebert et al., 1995, 1996, 1997; Daniels et al., 2003; Maggioni et al., 2005). In this paper, we have dissected the involvement of GT1 in the maturation of HA in its natural maturing environment using a mutant cell line that permitted isolation of the reglucosylation process. We found that HA can fold and oligomerize properly when lectin chaperone association is mediated solely through GT1 activity. The reglucosylation of HA was region-specific, with GT1 targeting glycans in the slow-folding stem domain of HA that associates with calnexin. Substrate glycoproteins were only reglucosylated posttranslationally after they had been released from the translocon environment. Transient nonnative conformations of HA trigger transitory GT1 reglucosylation, whereas terminal nonnative mutations induced long-term recognition. These results demonstrate that in the intact mammalian ER, GT1 acts posttranslationally to direct differential lectin chaperone binding through detection of slow folding and malformed regions of substrate glycoproteins.
Results
A cell-based reglucosylation assay
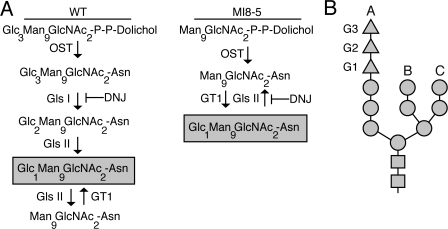
Studies of GT1 reglucosylation in the ER are hindered by the presence of two distinct pathways that can create monoglucosylated proteins. Monoglucosylated proteins can be generated by glucose addition to Man9GlcNAc2 glycans via GT1 or through the sequential trimming of newly added glycans by glucosidases I and II (Fig. 1 A, WT). To separate monoglucosylated glycans produced via GT1 activity from those generated by glucosidase activity, the CHO mutant cell line MI8-5 was used. The oligosaccharyltransferase of these cells transfers Man9GlcNAc2 glycans as opposed to triglucosylated high-mannose glycans because of a mutation in the alg6 gene (Fig. 1 A, MI8-5; Quellhorst et al., 1999). Alg6p is responsible for transferring glucose residues to the growing dolichol pyrophosphate precursor (Runge et al., 1984). Therefore, GT1 activity is the sole mechanism for producing monoglucosylated glycoproteins in MI8-5 CHO cells.
Figure 1.
Glycan processing in wild-type and MI8-5 cells. (A) Processing of the N-linked glycan in the ER of both wild-type and MI8-5 CHO cells. The glycoform generated by GT1 activity in each cell type is boxed and the point at which trimming is blocked by the presence of the glucosidase inhibitor DNJ is marked. OST, oligosaccharyltransferase; Gls I, glucosidase I; Gls II, glucosidase II; Glc, glucose; Man, Mannose; GlcNAc2, N-acetylglucosamine. (B) Schematic of a 14-member N-linked glycan. Glucoses are depicted as triangles, mannoses as circles, and N-acetylglucosamines as squares. The three mannose branches, A, B, and C, are marked. The three glucoses attached to the A branch are indicated as G3, G2, and G1.
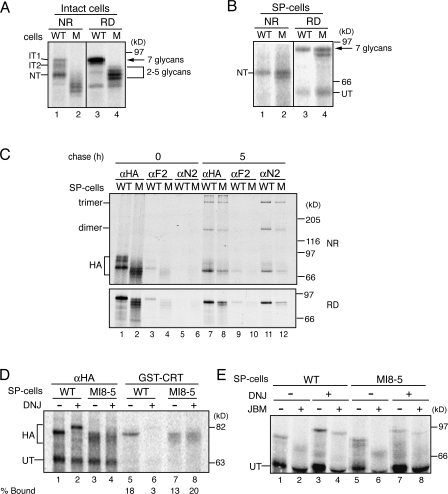
The cellular maturation pathway of HA is well defined; therefore, it is an ideal substrate to dissect the role of GT1 in glycoprotein maturation (Braakman et al., 1991, 1992b; Hebert et al., 1995; Daniels et al., 2003; Maggioni et al., 2005). HA was transiently expressed in MI8-5 and wild-type CHO cells using a modified vaccinia virus–T7 RNA polymerase expression system (Fuerst et al., 1986). 35S-labeled HA was immunoprecipitated with HA antisera and analyzed by nonreducing and reducing SDS-PAGE (Fig. 2 A). In contrast to wild type CHO cells, where HA received all seven of its N-linked glycans, HA expressed in MI8-5 cells formed four discrete species that likely possessed two to five glycans, accounting for their faster migration (Fig. 2 A, lanes 3 and 4; Hebert et al., 1997). Cell lines defective in assembling the complete dolichol precursor can frequently hypoglycosylate nascent proteins because of the inefficiency with which the oligosaccharyltransferase transfers the incompletely assembled glycan (Huffaker and Robbins, 1983).
Figure 2.
A cell-based reglucosylation assay. (A) For intact cells, HA was transiently transfected in both wild-type (WT) and MI8-5 (M) cells using the recombinant vaccinia virus–T7 RNA polymerase system and radiolabeled for 20 min in the presence of 0.5 mM DMJ at 34°C. The HA folding intermediates IT1, IT2, and NT and also the differences in glycosylation in wild-type and MI8-5 cells are indicated. (B) For SP cells, full-length HA mRNA was in vitro translated in the presence of wild-type or MI8-5 SP cells treated with 0.5 mM DMJ at 27°C. Radiolabeled HA was immunoprecipitated from lysates using a polyclonal antibody against HA. Untranslocated (UT) and native (NT) HA are marked. In A and B, black lines indicate that intervening lanes have been spliced out. (C) Full-length HA mRNA was translated in the presence of wild-type or MI8-5 SP cells for 60 min at 27°C. Samples were split, with half being chased for 5 h at 32°C. Samples were immunoprecipitated with HA antisera or the conformational antibodies F2 (IT2 and monomeric NT) and N2 (trimers). All samples were resolved via nonreducing (NR) and reducing (RD) 7.5% SDS-PAGE. (D) HA was translated in the presence of wild-type or MI8-5 SP cells in the presence or absence of DNJ and either immunoprecipitated with HA antisera or subjected to a GST-calreticulin pulldown. (E) HA was translated as in D. Radiolabeled HA treated with DNJ was alkylated with 20 mM NEM and immunoprecipitated with HA antisera. Radiolabeled HA produced in the absence of DNJ was subjected to an additional 3.5-h incubation with 1 mM cycloheximide to allow for complete glucose trimming. Samples were alkylated and subjected to immunoprecipitation by HA antisera. Samples were then split and treated with jack bean α-mannosidase (JBM) at 37°C for 18 h as indicated. All samples were resolved via 7.5% SDS-PAGE.
Slowing the translation rate can increase suboptimal glycosylation efficiencies. The rabbit reticulocyte lysate in vitro translation system has an ∼10-fold slower translation rate (∼0.5 amino acids per second) than what is observed in live mammalian cells (∼5 amino acids per second; Újvári et al., 2001). The reticulocyte lysate translation system can be coupled with semipermeabilized (SP) cells to support the targeting of in vitro translated proteins to intact ER membranes from various cell types (Wilson et al., 1995; Wang et al., 2005). SP cells are generated by treating cells with low concentrations of digitonin, which permeabilizes the cholesterol-rich plasma membranes while leaving the delimiting endomembranes of the cell, including the ER, intact.
To alleviate the hypoglycosylation of HA observed in MI8-5 cells, the in vitro translation of HA was coupled with MI8-5 SP cells. When 35S-labeled HA was translated in the presence of SP wild-type and MI8-5 cells, it received all seven of its glycans (Fig. 2 B, lanes 3 and 4). A subpopulation containing six glycans, which was easily discernible by SDS-PAGE, was also produced in MI8-5 SP cells.
Analysis by nonreducing SDS-PAGE indicated that HA matured properly in both wild-type and MI8-5 SP cells as it formed multiple disulfide bonds, which account for it attaining its native form (NT; Fig. 2 B, lanes 1 and 2; Braakman et al., 1991). HA synthesized in wild-type and MI8-5 SP cells was also probed with conformation-specific antibodies. 35S-labeled HA was translated for 1 h in the presence of wild-type or MI8-5 SP cells and either analyzed directly or chased for an additional 5 h before alkylation to block free sulfhydryls. The alkylated lysates were immunoprecipitated with anti-HA polyclonal antibodies or two different conformation-specific monoclonal antibodies termed F2, which recognizes the HA oxidative intermediate IT2 and the fully oxidized NT, or the N2 monoclonal antibody, which binds native trimeric HA (Fig. 2 C; Braakman et al., 1991). After 1 h of translation, similar levels of HA were immunoprecipitated with the F2 antibody from both wild-type and MI8-5 SP cells, and these levels decreased after 5 h of chase (Fig. 2 C, lanes 3, 4, 9, and 10). The decrease in F2 binding associated with increased N2 binding indicated that HA had properly folded and trimers had formed in both wild-type and MI8-5 SP cells (Fig. 2 C, lanes 11 and 12). These results indicate that HA matures correctly in MI8-5 SP cells, as it is properly glycosylated, oxidized, and assembled into native homotrimers.
To determine whether GT1 reglucosylates HA in MI8-5 SP cells, the presence of monoglucosylated glycans was probed by monitoring binding to a bacterial expressed and purified lectin fusion protein, GST-calreticulin. Treatment with the glucosidase inhibitor N-butyl deoxynojirimycin (DNJ) supports the accumulation of triglucosylated glycoproteins in wild-type SP cells, whereas in MI8-5 SP cells, monoglucosylated glycoproteins should accumulate if they have been targeted by GT1. As expected, GST-calreticulin efficiently bound HA from wild-type SP cells in a glucosidase-dependent manner (Fig. 2 D, lanes 5 and 6). In MI8-5 SP cells, GST-calreticulin efficiently bound to HA; however this binding was not inhibited by glucosidase inhibition (Fig. 2 D, lanes 7 and 8).
To verify that HA was reglucosylated in SP MI8-5 cells by GT1, 35S-labeled HA was subjected to jack bean α-mannosidase digestion. Terminal glucose residues present on N-linked glycans can prevent the cleavage of A-branch mannose residues by α-mannosidase but allow the digestion of the unglucosylated B and C branch mannoses (Fig. 1 B; Hammond et al., 1994; Hebert et al., 1995). Trapping HA in the triglucosylated state in wild-type SP cells with DNJ treatment resulted in the protection of HA glycans from α-mannosidase digestion when compared with HA that was chased to an unglucosylated form (Fig. 2 E, compare lanes 1 and 2 to 3 and 4). Inclusion of the glucosidase inhibitor during HA synthesis in MI8-5 SP cells resulted in a significant portion of HA glycans being protected against mannose removal when compared with the unglucosylated control, which is indicative of the presence of monoglucosylated glycans (Fig. 2 E, compare lanes 5, 6 and 7, and 8). Collectively, these results indicated that properly folded monoglucosylated HA was efficiently produced by GT1 reglucosylation in MI8-5 SP cells.
GT1 recognizes calnexin- but not calreticulin-associated glycans on HA
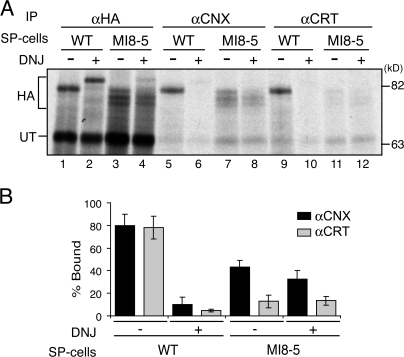
The lectin chaperones calnexin and calreticulin bind to monoglucosylated glycans (Hammond et al., 1994; Hebert et al., 1995; Kapoor et al., 2003). Calnexin associates with the membrane-proximal stem domain glycans of HA, whereas calreticulin binds to its top globular glycans (Hebert et al., 1997; Daniels et al., 2003). To determine whether GT1 reglucosylates a specific region of HA, the association of calnexin and calreticulin with HA was examined in MI8-5 SP cells and compared with wild-type SP cells.
35S-labeled HA was coimmunoprecipitated with anti-calnexin or -calreticulin antibodies after 1 h of translation. Both calnexin and calreticulin efficiently bound to HA from wild-type SP cells (Fig. 3, A and B; Hebert et al., 1995, 1996, 1997; Peterson et al., 1995; Molinari et al., 2002). This binding was abolished with the addition of the glucosidase inhibitor DNJ, which supported the accumulation of triglucosylated HA, indicating that the association was glycan-dependent. In contrast, only calnexin bound HA at significant levels in MI8-5 SP cells. DNJ treatment did not inhibit calnexin binding because DNJ promotes the accumulation of monoglucosylated substrates in MI8-5 SP cells (Fig. 3, A [lanes 7, 8, 11, and 12] and B). GT1 appears to recognize the membrane-proximal domain of HA, resulting in the reglucosylation of the glycans that interact with the integral membrane chaperone calnexin. The lack of observable calreticulin binding was likely caused by the different topologies of the lectin chaperones in their native environment rather than by differences in their substrate recognition because GST-calreticulin bound HA from SP cell lysates (Fig. 2 D). Therefore, GT1 specifically reglucosylates glycans on the slow-folding membrane-proximal stem domain of HA.
Figure 3.
HA glycans associated with calnexin are preferentially reglucosylated by GT1. (A) HA was translated in the presence of wild-type or MI8-5 SP cells treated with DMJ in the presence or absence of DNJ at 27°C for 60 min. After lysis of the SP cells, radiolabeled HA was immunoprecipitated with polyclonal HA, calnexin (αCNX), or calreticulin (αCRT) antisera. Samples were resolved via 7.5% SDS-PAGE. (B) Quantifications of A (n = 3). Error bars show SD.
Unfolded Protein Response (UPR) is partially activated in MI8-5 cells, but ER protein levels are unaffected
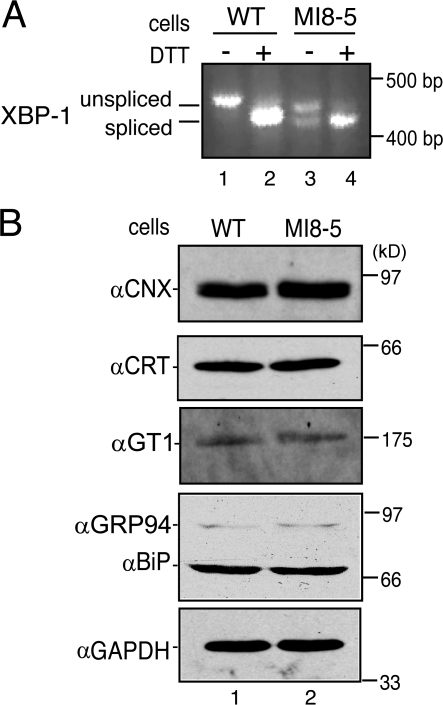
Inefficient glycan transfer can increase the level of misfolded proteins in the ER and potentially activate the UPR (Mori, 2000; Schroder and Kaufman, 2005). The UPR regulates the expression of a large number of ER proteins (Travers et al., 2000) and its activation could change the expression levels of calnexin and calreticulin, accounting for their observed binding profiles to HA within MI8-5 SP cells. UPR activation was investigated by analyzing transcripts of the transcription factor XBP-1 by RT-PCR. When the UPR is activated, the mRNA of XBP-1 is processed to an activated spliced form and can drive the expression of numerous ER proteins (Schroder and Kaufman, 2005). Wild-type CHO cells contained only the unspliced XBP-1 mRNA unless treated with the UPR inducer dithiothreitol (DTT; Fig. 4 A, lanes 1 and 2). However, ∼50% of the XBP-1 mRNA was found to be in the spliced form in MI8-5 cells, and this percentage increased to ∼100% after DTT treatment. Therefore, the UPR was constitutively active in MI8-5 cells compared with its parental wild-type CHO cells.
Figure 4.
Chaperone levels remain unchanged in MI8-5 cells despite UPR induction. (A) RT-PCR of XBP-1 RNA isolated from wild-type and MI8-5 cells incubated in the presence or absence of 5 mM DTT for 3 h. Primers were directed against XBP-1 cDNA. Samples were resolved on 2% agarose gel. Unspliced and spliced XBP-1 are designated. (B) Steady-state levels of ER proteins calnexin, calreticulin, GT1, GRP94, and BiP were immunoblotted from wild-type and MI8-5 cells. 50 μg of whole-cell lysate was loaded on each lane and analyzed via 10% SDS-PAGE. GAPDH serves as a non-ER loading control.
To determine if UPR activation altered the expression of ER chaperones, the steady-state levels of various chaperones were examined in both wild-type and MI8-5 cell lysates (Fig. 4 B). Immunoblotting for the ER chaperones calnexin, calreticulin, GT1, GRP94, and BiP revealed no changes in protein levels for any of the ER chaperones and folding enzymes analyzed, with respect to the non-ER protein control GAPDH. Although the UPR appeared to be activated in MI8-5 cells, the differential binding of calnexin and calreticulin to HA within these cells was not a consequence of alterations in chaperone expression levels.
GT1 does not reglucosylate translocon-associated nascent chains
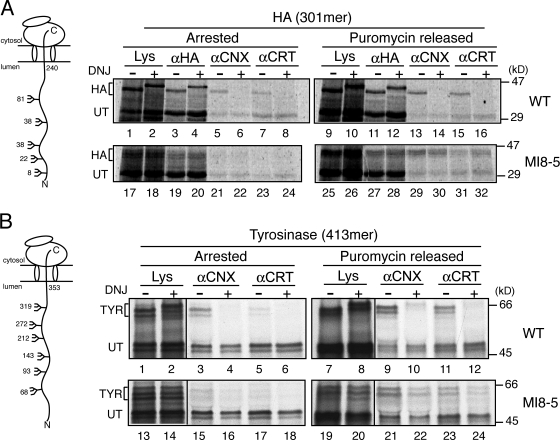
During HA translocation into the ER lumen, glucosidases I and II rapidly cleave HA glycans to a monoglucosylated state, supporting the cotranslational association of calnexin and calreticulin (Chen et al., 1995; Hebert et al., 1997; Molinari and Helenius, 2000; Daniels et al., 2003). To determine if GT1 can access and reglucosylate translocon-associated nascent chains to drive their cotranslational rebinding to calnexin and calreticulin, reglucosylation of ribosomal-arrested nascent chains was analyzed. Initially, a truncated HA mRNA (301mer) lacking a stop codon was transcribed and translated in the presence of wild-type and MI8-5 SP cells. Translation of the HA 301mer results in ribosome- and translocon-arrested chains with ∼240 lumenally residing residues containing five glycans (Fig. 5 A; Daniels et al., 2003). Both calnexin and calreticulin bound to the translocon-associated HA 301mer in wild-type SP cells (Fig. 5 A, lanes 5–8; Daniels et al., 2003). However in MI8-5 SP cells, neither calnexin nor calreticulin were found to interact with the translocon-associated HA 301mer (Fig. 5 A, lanes 21–24). In contrast, both calnexin and calreticulin binding to HA was observed in wild-type and MI8-5 SP cells when the protein was released from the ribosome and translocon with the protein synthesis inhibitor puromycin (Fig. 5 A, lanes 13–16 and 29–32). This indicated that GT1 reglucosylation of HA required the nascent chain to be dissociated from the ribosome and translocon and that calnexin and calreticulin binding to ribosome-arrested nascent chains occurred in a glucosidase-dependent manner and was not a result of GT1 reglucosylation.
Figure 5.
GT1-mediated reglucosylation occurs posttranslationally. (A) 301mer ribosome-arrested HA was translated in the presence of wild-type or MI8-5 SP cells with the addition of DMJ in the presence or absence of DNJ for 60 min at 27°C. Samples were then split and half were incubated with 1 mM puromycin for 45 min. Radiolabeled HA was directly analyzed or immunoprecipitated with polyclonal HA, calnexin (αCNX), or calreticulin (αCRT) antisera, as indicated. Samples were analyzed via 7.5–12% gradient reducing SDS-PAGE. (B) The 413mer ribosome-arrested tyrosinase was translated and analyzed as in A. Black lines indicate that intervening lanes have been spliced out.
To explore the generality of the requirement of GT1 for ribosome-released polypeptides, ribosome-arrested chains of the type-I membrane glycoprotein tyrosinase containing ∼350 lumenally exposed residues and 6 glycans were monitored for GT1-mediated lectin chaperone association (Fig. 5 B). As observed for HA, both calnexin and calreticulin bound to the tyrosinase 413mer in wild-type SP cells, which is in agreement with previous studies (Fig. 5 B, lanes 3–6; Wang et al., 2005). Similar to the results observed with HA, neither lectin chaperone was found to associate with the ribosome-arrested 413mer translated in the presence of MI8-5 SP cells (Fig. 5 B, lanes 15–18). Lectin chaperone binding to the tyrosinase 413mer was only detectable after the release of the nascent chains with puromycin (Fig. 5 B, lanes 21–24). This association in MI8-5 SP cells occurred in a glucosidase-independent manner because calnexin and calreticulin binding was not abolished with DNJ treatment (Fig. 5 B, compare lanes 9–12 and 21–24). Therefore, GT1-mediated reglucosylation of both HA and tyrosinase occurred after release of the nascent chains from the translocon-associated environment. It should be noted that the reglucosylation of the puromycin-released chains may be caused by the emergence of the C-terminal ribosome/translocon-associated regions in the ER lumen. If this is the case, the GT recognition signal for both HA and tyrosinase would have to be located in these occluded regions.
GT1 recognition is determined by the folding status of HA
During the productive folding of HA, glycans involved in calnexin interactions are targeted for reglucosylation by GT1, whereas glycans associated with calreticulin binding remain unmodified in MI8-5 cells (Fig. 3 A). Because calnexin and calreticulin interactions can only be mediated by GT1 activity in these cells, this implies that the membrane-proximal stem domain is the only region sufficiently structurally perturbed to attract GT1 during the folding process. To assess the folding sensor capabilities of GT1, the reglucosylation of HA was monitored under conditions where folding was disrupted with the reducing agent DTT followed by the posttranslational oxidation of HA (Fig. 6; Braakman et al., 1992a).
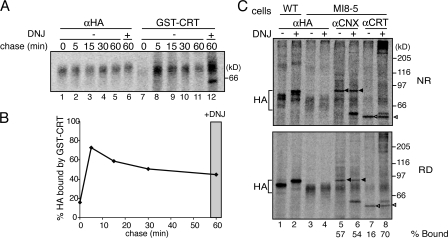
Figure 6.
Time course for GT1 reglucosylation of HA. (A) HA was transiently overexpressed in wild-type or MI8-5 cells. Transfected cells were pulse labeled in the presence of 4 mM DTT for 5 min and chased under nonreducing conditions for the indicated times. 0.5 mM DNJ was present throughout the experiment for lanes 6 and 12. Radiolabeled HA was isolated by either immunoprecipitation with HA antisera or GST-calreticulin (GST-CRT) pulldown and resolved via 7.5% SDS-PAGE. (B) Quantifications of A. Fraction bound by GST-calreticulin pulldown versus total immunoprecipitated HA, normalized to maximum reglucosylation observed in the presence of DNJ. (C) HA was transiently overexpressed as in A. Transfected cells were pulse labeled in the presence of 4 mM DTT for 30 min, washed extensively to remove the reductant, and chased under nonreducing conditions for 30 min before alkylation and immunoprecipitation. 0.5 mM DNJ was present throughout the starvation, pulse, and chase period as indicated. Radiolabeled HA was immunoprecipitated with polyclonal HA, calnexin (αCNX), or calreticulin (αCRT) antisera. Radiolabeled calnexin (closed triangle) and calreticulin (open triangle) are indicated. Samples were resolved via 7.5% nonreducing (top) and reducing (bottom) SDS-PAGE.
Cells were transfected with HA, radiolabeled in the presence of DTT, and chased under oxidizing conditions. To analyze the time course of HA reglucosylation during delayed disulfide bond formation, GST-calreticulin was used as a probe for monoglucosylated glycans in MI8-5 cells. Interestingly, HA received all seven glycans during reducing pulse conditions (Fig. 6 C, RD, lanes 1 and 3). However, GST-calreticulin did not bind HA, indicating that reduced HA was not reglucosylated (Fig. 6 A, lane 7). Reductant has been previously shown to have no effect on the reglucosylation capabilities of Schizosaccharomyces pombe GT1 even though it possesses multiple Cys residues (Fernandez et al., 1998). Therefore, the lack of GST-calreticulin binding to reduced HA may be caused by the inability of GT1 to modify severely misfolded substrates or by the inactivation of the hamster GT1 by DTT.
Upon restoration of oxidizing conditions, HA was efficiently reglucosylated in MI8-5 cells, reaching a maximum level of 73% GST-calreticulin binding after 5 min of oxidation. This level gradually decreased in a time-dependent manner as the protein oxidized to its native form (Fig. 6 A, lanes 8–11; and not depicted). The inclusion of DNJ in the chase media trapped the reglucosylated species, which is representative of the maximal level of GST-calreticulin binding (Fig. 6 A, lane 12). As previously observed in CHO15B cells, HA maturation initiated under posttranslational disulfide bond formation conditions in MI8-5 cells matured to native trimers, as indicated by immunoprecipitation with the trimer-specific HA antibody (Copeland et al., 1988; Braakman et al., 1992a; unpublished data). Therefore, GT1 reglucosylation of HA was transient and the extent of reglucosylation was dictated by the degree of the folding disruption.
To determine which domains on HA were reglucosylated, chaperone binding to the posttranslationally oxidized HA was monitored after 30 min of oxidation (Fig. 6 C). In MI8-5 cells, calnexin binding was observed in the absence and presence of the glucosidase inhibitor DNJ (Fig. 6 C, lanes 5 and 6). In contrast, calreticulin binding to posttranslationally oxidized HA dramatically increased when glucosidase activity was inhibited by DNJ treatment (Fig. 6 C, lane 8, 70% bound). This indicated that the head domain glycans of HA were transiently reglucosylated during the posttranslational oxidation of HA. This is in sharp contrast to the cotranslational oxidation SP cell program, for which only the stem domain calnexin binding glycans were reglucosylated during the maturation of HA, leaving the head domain glycans unmodified. The reglucosylation of the calreticulin binding glycans was transient because calreticulin binding was only weakly observed in the absence of DNJ (Fig. 6 C, lane 7, 16% bound). Collectively, these results indicated that GT1 transiently reglucosylated HA as it folded and that the level of reglucosylation was modulated by the degree that HA was in nonnative conformations.
GT1 persistently reglucosylates terminally nonnative HA
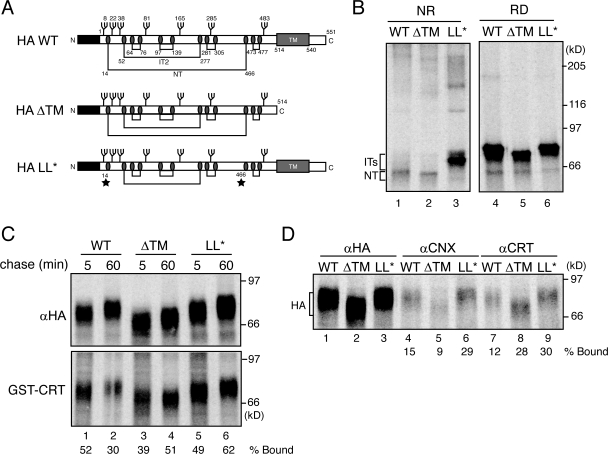
GT1 can act as a folding sensor and target slow-folding regions of HA for reglucosylation when disulfide bond formation is delayed. To expand upon the role of the folding sensor function of GT1, mutant forms of HA terminally, rather than transiently, impaired in their maturation pathway were probed for their recognition by GT1. Mutant substrates used in the reglucosylation assay included an oxidation mutant missing both Cys residues that form the large loop disulfide (Cys14 and Cys466, termed HA-LL*; Fig. 7 A) and a truncated HA mutant, which lacks the C-terminal transmembrane segment (HA-ΔTM; Fig. 7 A). HA-LL* is missing the critical disulfide bond, which stabilizes the membrane-proximal stem domain and is responsible for the large shift observed for NT formation by nonreducing SDS-PAGE. In contrast, HA-ΔTM is soluble and unable to oligomerize, resulting in its delayed trafficking to the cell surface; however, all of its native disulfide bonds form efficiently (Singh et al., 1990).
Figure 7.
GT1 reglucosylates terminally nonnative HA. (A) Schematic of WT HA, HA with a premature stop codon at aa 514 (HA-ΔTM), and HA Cys14Ser/Cys466Ala (HA-LL*). All three constructs contain seven N-linked glycosylation sites. Both HA WT and HA-ΔTM have the capacity to form six disulfide bonds and the three oxidative forms of HA (IT1, IT2, and NT), whereas, HA-LL* is lacking the Cys (ovals) involved in the large loop disulfide (stars). (B) HA constructs were transiently overexpressed in wild-type CHO cells and radiolabeled for 10 min. HA was immunoprecipitated from cell lysates with HA antisera and analyzed via both nonreducing and reducing SDS-PAGE. (C) HA was expressed, as in B, in MI8-5 cells. Cells were radiolabeled for 10 min in the presence of 4 mM DTT and chased for the indicated time periods. HA was isolated by immunoprecipitation with HA antisera or by GST-calreticulin (GST-CRT) pulldown. Quantifications represent the percent bound to GST-calreticulin relative to the total immunoprecipitated HA. (D) HA was analyzed as in C. Cells were radiolabeled for 10 min in the presence of 4 mM DTT and chased for 60 min under nonreducing conditions. HA was isolated by immunoprecipitation with HA, calnexin (αCNX), or calreticulin (αCRT) antisera. Samples were resolved by 7.5% reducing SDS-PAGE and visualized by phosphorimaging. Quantifications represent the percent binding of the total immunoprecipitated HA to the lectin chaperones.
The native oxidized form of HA accumulated for both HA wild type and ΔTM after 10 min of oxidation (Fig. 7 B, lanes 1 and 2, NT). HA-ΔTM displayed an increased mobility caused by its truncated C terminus (Fig. 7 B, lane 5). HA-LL* could not form NT but was trapped as oxidative intermediates IT1 and IT2, indicating that disulfide formation was deficient in the large loop deletion mutant (Fig. 7 B, lane 3). HA wild type and LL* migrated at the same position by reducing SDS-PAGE (Fig. 7 B, lanes 4 and 6).
To investigate the reglucosylation of these HA mutants at late stages of their maturation in MI8-5 cells, proteins were synthesized in the presence of reductant and allowed 5 or 60 min for oxidation (Fig. 7 C). The total reglucosylated fraction of HA was isolated by GST-calreticulin pulldowns. The maximal level of reglucosylation was observed for HA wild type at 5 min. The total fraction of reglucosylated HA wild type decreased by close to half after 60 min of oxidation (Fig. 7 C, lane 2). Interestingly, a greater fraction of HA-ΔTM and HA-LL* were associated with GST-calreticulin after 60 min of oxidation (51 and 62%, respectively; Fig. 7 C, lanes 4 and 6), indicating that the mutant form of HA that was not undergoing productive maturation was persistently more efficiently reglucosylated with time.
To determine which chaperones bound the reglucosylated proteins, calnexin and calreticulin-associated HA species were isolated by coimmunoprecipitation with antibodies against either calnexin or calreticulin. A similar level of wild-type 35S-labeled HA was coimmunoprecipitated with calnexin and calreticulin antisera (Fig. 7 D, lanes 4 and 7). Both these levels substantially increased with HA-LL*, which is indicative of more efficient recognition by GT1 of glycans that support both calnexin and calreticulin binding for the folding-defective mutant (Fig. 7 D, lanes 6 and 9). For the oligomerization mutant HA-ΔTM, weak binding by calnexin was observed, which is in agreement with previously published studies (Hebert et al., 1997). Interestingly, calreticulin was found efficiently associated with HA-ΔTM, even after 60 min of oxidation, which could be because of persistent reglucosylation of the soluble protein which is more lumenally accessible (Fig. 7 D, lane 8). Collectively, these results indicated that not only can GT1 act as a folding sensor on HA that is temporarily disrupted in its folding pathway but it can also efficiently recognize permanently defective HA mutants.
Discussion
The reglucosylation by GT1 of a maturing glycoprotein in the mammalian ER was analyzed to provide insight into the role of this key quality control enzyme in the early secretory pathway. We found that GT1 targets slow-folding domains of a glycoprotein in a region-specific manner to direct chaperone binding to immature regions. GT1-directed differential chaperone binding is modulated by transient as well as permanent structural defects associated with terminally misfolded or unassembled substrates. GT1 acts posttranslationally after clearance of the nascent chain from the translocon environment, indicating that the lectin chaperone binding cycle is initiated by GT1 at a later stage in maturation.
The lack of mammalian cellular data concerning reglucosylation in the ER of a maturing substrate can be attributed to the difficulty in following its activity in live cells. The use of the CHO MI8-5 mutant cell line permits the isolation of the reglucosylation process because they assemble Man9GlcNAc2-P-P-dolichol rather than the fully glucosylated Glc3Man9GlcNAc2-P-P-dolichol because of a defect in the alg6 gene (Quellhorst et al., 1999). Pharmacological inhibition of glucosidase activity in MI8-5 cells traps reglucosylated side chains by inhibiting glucosidase removal of the reglucosylated glycan. A similar approach has been used to study GT1 activity in unicellular organisms, including Trypanosoma cruzi, that naturally transfer Man9GlcNAc2 and mutant S. pombe (gls2/alg6 double mutant; Ganan et al., 1991; Fernandez et al., 1998). In contrast to these studies, which characterized the global effect of reglucosylation, our study provides a detailed analysis of the reglucosylation of a single dynamic maturing glycoprotein in mammalian cells.
In MI8-5 cells, HA associated with calnexin and not calreticulin when under normal physiological conditions where lectin chaperone binding could only be mediated through reglucosylation. Previous studies have revealed that calnexin interacts with the membrane-proximal glycans of HA, whereas calreticulin binds to the globular head domain glycans that extend deeper into the ER lumen (Hebert et al., 1997; Daniels et al., 2003). The head domain of HA folds extensively cotranslationally and is the first native domain of HA formed as probed by conformation-specific antibodies and disulfide bond formation (Braakman et al., 1991, 1992a; Chen et al., 1995). Completion of the membrane-proximal stem domain involves the posttranslational formation of large loop disulfide Cys14-Cys466. It is the last domain to form before HA trimerization. HA is an obligatory substrate of calnexin, but not calreticulin, emphasizing the requirement of efficient folding in the stem domain to attain its native state (Molinari et al., 2004). Taylor et al., (2003) found that purified GT1 preferentially reglucosylates glycopeptides possessing a hydrophobic patch C-terminal to the carbohydrate. For HA, stem domain glycans at Asn8 and Asn22 both possess these signature hydrophobic patches (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200712068/DC1). This is in agreement with recent studies where reglucosylation only occurs locally in relationship to the protein lesion (Ritter and Helenius, 2000; Ritter et al., 2005). However, a conflicting study found that GT1 was able to reglucosylate a glycan positioned 40 Å from the protein defect (Taylor et al., 2004).
Transient reglucosylation of HA was observed when disulfide bond formation was delayed by the inclusion of reductant in the radioactive pulse followed by an oxidative chase period. This procedure prevented cotranslational disulfide bond formation and supported efficient recognition of the glycosylation consensus sites by the oligosaccharyltransferase in live cells. An active cycle of HA reglucosylation existed that peaked shortly after the initiation of oxidation. Interestingly, GT1 activity was not observed on completely reduced HA, likely because of the severity of the unfoldedness of the substrate or inactivation of GT1 by reduction. The starting material in the posttranslational oxidation reaction had all disulfide bonds reduced, including linkages found in both the top globular and the membrane-proximal domains. GT1 appeared to modify all nonnative domains on HA upon restoration of oxidizing conditions because both calnexin and calreticulin binding was observed. The reglucosylation of the head domain was extremely transient, indicating that the activity of GT1 toward this domain was diminished as disulfide bonds were rapidly formed. The time-dependent decrease in reglucosylation observed corresponded to an increase in oxidation and folding of HA.
The folding sensor abilities of GT1 were further analyzed using mutants of HA that possessed maturation defects. Reglucosylation supported both calnexin and calreticulin binding when the oxidation of HA was inhibited by mutating the two Cys involved in the formation of the large loop disulfide that completes its oxidation (Cys14 and Cys466; HA-LL*). The loss of a single stabilizing disulfide bond in RNase B led to its in vitro reglucosylation at a position local to the structural perturbation (Ritter et al., 2005). As the glycan consensus sequence was moved distal to the mutated Cys residue, reglucosylation efficiency decreased. Collectively, these results support the hypothesis that GT reglucosylates aberrant domains.
HA persists in its association with calnexin until it oligomerizes in the ER (Hebert et al., 1995, 1996). The removal of the transmembrane region of HA (HA-ΔTM) generates a mutant that can be oxidized to its native form but cannot oligomerize (Singh et al., 1990). The membrane anchor deletion mutant continued to be reglucosylated, supporting its binding to calreticulin. In contrast to the wild-type membrane-anchored fully oxidized monomeric protein that only associated with calnexin, persistent calreticulin binding was observed with the HA-ΔTM, likely because of its soluble characteristics increasing its access to the ER lumen (Hebert et al., 1997). GT1 possibly recognizes a hydrophobic surface on the unassembled protein that is hidden within the oligomeric interface. A hydrophobic patch is found at the trimer interface in the stem region of HA. These results are consistent with in vitro studies using the multimeric soybean agglutinin (Keith et al., 2005). GT1 appears to play a role in the retention of orphan subunits in the ER, ensuring that oligomerization occurs before exiting the ER (Gardner and Kearse, 1999). Together, the observed reglucosylation of HA-LL* and -ΔTM supporting the recruitment of both calnexin and calreticulin demonstrate that GT1 is capable of recognizing terminally defective oxidation and oligomerization species.
Lectin chaperone binding initiated by GT1 did not occur cotranslationally or for ribosomal- and translocon-associated nascent chains. Only full-length chains or puromycin-released ribosomal-arrested chains were reglucosylated for both HA and tyrosinase. These results are supportive of the localization of GT1 to late or smooth regions of the ER, as observed by electron microscopy and proteomic studies (Cannon and Helenius, 1999; Zuber et al., 2001; Gilchrist et al., 2006). Cross-linking studies have uncovered a large multiprotein ER complex containing GT1, BiP, GRP94, CaBP1, PDI, ERdj3, cyclophilin B, ERp72, GRP170, and SDF2-L1 (Meunier et al., 2002). Interestingly, the lectin chaperones and glucosidases are absent from this complex. The posttranslational recognition of later folding intermediates and uncomplexed subunits by GT1 may also be dictated by its localization within complexes in the ER.
HA folding and oxidation appear to take place after the maturing substrate is released from the lectin chaperones because when release was inhibited, so was oxidation (Hebert et al., 1995, 1996). The posttranslational involvement of GT1 provides a window for folding to occur by initially having a single round of monoglucosylated side chains created through deglucosylation supporting the first round of binding to calnexin and calreticulin. If a domain has not folded completely or oligomerized successfully after this first round of binding by the lectin chaperones, the calnexin cycle would then be elicited through GT1 recognition. In addition, BiP has been shown to sequester irreparably misfolded proteins from calnexin and calreticulin, precluding GT1 recognition (Molinari et al., 2005). There has been debate on the tenacity of the GT1-directed calnexin cycle. A recent study in GT1−/− cells has revealed variability in calnexin binding to glycoproteins in the absence of reglucosylation (Solda et al., 2007). Glycoproteins that are likely unrecognized by GT1 display an unchanged calnexin binding profile, whereas other substrates were released earlier from the lectin chaperones. Surprisingly, HA exhibited prolonged calnexin binding and severe delays in its trafficking out of the ER in the GT1−/− cells, demonstrating that GT1 may also somehow provide cues for release of substrates from the lectin chaperones.
GT1 is a sensitive and efficient sensor of conformational instability in the ER. It displays an ability to recognize specific nonnative regions within glycoproteins during their maturation and reglucosylate glycans local to these structural imperfections. GT1 recognizes and reglucosylates folding intermediates, orphan subunits of multimeric complexes, and terminally aberrant or misfolded proteins. These reglucosylated substrates are retained in the ER through their binding to the lectin chaperones. The reliance of a maturing glycoprotein on GT1 is dependent on the properties of the substrate (Solda et al., 2007). Mutations in the Arabidopsis thaliana GT1 were recently found to permit the release from the ER of a defective cell surface receptor (Jin et al., 2007). In this paper, we demonstrated that the extent of the response by GT1 to a folding defect was directly related to the maturation capabilities of the substrate. Reglucosylation occurred after the protein was released from the translocon, supporting GT1 involvement in monitoring the later stages of maturation in the ER. We analyzed the reglucosylation of the model glycoprotein HA to further our knowledge of the cellular activities of GT1. A complete characterization of the activity of GT1 is essential to understand the basis for the growing number of ER storage diseases, which involve the ER retention and subsequent degradation of proteins evaluated as aberrant by the ER quality control system.
Materials and methods
Reagents
Wild-type and MI8-5 CHO cells were a gift from S. Krag (Johns Hopkins University, Baltimore, MD). DME, α-MEM, penicillin-streptomycin, FCS, and Lipofectamine 2000 were purchased from Invitrogen. The T7 mMessage mMachine kit was obtained from Ambion. The RNeasy kit and EasyTag [35S]Cys/Met were purchased from QIAGEN and PerkinElmer, respectively. Components of the SP cell translation assays were acquired from Promega. Calreticulin antisera (PA3-900) and reduced glutathione Sepharose 4B beads were purchased from Affinity BioReagents and GE Healthcare, respectively. The GST-calreticulin pGEX-3X plasmid, HA antibodies, and GT1 antisera were gifts from M. Michalak (University of Alberta, Edmonton, Alberta), A. Helenius (Eidgenössische Technische Hochschule Zürich, Zurich, Switzerland), and A. Parodi (Leloir Institute Foundation, Buenos Aires, Argentina), respectively. All other reagents were purchased from Sigma-Aldrich.
Plasmids and in vitro transcription
HA cDNA was cloned into pBluescript SK(−). The 301mer cDNA is a Met-enhanced construct with six extra Met residues placed at positions 100–102 (Tyr-Asp-Val) and 136–137 (Ser-Asn-Ala; Daniels et al., 2003). The full-length HA mRNA was generated by linearization of pBluescript containing wild-type HA cDNA, followed by run-off transcription, and the mRNA was purified with the RNeasy kit. The 301mer HA mRNA and the 413mer tyrosinase mRNA were produced as described previously (Daniels et al., 2003; Wang et al., 2005).
Preparation of SP cells and translation
SP CHO cells were prepared from near confluent dishes (∼1 × 107 cells) as previously described (Wilson et al., 1995; Francis et al., 2003; Wang et al., 2005). Full-length and truncated HA and tyrosinase mRNAs were translated and translocated into digitonin-permeabilized cells as previously described (Újvári et al., 2001; Wang et al., 2005). Samples were prewarmed at 27°C for 10 min before the addition of mRNA, when glucosidase inhibitors were used. After translation at 27°C, samples were alkylated with 20 mM N-ethylmaleimide (NEM) to block free sulfhydryls. For the release of arrested chains from the ribosome, 1 mM puromycin was added, where indicated, to the translation mix and incubated for an additional 45 min. Jack bean α-mannosidase digestion was performed as previously described (Hebert et al., 1995).
Transfection of CHO cells and pulse-chase analysis
CHO cells were infected with recombinant vaccinia virus expressing T7 RNA polymerase (Fuerst et al., 1986) and transfected with Lipofectamine 2000 according the manufacturer's instructions, and synthesized HA was radiolabeled as previously described (Braakman et al., 1991). In brief, cells were starved in Cys/Met-free DME containing 0.5 mM 1-deoxymannojirimycin (DMJ) and 0.5 mM DNJ, where indicated, for 1 h and then labeled with 220 μCi of [35S]Cys/Met for the indicated time periods at 34°C. Immediately after the pulse period, cells were washed twice with cold PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4) containing 20 mM NEM, pelleted, and lysed with either MNT (20 mM 2-[N-morpholino] ethanesulfate, 30 mM Tris-Cl, pH 7.5, and 0.5% Triton X-100) containing 20 μM leupeptin, 1.5 μM aprotinin, 1 μM pepstatin A, 100 μM PMSF, 50 μM LLnL, and 20 mM NEM or 2% CHAPS (3-[(cholamidopropyl)-dimethylammonio]-1-propanesulfonate)/HBS (200 mM NaCl, 50 mM Hepes, pH 7.5) with the appropriate protease inhibitors when chaperone coimmunoprecipitations were performed to maintain protein–protein interactions. For the reducing pulse-chase experiments, pulse media contained 4 mM DTT, and the culture dishes were extensively washed with chase media (α-MEM, 5% FCS) to remove the reductant before the chase period.
Immunoprecipitation and SDS-PAGE
Anti-HA immunoprecipitations were performed as described previously (Hebert et al., 1995; Daniels et al., 2003). Anti-calnexin and anti-calreticulin precipitations were also performed as previously described (Daniels et al., 2003; Wang et al., 2005). Radiolabeled samples were resolved by SDS-PAGE using standard protocols and scanned in a phosphoimager (FLA-500; Fuji). Data were analyzed and quantified using Muti Gauge v. 2.02 (Fuji).
GST-calreticulin pulldown assay
Recombinant GST-calreticulin was expressed and purified as previously described (Baksh and Michalak, 1991). After cell lysis, the postnuclear supernatant was incubated with 8 μg of purified GST-calreticulin prebound to reduced glutathione Sepharose 4B beads and rotated end-over-end overnight at 4°C. After centrifugation at 1,000 g for 5 min, samples were washed twice with 0.5% CHAPS/HBS and resuspended in gel loading buffer.
RT-PCR and immunoblotting
Cytoplasmic RNA was isolated from wild-type or MI8-5 CHO cells incubated in the presence or absence of 5 mM DTT using the RNeasy kit according to the manufacturer's instructions. The resulting templates were used to detect XBP-1 mRNA using the Superscript First-Strand synthesis system for RT-PCR (Invitrogen), according to the manufacturer's instructions, using specific primers for mouse XBP-1 (Lee et al., 2002). PCR-amplified cDNA products were resolved on 2% agarose gels and visualized with BioMax software (Kodak).
For immunoblots, confluent cells on 10-cm tissue culture dishes were washed three times in PBS and detached with 3 ml of 0.25% trypsin/EDTA. Cells were collected in PBS containing 100 μg/ml of soybean trypsin inhibitor and centrifuged at 250 g for 5 min at 4°C. After cell lysis, samples were vortexed for 10 min at 4°C and centrifuged at 18,000 g to isolate the whole-cell lysate. 50 μg of total protein was loaded per lane and resolved by SDS-PAGE. Samples were then transferred to PVDF membranes and immunoblotted according to standard procedures.
Online supplemental material
Fig. S1 depicts the hydropathic profiles of HA glycan regions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200712068/DC1.
Supplemental Material
Acknowledgments
We are grateful to Dr. S. Krag for her generous gift of wild-type and MI8-5 CHO cell lines. We would also like to thank Drs. A. Helenius, A. Parodi, and M. Michalak for providing HA antisera, GT1 antisera, and the GST-calreticulin expression plasmid, respectively. We would like to acknowledge members of the Hebert laboratory, including J. Cormier and Dr. R. Daniels, for thoughtful discussions.
This work was supported by US Public Health grant CA79864 (to D.N. Hebert). B.R. Pearse was partially supported by an National Institutes of Health Chemistry-Biology Interface training grant (T32GM00815) and a predoctoral University fellowship from the University of Massachusetts.
B.R. Pearse and L. Gabriel contributed equally to this paper.
Abbreviations used in this paper: DMJ, 1-deoxymannojirimycin; DNJ, N-butyl deoxynojirimycin; DTT, dithiothreitol; GT1, UDP-glucose: glycoprotein glucosyltransferase; HA, hemagglutinin; NEM, N-ethylmaleimide; SP, semipermeabilized; UPR, unfolded protein response.
References
- Baksh, S., and M. Michalak. 1991. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J. Biol. Chem. 266:21458–21465. [PubMed] [Google Scholar]
- Braakman, I., H. Hoover-Litty, K.R. Wagner, and A. Helenius. 1991. Folding of influenza hemagglutinin in the endoplasmic reticulum. J. Cell Biol. 114:401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman, I., J. Helenius, and A. Helenius. 1992. a. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 11:1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman, I., J. Helenius, and A. Helenius. 1992. b. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 356:260–262. [DOI] [PubMed] [Google Scholar]
- Bukau, B., and A.L. Horwich. 1998. The hsp70 and hsp60 chaperone machines. Cell. 92:351–366. [DOI] [PubMed] [Google Scholar]
- Cannon, K.S., and A. Helenius. 1999. Trimming and readdition of glucose to N-linked oligosaccharides determines calnexin association of a substrate glycoprotein in living cells. J. Biol. Chem. 274:7537–7544. [DOI] [PubMed] [Google Scholar]
- Caramelo, J.J., O.A. Castro, L.G. Alonso, G. de Prat-Gay, and A.J. Parodi. 2003. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc. Natl. Acad. Sci. USA. 100:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo, J.J., O.A. Castro, G. de Prat-Gay, and A.J. Parodi. 2004. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J. Biol. Chem. 279:46280–46285. [DOI] [PubMed] [Google Scholar]
- Chen, W., J. Helenius, I. Braakman, and A. Helenius. 1995. Cotranslational folding and calnexin binding of influenza hemagglutinin in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 92:6229–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland, C.S., K.-P. Zimmer, K.R. Wagner, G.A. Healey, I. Mellman, and A. Helenius. 1988. Folding, trimerization, and transport are sequential events in the biogenesis of Influenza virus hemagglutinin. Cell. 53:197–209. [DOI] [PubMed] [Google Scholar]
- Daniels, R., B. Kurowski, A.E. Johnson, and D.N. Hebert. 2003. N-linked glycans direct the cotranslational maturation of influenza hemagglutinin. Mol. Cell. 11:79–90. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., M. Molinari, and A. Helenius. 1999. Setting the standards: quality control in the secretory pathway. Science. 286:1882–1888. [DOI] [PubMed] [Google Scholar]
- Fernandez, F., C. D'Alessio, S. Fanchiotti, and A.J. Parodi. 1998. A misfolded protein conformation is not a sufficient condition for in vivo glucosylation by the UDP-Glc:glycoprotein glucosyltransferase. EMBO J. 17:5877–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, E., N. Wang, H. Parag, R. Halaban, and D.N. Hebert. 2003. Tyrosinase maturation and oligomerization in the endoplasmic reticulum requires a melanocyte specific factor. J. Biol. Chem. 278:25607–25617. [DOI] [PubMed] [Google Scholar]
- Fuerst, T.R., E.G. Niles, F.W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA. 83:8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganan, S., J.J. Cazzulo, and A.J. Parodi. 1991. A major proportion of the N-linked glycoproteins are transiently glucosylated in the endoplasmic reticulum. Biochemistry. 30:3098–3104. [DOI] [PubMed] [Google Scholar]
- Gardner, T.G., and K.P. Kearse. 1999. Modification of the T cell antigen receptor (TCR) complex by UDP-glucose:glycoprotein glucosyltransferase. TCR folding is finalized convergent with formation of alpha beta delta epsilon gamma epsilon complexes. J. Biol. Chem. 274:14094–14099. [DOI] [PubMed] [Google Scholar]
- Gilchrist, A., C.E. Au, J. Hiding, A.W. Bell, J. Fernandez-Rodriguez, S. Lesimple, H. Nagaya, L. Roy, S.J. Gosline, M. Hallett, et al. 2006. Quantitative proteomics analysis of the secretory pathway. Cell. 127:1265–1281. [DOI] [PubMed] [Google Scholar]
- Hammond, C., I. Braakman, and A. Helenius. 1994. Role of N-linked oligosaccharides, glucose trimming and calnexin during glycoprotein folding in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 91:913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, D.N., and M. Molinari. 2007. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87:1377–1408. [DOI] [PubMed] [Google Scholar]
- Hebert, D.N., B. Foellmer, and A. Helenius. 1995. Glucose trimming and reglucosylation determines glycoprotein association with calnexin. Cell. 81:425–433. [DOI] [PubMed] [Google Scholar]
- Hebert, D.N., B. Foellmer, and A. Helenius. 1996. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- Hebert, D.N., J.-X. Zhang, W. Chen, B. Foellmer, and A. Helenius. 1997. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J. Cell Biol. 139:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, D.N., S.C. Garman, and M. Molinari. 2005. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 15:364–370. [DOI] [PubMed] [Google Scholar]
- Helenius, A. 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell. 5:253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019–1049. [DOI] [PubMed] [Google Scholar]
- Huffaker, T.C., and P.W. Robbins. 1983. Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. USA. 80:7466–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., Z. Yan, K.H. Nam, and J. Li. 2007. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell. 26:821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, M., H. Srinivas, E. Kandiah, E. Gemma, L. Ellgaard, S. Oscarson, A. Helenius, and A. Surolia. 2003. Interactions of substrate with calreticulin, an endoplasmic reticulum chaperone. J. Biol. Chem. 278:6194–6200. [DOI] [PubMed] [Google Scholar]
- Keith, N., A.J. Parodi, and J.J. Caramelo. 2005. Glycoprotein tertiary and quaternary structures are monitored by the same quality control mechanism. J. Biol. Chem. 280:18138–18141. [DOI] [PubMed] [Google Scholar]
- Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631–664. [DOI] [PubMed] [Google Scholar]
- Labriola, C., J.J. Cazzulo, and A.J. Parodi. 1995. Retention of glucose units added by the UDP-GLC:glycoprotein glucosyltransferase delays exit of glycoproteins from the endoplasmic reticulum. J. Cell Biol. 130:771–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labriola, C., J.J. Cazzulo, and A.J. Parodi. 1999. Trypanosoma cruzi calreticulin is a lectin that binds monoglucosylated oligosaccharides but not protein moieties of glycoproteins. Mol. Biol. Cell. 10:1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, and R.J. Kaufman. 2002. IRE1- mediated unconventional mRNA splicing and S2p-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggioni, M.C., I.M. Liscaljet, and I. Braakman. 2005. A critical step in the folding of influenza virus HA determined with a novel folding assay. Nat. Struct. Mol. Biol. 12:258–263. [DOI] [PubMed] [Google Scholar]
- Meunier, L., Y.-K. Usherwood, K.T. Chung, and L.M. Hendershot. 2002. A subset of chaperones and folding enzymes from multiprotein complexes in the endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 13:4456–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari, M., and A. Helenius. 2000. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 288:331–333. [DOI] [PubMed] [Google Scholar]
- Molinari, M., C. Galli, V. Piccaluga, M. Pieren, and P. Paganetti. 2002. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J. Cell Biol. 158:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari, M., K.K. Eriksson, V. Calanca, C. Galli, P. Cresswell, M. Michalak, and A. Helenius. 2004. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol. Cell. 13:125–135. [DOI] [PubMed] [Google Scholar]
- Molinari, M., C. Galli, O. Vanoni, S.M. Arnold, and R.J. Kaufman. 2005. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol. Cell. 20:503–512. [DOI] [PubMed] [Google Scholar]
- Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 101:451–454. [DOI] [PubMed] [Google Scholar]
- Oliver, J.D., F.J. van der Wal, N.J. Bulleid, and S. High. 1997. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 275:86–88. [DOI] [PubMed] [Google Scholar]
- Ou, W.J., P.H. Cameron, D.Y. Thomas, and J.J.M. Bergeron. 1993. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 364:771–776. [DOI] [PubMed] [Google Scholar]
- Peterson, J.R., A. Ora, P. Nguyen Van, and A. Helenius. 1995. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol. Biol. Cell. 6:1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quellhorst, G.J., J.L. O'Rear, R. Cacan, A. Verbert, and S.S. Krag. 1999. Nonglucosylated oligosaccharides are transferred to protein in MI8-5 Chinese hamster ovary cells. Glycobiology. 9:65–72. [DOI] [PubMed] [Google Scholar]
- Ritter, C., and A. Helenius. 2000. Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase. Nat. Struct. Biol. 7:278–280. [DOI] [PubMed] [Google Scholar]
- Ritter, C., K. Quirin, M. Kowarik, and A. Helenius. 2005. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 24:1730–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge, K.W., T.C. Huffaker, and P.W. Robbins. 1984. Two yeast mutations in glucosylation steps of the asparagine glycosylation pathway. J. Biol. Chem. 259:412–417. [PubMed] [Google Scholar]
- Schroder, M., and R.J. Kaufman. 2005. The Mammalian unfolded protein response. Annu. Rev. Biochem. 74:739–789. [DOI] [PubMed] [Google Scholar]
- Singh, I., R.W. Doms, K.R. Wagner, and A. Helenius. 1990. Intracellular transport of soluble and membrane-bound glycoproteins: folding, assembly and secretion of anchor-free influenza hemagglutinin. EMBO J. 9:631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda, T., C. Galli, R.J. Kaufman, and M. Molinari. 2007. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol. Cell. 27:238–249. [DOI] [PubMed] [Google Scholar]
- Sousa, M., and A.J. Parodi. 1995. The molecular basis for the recognition of misfolded glycoproteins by the UDP-Glc: glycoprotein glucosyltransferase. EMBO J. 14:4196–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, M.C., M.A. Ferrero-Garcia, and A.J. Parodi. 1992. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 31:97–105. [DOI] [PubMed] [Google Scholar]
- Taylor, S.C., P. Thibault, D.C. Tessier, J.J. Bergeron, and D.Y. Thomas. 2003. Glycopeptide specificity of the secretory protein folding sensor UDP-glucose glycoprotein:glucosyltransferase. EMBO Rep. 4:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.C., A.D. Ferguson, J.J. Bergeron, and D.Y. Thomas. 2004. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat. Struct. Mol. Biol. 11:128–134. [DOI] [PubMed] [Google Scholar]
- Travers, K.J., C.K. Patil, L. Wodicka, D.J. Lockhart, J.S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between unfolded protein response and ER-associated degradation. Cell. 101:249–258. [DOI] [PubMed] [Google Scholar]
- Trombetta, E.S., and A. Helenius. 2000. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J. Cell Biol. 148:1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta, E.S., and A.J. Parodi. 2003. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19:649–676. [DOI] [PubMed] [Google Scholar]
- Újvári, A., R. Aron, T. Eisenhaure, E. Cheng, H.A. Parag, Y. Smicun, R. Halaban, and D.N. Hebert. 2001. Translation rate of human tyrosinase determines its N-Linked glycosylation level. J. Biol. Chem. 276:5924–5931. [DOI] [PubMed] [Google Scholar]
- Vassilakos, A., M.F. Cohen-Doyle, P.A. Peterson, M.R. Jackson, and D.B. Williams. 1996. The molecular chaperone calnexin facilitates folding and assembly of class I histocompatibility molecules. EMBO J. 15:1495–1506. [PMC free article] [PubMed] [Google Scholar]
- Wada, I., M. Kai, S. Imai, F. Sakane, and H. Kanoh. 1997. Promotion of transferrin folding by cyclic interactions with calnexin and calreticulin. EMBO J. 16:5420–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., R. Daniels, and D.N. Hebert. 2005. The cotranslational maturation of the type I membrane glycoprotein tyrosinase: the heat shock protein 70 system hands off to the lectin-based chaperone system. Mol. Biol. Cell. 16:3740–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R., A.J. Allen, J. Oliver, J.L. Brookman, S. High, and N.J. Bulleid. 1995. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem. J. 307:679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun, A., N.J. Darby, D.C. Tessier, M. Michalak, J.J. Bergeron, and D.Y. Thomas. 1998. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J. Biol. Chem. 273:6009–6012. [DOI] [PubMed] [Google Scholar]
- Zuber, C., J.Y. Fan, B. Guhl, A. Parodi, J.H. Fessler, C. Parker, and J. Roth. 2001. Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc. Natl. Acad. Sci. USA. 98:10710–10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.