Abstract
The cellular events that precede myelination in the peripheral nervous system require rapid and dynamic morphological changes in the Schwann cell. These events are thought to be mainly controlled by axonal signals. But how signals on the axons are coordinately organized and transduced to promote proliferation, migration, radial sorting, and myelination is unknown. We describe that the axonal signal neuregulin-1 (NRG1) controls Schwann cell migration via activation of the atypical Dock180-related guanine nucleotide exchange factor (GEF) Dock7 and subsequent activation of the Rho guanine triphosphatases (GTPases) Rac1 and Cdc42 and the downstream c-Jun N-terminal kinase. We show that the NRG1 receptor ErbB2 directly binds and activates Dock7 by phosphorylating Tyr-1118. Dock7 knockdown, or expression of Dock7 harboring the Tyr-1118–to–Phe mutation in Schwann cells, attenuates the effects of NRG1. Thus, Dock7 functions as an intracellular substrate for ErbB2 to promote Schwann cell migration. This provides an unanticipated mechanism through which ligand-dependent tyrosine phosphorylation can trigger the activation of Rho GTPase-GEFs of the Dock180 family.
Introduction
During development, Schwann cells proliferate, migrate, sort, and ensheath individual axons, all of which contributes to the proper formation of myelin. Throughout these events, Schwann cells are in constant contact with axons, suggesting that glial-neuronal communication is essential for regulating these processes (Bunge, 1993). One such factor involved in Schwann cell proliferation and migration is neuregulin-1 (NRG1), which is expressed primarily by neurons and signals through the cognate ErbB family of receptors expressed by Schwann cells (Bunge, 1993; Mahanthappa et al., 1996; Garratt et al., 2000; Citri et al., 2003). Additionally, growing evidence clearly illustrates that NRG1 type III, a membrane-bound form, plays a critical role in the ensheathment and myelination of axons (Nave and Salzer, 2006). Although all of these processes are clearly distinct, they all require rapid and dynamic morphological changes in the Schwann cell, initiated by the activation of the ErbB2 and ErbB3 heterodimer of the receptor tyrosine kinase family (Garratt et al., 2000; Citri et al., 2003). How can the same ligand and receptor complex, namely NRG1 via ErbB2/3, regulate multiple cellular processes that are thought to be distinct and highly controlled in a temporal and spatial manner? How is specificity conferred to generate the proper number of Schwann cells to appropriately match the number of axons and then to determine which axons should be myelinated?
It is well established that the ability of cells to respond to extracellular signals to change cell morphology is controlled in part by the Rho family of small GTPases, including Rac1, Cdc42, and RhoA (Schmidt and Hall, 2002). In fact, recent studies identify Rac1 as the downstream effector molecule responsible for process extension and lamellipodia formation in Schwann cells, allowing for proper radial sorting and myelination (Benninger et al., 2007; Nodari et al., 2007). The Rho GTPases are active when bound to GTP and are inactive when bound to GDP. Guanine nucleotide exchange factors (GEFs) catalyze the replacement of GDP with free cytoplasmic GTP to generate active GTPases, whereas GTPase-activating proteins (GAPs) accelerate the intrinsic GTPase activity to inactivate the GTPases. Therefore, the rate-limiting step and the specificity in the activation of the distinct GTPases lie in the expression and activation of both the GEFs and GAPs.
The GEFs are largely divided into two major categories (Rossman et al., 2005). The first category is composed of ∼80 genes related to the protooncogene Dbl. These gene products share a catalytic Dbl homology (DH) domain, which was first identified over a decade ago (Schmidt and Hall, 2002; Rossman et al., 2005). The second category consists of at least 11 GEFs, which contain a catalytic domain that is structurally distinct from the DH domain. This catalytic domain is named the Dock homology region (DHR)-2 (also called Caenorhabditis elegans Ced-5/mammalian Dock180/Drosophila melanogaster Mbc-zizimin homology domain 2 or Docker; Rushton et al., 1995; Hasegawa et al., 1996; Wu and Horvitz, 1998; Brugnera et al., 2002; Côté and Vuori, 2002; Meller et al., 2002; Rossman et al., 2005) and, until recently, little was known about the mechanisms that regulate the activities of the Dock180 (also called Dock1)-related GEFs. In this paper, we report the role of the atypical Dock180-related GEF Dock7 and the subsequent GTPase signaling cascade in NRG1-induced migration of primary Schwann cells. Furthermore, we identify Dock7 as the functional intracellular substrate of ErbB2, suggesting the importance of Dock7 in early peripheral nervous system development.
Results
NRG1 activation of ErbB2 and 3 promotes Schwann cell migration through Rho GTPases Rac1 and Cdc42 and the downstream JNK
Using Boyden chambers, we previously demonstrated that dorsal root ganglion (DRG) neurons secrete various growth factors, including neurotrophin-3 (NT3) and brain-derived neurotrophic factor, to regulate Schwann cell migration (Yamauchi et al., 2004). We placed primary Schwann cells onto filters of Boyden chambers coated with axonal membranes from DRG neurons and allowed them to migrate into the lower compartment along a concentration gradient of the neuronal conditioned medium for 6 h. When we added the NRG1 scavenger ErbB3-Fc to the DRG conditioned medium, migration was significantly diminished. Removal of the NRG1-like activity inhibited migration by ∼80% (Fig. 1, A and B), implicating NRG1 as a positive regulator of Schwann cell migration. We next examined the effects of extracellular matrix proteins on Schwann cell migration induced by the conditioned medium. Filters in the Boyden chambers were coated with collagen (type I or IV), fibronectin, or laminin (Fig. 1, C and D). The conditioned medium stimulated migration on any of the extracellular matrices, although fibronectin and laminin enhanced Schwann cell migration in the absence of the conditioned medium. Therefore, it is possible that extracellular matrices act cooperatively with soluble factors to control migration. In contrast, the collagen-coated filters were similar to the filters coated with DRG axonal membranes. Because collagen modestly stimulates migration as compared with fibronectin and laminin, collagen (type I)-coated filters were used in the subsequent experiments. Next, we explored whether ErbB3-Fc is specific for the NRG1-like activity in the conditioned medium. As shown in Fig. 1 (E–G), ErbB3-Fc specifically inhibits migration induced by NRG1 but not by NT3 or insulin-like growth factor (IGF)-I, which stimulates Schwann cell migration (Cheng et al., 2000 Yamauchi et al., 2004). These results again suggest that the DRG conditioned medium contains NRG1-like activity, which can enhance Schwann cell migration. We further investigated the effect of the NRG1-like activity on migration of reaggregated Schwann cells on live DRG axons to mimic physiological conditions. The Schwann cell reaggregates initially spread out slowly and then begin to migrate out of the reaggregates along axons. Addition of ErbB3-Fc to the culture medium of the DRG neurons inhibits Schwann cell migration from the reaggregates (Fig. 1, H and I). This observation is consistent with the results from our Boyden chamber assay and, in fact, ErbB3-Fc has a greater initial inhibitory effect on migration from the reaggregates. It is important to note that after a longer time course the effects on migration distances in the presence or absence of ErbB3-Fc were diminished. Thus, it is likely that the effect of ErbB3-Fc delays migration rather than completely inhibiting it.
Figure 1.
NRG1 promotes Schwann cell migration through the ErbB2 and 3 heterodimer. (A and B) The migration of primary Schwann cells was measured by using Boyden chambers. Filters were coated with DRG axonal membranes. After incubation for 6 h with normal or conditioned medium from DRG neurons containing 5 μg/ml of control IgG or ErbB3-Fc, Schwann cells were stained with Giemsa solution and the number of migrating cells was counted (16 independent fields). Bar, 50 μm. (C and D) Filters in Boyden chambers were coated with DRG axonal membranes, collagen (type I or IV), fibronectin, or laminin. Schwann cell migration was measured in the presence of normal medium or conditioned medium (eight independant fields). (E–G) In the presence of control IgG or ErbB3-Fc, Schwann cells were incubated with or without 20 ng/ml of NRG1, NT3, or IGF-I in Boyden chambers (eight independent fields). Filters were coated with collagen (type I). (H and I) Schwann cell reaggregates were placed onto DRG neurons and control IgG or ErbB3-Fc was added. After 6 h, DRG axons were stained with an antineurofilament antibody (red), and Schwann cells were stained with an anti-S100β antibody (green). The distance of migration was measured (n = 16). Bar, 100 μm. (J and K) Schwann cells were pretreated in the presence or absence of 10 μM AG825 and then incubated with or without 20 ng/ml NRG1 in Boyden chambers. The number of migrating cells was counted (16 independent fields). Bar, 50 μm. (L and M) Schwann cells were transfected with control, ErbB2, or ErbB3 siRNA and incubated with or without NRG1 in Boyden chambers (16 independent fields). To confirm the effects of siRNAs, the lysates of transfected cells were immunoblotted with an anti-ErbB2, ErbB3, or actin antibody. Error bars show ±SD. Data were evaluated by using one-way ANOVA (*, P < 0.01; ***, P < 0.02).
We examined whether migration of Schwann cells requires the tyrosine kinase activity of ErbB2. Pretreatment with AG825, an inhibitor of the ErbB2 tyrosine kinase (Tsai et al., 1996), reduced NRG1-induced migration by ∼80% (Fig. 1, J and K). In addition, we transfected siRNA oligonucleotides for both ErbB2 and 3 in Schwann cells. Expression of ErbB2 and 3 was specifically down-regulated after transfection with the siRNA, whereas expression of control proteins was unaffected, as revealed by immunoblotting (Fig. 1 M). Knockdown of ErbB2 or ErbB3 attenuated migration induced by NRG1. Collectively, the ErbB2 and 3 heterodimer responds to NRG1, and the tyrosine kinase activity of ErbB2 is important for Schwann cell migration.
Next, we tested whether the Rho GTPases Rac1 and Cdc42 are involved in the NRG1-induced migration of Schwann cells, as previously seen in NT3-induced migration (Yamauchi et al., 2004). Pretreatment of Clostridium difficile Toxin B, which glycosylates and blocks the functions of Rho GTPases such as RhoA, Rac1, and Cdc42 (Just et al., 1995), inhibited the NRG1 effect by ∼70% (Fig. 2 A). In contrast, C3 exoenzyme, which ADP ribosylates RhoA and blocks its function (Hirose et al., 1998), did not have any obvious effect. Furthermore, we transfected a siRNA for Rac1 or Cdc42 into Schwann cells. Knockdown of Rac1 inhibited the NRG1-induced migration in Boyden chambers by ∼25% (Fig. S1 D, available at http://www.jcb.org/cgi/content/full/jcb.200709033/DC1) as well as migration from reaggregates on DRG axons (compare videos 1–4 for cells transfected with control siRNA with videos 5 and 6 for cells transfected with Rac1 siRNA, available at http://www.jcb.org/cgi/content/full/jcb.200709033/DC1), which is consistent with recent studies (Benninger et al., 2007; Nodari et al., 2007). Transfection with nonoverlapping siRNA, Cdc42-1 or Cdc42-2, decreased the migration in Boyden chambers by ∼15 and 25%, respectively (Fig. S1, E and F), as well as decreasing the migration from reaggregates on DRG axons (videos 7 and 8 for cells transfected with Cdc42-2 siRNA). However, because the effect after knockdown of Rac1 or Cdc42 is weaker than that of Toxin B, it is possible that Rac1 activity transduces an intracellular signal from NRG1 together with Cdc42 and that Rac1 and Cdc42 may share a common downstream signaling pathway. To examine whether NRG1 directly activates Cdc42 and Rac1, we performed affinity precipitation using the Rac1-GTP and Cdc42-GTP binding domain of Pak1. The activities of Rac1 and Cdc42 reached maximum levels at 60–120 min after stimulation with NRG1 and remained activated for at least 360 min (Fig. 2, B–I). Therefore, NRG1 activation of ErbB2 and 3 can stimulate the increase of Rac1-GTP and Cdc42-GTP to enhance Schwann cell migration.
Figure 2.
NRG1-induced migration of Schwann cells is dependent on the activation of the Rho GTPases Rac1 and Cdc42. (A) Schwann cells were pretreated with or without 2 ng/ml C. difficile Toxin B or 2 μg/ml C3 exoenzyme, and migration was assayed in Boyden chambers (12 independent fields). (B and C) After the addition of NRG1 for 0–120 min, endogenous Rac1-GTP in the lysates of Schwann cells was affinity precipitated with GST-Pak1-CRIB and immunoblotted with an anti-Rac1 antibody. The levels of Rac1-GTP were normalized to the amount of total Rac1 (n = 3). (D and E) The Rac1 activities were measured at 0–360 min (n = 5). (F–I) Endogenous Cdc42-GTP in the cell lysates was affinity precipitated with GST-Pak1-CRIB. The Cdc42-GTP levels were normalized to the amount of total Cdc42 (n = 3). Error bars show ±SD. Data were evaluated by using one-way ANOVA (*, P < 0.01).
We previously reported that JNK acts downstream of Rac1 and Cdc42 in NT3-induced migration of Schwann cells (Yamauchi et al., 2005a,b). In addition, the JNK cascade is the direct target of Rho GTPases in many other types of cells (Schmidt and Hall, 2002). Therefore, we investigated the effect of structurally unrelated JNK inhibitors SP600125 and JNK inhibitor I (Heo et al., 2004) on NRG1-induced migration. SP600125 and JNK inhibitor I inhibited the NRG1 effect by ∼80 and 70%, respectively (Fig. 3 A). We next immunoblotted with an antiphosphorylated JNK antibody that recognizes the phosphorylated or active state of JNK. JNK phosphorylation was detected in a time-dependent manner, and the level of phosphorylation reached maximum at 120 min and gradually decreased afterward (Fig. 3, B–E). In addition, C. difficile Toxin B inhibited the JNK phosphorylation by ∼70% (Fig. 3 F), indicating that JNK is one of the transducers acting downstream of Rac1 and Cdc42 in Schwann cell migration.
Figure 3.
JNK acts downstream of Rho GTPases to promote Schwann cell migration. (A) Schwann cells were pretreated in the presence or absence of 10 μM SP600125 or 20 μM JNK inhibitor I and incubated with or without 20 ng/ml NRG1 in Boyden chambers (12 independent fields). (B and C) Schwann cells were stimulated with NRG1 for 0–120 min. The cell lysates were immunoblotted with an anti-(pThr183/pTyr185) JNK antibody that recognizes active JNK. The cell lysates were also immunoblotted with an anti-JNK antibody. The levels of phosphorylated forms were normalized to the amount of total JNK (n = 3). (D and E) JNK phosphorylation was measured at 0–360 min (n = 3). (F) Cells were pretreated with or without 2 ng/ml C. difficile Toxin B. After incubation with NRG1 for 120 min, JNK phosphorylation was assayed (n = 5). Error bars show ±SD. Data were evaluated by using one-way ANOVA (*, P < 0.01).
The atypical GEF Dock7 mediates NRG1-induced Schwann cell migration
To identify the specific GEFs involved in the NRG1 regulation of Rac1 and Cdc42, we designed siRNA against Tiam1 and Dbs, major GEFs of the Dbl family for Rac1 and Cdc42, respectively, in Schwann cells (Yamauchi et al., 2005a,b). Neither Tiam1 nor Dbs knockdown had any effect on the NRG1-induced migration of Schwann cells (Fig. S1, A and B). Therefore, we examined the involvement of Dock7, a Dock180-related GEF expressed abundantly in Schwann cells (Fig. S1 C). Transfection of nonoverlapping siRNA, Dock7-1 or Dock7-2, into Schwann cells knocked down the expression of endogenous Dock7, as revealed by immunoblotting with an anti-Dock7 antibody (Fig. S2, A–C). Dock7-1 or -2 siRNA inhibited the NRG1-induced migration in Boyden chambers by ∼70 and 50%, respectively (Fig. 4, A and B), as well as inhibiting migration from reaggregates on DRG axons (videos 9 and 10 for cells transfected with Dock7-1 siRNA, available at http://www.jcb.org/cgi/content/full/jcb.200709033/DC1). Similarly, knockdown of Dock7 by Dock7-1 siRNA reduced the NRG1 activation of Rac1 by ∼80%, Cdc42 by 80%, and JNK phosphorylation by 60% (Fig. 4, C–E). Despite the potential involvement of other GEFs in these signaling pathways, our results hint at the possible role for Dock7 in Schwann cell migration and provide the rationale and impetus for the subsequent experiments.
Figure 4.
Dock7 is required for migration and the activation of Rac1, Cdc42, and JNK induced by NRG1 in Schwann cells. (A and B) Schwann cells were transfected with control, Dock7-1, or Dock7-2 siRNA and incubated with or without NRG1 in Boyden chambers (12 independent fields). To confirm the effects of siRNA, the lysates of transfected cells were immunoblotted with an anti-Dock7, Rac1, Cdc42, or actin antibody. (C and D) Schwann cells were transfected with control or Dock7-1 siRNA and stimulated with NRG1 for 60 min. The activities of Rac1 and Cdc42 were assayed by affinity precipitation with GST-Pak1-CRIB (n = 3). (E) Cells were transfected with control or Dock7-1 siRNA and JNK phosphorylation was measured (n = 3). Error bars show ±SD. Data were evaluated by using one-way ANOVA (*, P < 0.01).
ErbB2 directly binds and activates Dock7 by phosphorylating Tyr-1118 to regulate Schwann cell migration
Because the DHR-2 domain of the Dock180-related GEFs shows catalytic activity (Brugnera et al., 2002; Côté and Vuori, 2002; Meller et al., 2002), we tested whether Rho GTPases could be activated by the DHR-2 domain of Dock7. The purified DHR-2 domain (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200709033/DC1) promoted the exchange, binding, and release of the guanine nucleotide for Rac1 and Cdc42 in a time-dependent manner (Fig. 5, A, B, D, and E), whereas no effect was observed for RhoA (Fig. 5, C and F). Catalytically active GEFs preferentially interact with guanine nucleotide–free forms of the small GTPases (Arthur et al., 2002; Schmidt and Hall, 2002; Rossman et al., 2005). A Gly-to-Ala mutation in the P loop of the small GTPases decreases their guanine nucleotide binding activities (Arthur et al., 2002). We performed an affinity precipitation of the DHR-2 domain of Dock7 with guanine nucleotide–free Rac1G15A, Cdc42G15A, or RhoAG17A as well as wild-type Rac1, Cdc42, or RhoA. DHR-2 specifically coprecipitated with Rac1G15A (Fig. 5 G) and Cdc42G15A (Fig. 5 I) but not with RhoAG17A (Fig. 5 K), which is consistent with the results from the guanine nucleotide exchange assays. In contrast, the DH and pleckstrin homology (PH) domains of Dbs affinity precipitated with Rac1G15A (Fig. 5 H) or Cdc42G15A (Fig. 5 J) as well as with RhoAG17A (Fig. 5 L). Similarly, the affinity precipitation with wild-type Rac1 or Cdc42 also showed binding to DHR-2 but was slightly weaker than the precipitation with each GTPase harboring the Gly-to-Ala mutation. To investigate whether NRG1 activation of the ErbB2 and 3 heterodimer stimulates the GEF activity of Dock7, we cotransfected the plasmids encoding wild-type Dock7, ErbB2, and ErbB3 into 293T cells and measured the exchange of the guanine nucleotide from immunoprecipitated Dock7 for Rac1 and Cdc42. The activity of wild-type Dock7 was significantly increased after stimulation with NRG1 (Fig. 5, M and O). Similarly, NRG1 promoted the affinity-precipitation of Dock7 with Rac1G15A or Cdc42G15A (Fig. 5, N and P).
Figure 5.
NRG1 activation of the ErbB2 and 3 heterodimer stimulates the GEF activity of Dock7. (A–C) 125 ng of immobilized FLAG-Dock7-DHR-2 was incubated with 16 ng/μl GST-Rac1, Cdc42, or RhoA and 3 μM [3H]GDP in 30 μl of reaction buffer for 0–30 min, and the guanine nucleotide binding activities were measured (n = 10). (D–F) The release of [3H]GDP from GST-Rac1-[3H]GDP, Cdc42-[3H]GDP, or GST-RhoA-[3H]GDP by FLAG–Dock7–DHR-2 was measured (n = 10). Immunoprecipitated FLAG-Dbs-DHPH was used as the positive control for the RhoA-GEF. Dock7–DHR-2, closed circle; control, open circle; Dbs-DHPH, closed square. (G–L) 293T cells were transfected with pCMV–FLAG–Dock7–DHR-2 or pCMV-FLAG-Dbs-DHPH. The cell lysates were affinity precipitated with 20 μg each of nucleotide-free GST-Rho GTPase (Rac1G15A, Cdc42G15A, or RhoAG17) or the wild type (Rac1, Cdc42, or RhoA) and immunoblotted with an anti-FLAG antibody. The total FLAG–Dock7–DHR-2 or FLAG-Dbs-DHPH is also shown. Each GST-Rho GTPase was immobilized in the same experimental conditions, subjected to SDS-PAGE, and stained with Coomassie brilliant blue. (M and O) 293T cells were cotransfected with pCMV-FLAG-Dock7, pCMV-ErbB2, and pCMV-ErbB3 and stimulated with or without NRG1 for 30 min. The expression of ErbB2 and 3 in 293T cells was below the detection level of immunoblotting (not depicted). The release of [3H]GDP from GST-Rac1-[3H]GDP or Cdc42-[3H]GDP by immunoprecipitated FLAG-Dock7 was measured (n = 3). (N and P) Cells were cotransfected with pCMV-FLAG-Dock7, pCMV-ErbB2, and pCMV-ErbB3. The affinity precipitation of the cell lysates with GST-Rac1G15A or Cdc42G15A was performed. The total FLAG-Dock7 is also shown. Error bars show ±SD. Data were evaluated by using one-way ANOVA (*, P < 0.01).
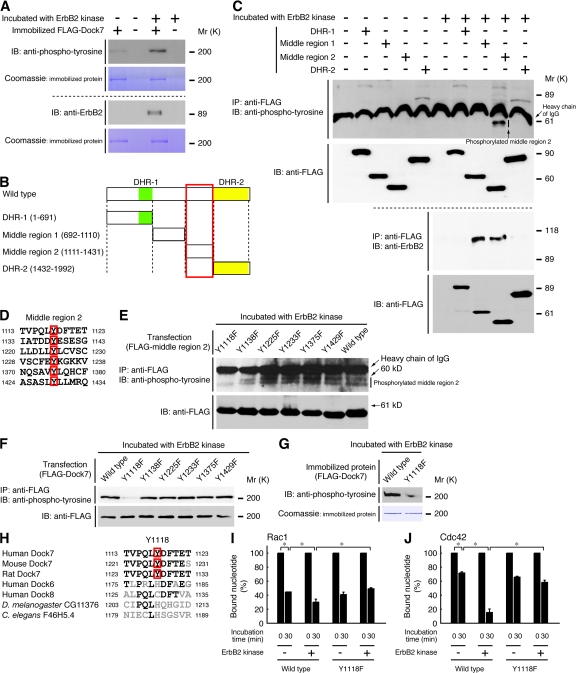
We asked if ErbB2 could directly phosphorylate Dock7 because Dock7 is stimulated after the activation of the ErbB2 and 3 heterodimer and apparently possesses various tyrosine phosphorylation sites. The recombinant intracellular kinase domain of ErbB2 (HTScan ErbB2 kinase) phosphorylated the purified wild-type Dock7 protein (Fig. S3 B) and coprecipitated with Dock7 in vitro (Fig. 6 A). To investigate the potential sites on Dock7 that could be tyrosine phosphorylated by ErbB2, we transfected a plasmid encoding the N-terminal region containing DHR-1 (aa 1–691), middle region 1 (aa 692–1110), middle region 2 (aa 1111–1431), or DHR-2 (aa 1432–1992) into 293T cells (Fig. 6 B). Because the amino acid sequence positioned between DHR-1 and -2 is quite extensive, it was divided into two regions. The ErbB2 kinase coprecipitated with both middle regions and phosphorylated the middle region 2 (Fig. 6 C). These results are also supported by findings that the middle region 2, acting as a specific substrate for ErbB2, has a dominant-negative effect on NRG1-induced migration (Fig. S4, A–D, available at http://www.jcb.org/cgi/content/full/jcb.200709033/DC1) and activation of Rac1 (Fig. S4, E, F, and I) and Cdc42 (Fig. S4, G, H, and J). Because the middle region 2 of Dock7 contains six tyrosine residues that may be phosphorylated by ErbB2 (Fig. 6 D), we made a series of constructs harboring Tyr-to-Phe mutations in the middle region 2. The ErbB2 kinase had the ability to phosphorylate the Y1138F, Y1225F, Y1233F, Y1375F, and Y1429F mutants but could not effectively phosphorylate the Y1118F mutant (Fig. 6 E). The Y1138F mutation of the middle region 2 modestly inhibited phosphorylation by the ErbB2 kinase. Thus, to ascertain whether Tyr-1118 is the major phosphorylation site of Dock7, we made the Y1118F, Y1138F, Y1225F, Y1233F, Y1375F, and Y1429F mutants of the full-length Dock7 and analyzed the phosphorylation by the ErbB2 kinase. In Fig. 6 F, the Y1118F mutation greatly reduced the phosphorylation of Dock7 by the ErbB2 kinase, whereas the Y1138F, Y1225F, Y1233F, Y1375F, and Y1429F mutations did not possess inhibitory effects on phosphorylation. Similarly, the purified Y1118F Dock7 (Fig. S3 C) reduced phosphorylation by the ErbB2 kinase (Fig. 6 G). Amino acid sequences surrounding Tyr-1118 in Dock7 are uniquely conserved among mammalian Dock7 proteins but not in the Dock180-related GEFs homologous to Dock7 and those from other species (Fig. 6 H).
Figure 6.
ErbB2 directly binds and activates Dock7 by phosphorylating Tyr-1118. (A) 250 ng of immobilized full-length FLAG-Dock7 protein was incubated in 30 μl of reaction buffer containing 20 μM of cold ATP in the presence or absence of 100 ng ErbB2 kinase for 30 min, washed, and immunoblotted with an anti-pTyr or ErbB2 antibody. Immobilized FLAG-Dock7 was also stained with Coomassie brilliant blue. (B) The schematic structures of Dock7 and the domains are illustrated. Red rectangle, Tyr in the middle region 2. (C) 293T cells were transfected with the plasmid encoding DHR-1, middle region 1, middle region 2, or DHR-2 of Dock7. The lysates of transfected cells were immunoprecipitated with an anti-FLAG antibody, incubated with ErbB2 kinase and ATP, and immunoblotted with an anti-pTyr or ErbB2 antibody. A shift in the mobility of the bands for the tyrosine-phosphorylated protein was observed. The cell lysates were also immunoblotted with an anti-FLAG antibody. (D) The amino acid sequences containing six tyrosine residues in the middle region 2 are shown. (E) Cells were transfected with the plasmid encoding each middle region 2 containing one Tyr-to-Phe mutation. The samples, immunoprecipitated with the anti-FLAG antibody, were incubated with ErbB2 kinase and ATP. A shift in the mobility was observed in bands of the tyrosine-phosphorylated protein. The tyrosine phosphorylation of the constructs and their expression are also shown. (F) Cells were transfected with each full-length Dock7 harboring one Tyr-to-Phe mutation in the middle region 2, immunoprecipitated with anti-FLAG antibody, and incubated with ErbB2 kinase and ATP. The tyrosine phosphorylation of the constructs and their expression are also shown. (G) 250 ng of immobilized full-length FLAG-Dock7 or FLAG-Dock7Y1118F was incubated with ErbB2 kinase and ATP. (H) A comparison of the amino acid sequences surrounding the ErbB2 phosphorylation sites (red squares) of mammalian Dock7 with other homologous proteins is shown. Black, conserved amino acids; grey, nonconserved amino acids. (I and J) Immobilized FLAG-Dock7 or the Y1118F mutant was incubated in 30 μl of reaction buffer containing 20 μM of cold ATP in the presence or absence of ErbB2 kinase and washed. The release of [3H]GDP from GST-Rac1–[3H]GDP or Cdc42–[3H]GDP by immobilized proteins was measured (n = 3). Error bars show ±SD. Data were evaluated by using one-way ANOVA (*, P < 0.01).
To clarify whether ErbB2 directly activates Dock7 and whether Tyr-1118 is critical for ErbB2-dependent GEF activity, we incubated the purified wild-type or Y1118F Dock7 with the ErbB2 kinase and performed guanine nucleotide release assays in vitro. ErbB2 stimulated guanine nucleotide release of Dock7 for Rac1 and Cdc42, whereas the Y1118F mutation in Dock7 abolished the release (Fig. 6, I and J). These results provide evidence that ErbB2 directly binds to Dock7 and phosphorylates the Tyr-1118 position to activate Rac1 and Cdc42 in vitro.
To explore whether ErbB2 phosphorylates endogenous Dock7 in Schwann cells, we made an antibody specific to phosphorylated Tyr-1118 of Dock7 (Fig. S2, D and E). Stimulation with NRG1 dramatically enhanced the phosphorylation of Tyr-1118, and this phosphorylation was reduced by AG825 or ErbB3-Fc (Fig. 7 A). NT3 or IGF-1 did not enhance the phosphorylation of Dock7, indicating that the Tyr-1118 position is specific for NRG1. In addition, Dock7 was immunoprecipitated with ErbB2, and this interaction was enhanced by stimulation with NRG1 (Fig. 7 B). Stimulation with NRG1 also increased the colocalization of Dock7 with ErbB2 (Fig. 7 C, bottom) and phosphorylated Dock7 at Tyr-1118 with ErbB2 (Fig. 7 C, middle) in cell bodies and in processes. In addition, after stimulation with NRG1, colocalization of phosphorylated Dock7 with phosphorylated ErbB2 was observed in punctate structures (Fig. 7 C, top). Furthermore, NRG1 induced an affinity precipitation of Dock7 with Rac1G15A or Cdc42G15A (Fig. 7, D and E). Thus, stimulation with NRG1 can promote the binding of Dock7 with ErbB2, phosphorylate Dock7 at the Tyr-1118 position, and regulate the Dock7 activity in Schwann cells.
Figure 7.
NRG1, acting through the phosphorylation of Dock7 at Tyr-1118, regulates Schwann cell migration. (A) After stimulation with vehicle or 20 ng/ml of NRG1, NT3, or IGF-I for 30 min, Schwann cells were lysed, immunoprecipitated with an anti-Dock7 antibody, and immunoblotted with an anti-(pTyr1118)Dock7 antibody. In some experiments, cells were treated with or without AG825 or ErbB3-Fc. The cell lysates were also immunoblotted with an anti-Dock7 antibody. (B) After stimulation with vehicle or NRG1, immunoprecipitated Dock7 was immunoblotted with an anti-ErbB2 antibody. Immunoblots for ErbB2, ErbB3, and Dock7 are shown. (C) After stimulation with vehicle or NRG1, Schwann cells were costained with the following antibodies: anti-(pTyr1118)Dock7 (green; top) and anti-(pTyr1112)ErbB2 (red; top), anti-(pTyr1118)Dock7 (green; middle) and anti-ErbB2 (red; middle), and anti-Dock7 (green; bottom) and anti-ErbB2 (red; bottom). After stimulation with vehicle or NRG1, increased colocalization (indicated by arrows) was observed (Bar, 25 μm). a–d are magnifications of the boxed areas as indicated (Bar, 10 μm). Dotted lines indicate the outlines of cells. (D and E) Affinity precipitation with GST-Rac1G15A or Cdc42G15A was performed and immunoblotted with an anti-Dock7 antibody. Immunoblots for Dock7 are shown.
Next, we investigated the role of the phosphorylation of Dock7 at the Tyr-1118 position in Schwann cell migration. We cotransfected a plasmid encoding Dock7-1 siRNA-resistant wild-type or Y1118F Dock7 together with a control or Dock7-1 siRNA into Schwann cells. Expression of siRNA-resistant wild-type Dock7 reversed the Dock7-1 siRNA-mediated inhibition of NRG1-induced migration in Boyden chambers, whereas Y1118F Dock7 failed to rescue Dock7-1 siRNA-mediated inhibition of migration (Fig. 8, A and B). Because there is the possibility that the Y1118F mutation has an effect on the protein conformation of Dock7, we tested the effects of the other mutants, Y1138F, Y1225F, Y1233F, Y1375F, and Y1429F, on migration. The Y1138F, Y1225F, Y1233F, or Y1375F mutant rescued siRNA-mediated inhibition of migration at the same level as that of the wild type (Fig. 8, D and E), indicating that the Y1118F mutation mimics the nonphosphorylated form and that the phosphorylation at the Tyr-1118 position is required for migration. The Y1429F mutant could rescue siRNA-mediated inhibition of migration but did not completely. The reason may be that Tyr-1429 interacts functionally with the catalytic DHR-2 because it is adjacent to DHR-2. Alternatively, because the Tyr-1429 position is contained in the canonical phosphatidylinositol-3-kinase binding motif Tyr-X-X-Met (Fig. 5 D; Ponzetto et al., 1993), the binding may partially affect Dock7 activation (Côté et al., 2005). Expression of Dock7-1 siRNA-resistant constructs was not down-regulated by cotransfection with Dock7-1 siRNA, which specifically reduced expression of native siRNA-sensitive nucleotide sequence of Dock7 (Fig. 8 C).
Figure 8.
Effects of the Tyr-to-Phe mutations in the middle region 2 of Dock7 on NRG1-induced migration of Schwann cells. (A and B) pEGFP, pEGFP-siRNA–-resistant wild-type Dock7, or pEGFP-siRNA–resistant Dock7Y1118F was cotransfected with control or Dock7-1 siRNA into Schwann cells. The number of GFP-fluorescent migrating Schwann cells in Boyden chambers was counted. Bar, 100 μm. (C) Expression of GFP-tagged siRNA-sensitive wild-type Dock7 or Dock7-1 siRNA-resistant Dock7 (wild type, Y1118F, Y1138F, Y1225F, Y1233F, Y1375F, or Y1429F) in Schwann cells are shown in the immunoblots. The cell lysates were immunoblotted with an anti-GFP or -actin antibody. (D and E) Schwann cells were cotransfected with pEGFP-siRNA–resistant wild-type or mutated Dock7 together and Dock7-1 siRNA. The number of GFP-fluorescent migrating Schwann cells was counted. Bar, 100 μm. Error bars show ±SD. Data were evaluated by using one-way ANOVA (n = 16; *, P < 0.01; ***, P < 0.02).
Consistent with the results in the previous paragraph, expression of Dock7-1 siRNA-resistant wild-type Dock7 reversed Dock7-1 siRNA-mediated inhibition of NRG1-induced migration from reaggregates on DRG axons (Fig. 9, A and B). In contrast, expression of siRNA-resistant Y1118F Dock7 did not rescue siRNA-mediated inhibition of migration. These results indicate again that the phosphorylation of Dock7 at the Tyr-1118 position by ErbB2 plays a key role in promoting Schwann cell migration.
Figure 9.
Tyr-1118 of Dock7 is essential for NRG1-induced migration of reaggregated Schwann cells. (A and B) pEGFP or pEGFP-siRNA–resistant wild-type Dock7 or Dock7Y1118F was cotransfected with control or Dock7-1 siRNA into Schwann cells. Cells were fixed with PFA, blocked, and stained with an anti-neurofilament antibody (red). The immunofluorescence images were merged. Bar, 100 μm. Error bars show ±SD. Data were evaluated by using one-way ANOVA (n = 16; *, P < 0.01; **, P < 0.015; ***, P < 0.02).
Discussion
Each stage in Schwann cell development involves characteristic morphological changes regulated by reciprocal and complex glial–neuronal interactions. Membrane-bound NRG1, expressed primarily on axons, represents an essential determinant in controlling myelination by Schwann cells (Nave and Salzer, 2006). In this paper, we demonstrate that NRG1 binding to the ErbB2 and 3 heterodimer promotes migration of premyelinating Schwann cells and that this effect is mediated by the direct activation of the Dock180-related GEF Dock7 and the subsequent Rho GTPase cascade. This conclusion is supported by the findings that blocking the signaling molecules coupling the ErbB receptor to the Rho GTPase cascade results in the attenuation of migration. Importantly, we identify Dock7 as the functional intracellular substrate for the ErbB2 receptor. ErbB2 directly binds to Dock7 and promotes the GEF activities for Rho GTPases by phosphorylating Tyr-1118 in vitro. Stimulation with NRG1 in Schwann cells leads to the phosphorylation of Dock7 at Tyr-1118 and activation. Transfection of Dock7 harboring the Tyr-1118–to–Phe mutation inhibits the NRG1-induced migration. These results demonstrate that NRG1 activation of the ErbB2 and ErbB3 heterodimer induces Schwann cell migration through an unexplored mechanism, namely that a receptor-mediated tyrosine phosphorylation event triggers the activation of Dock7. Dock7 has a GEF activity that is preferential for Cdc42 rather than Rac1; however, Schwann cell migration by NRG1 requires both Rac1 and Cdc42. Because Schwann cells modestly express Dock3, 4, and 5 of the Rac1-specific Dock180-related GEFs, they may cooperatively support the remaining NRG1-dependent Rac1 activity.
Possible alternative regulation of Dock7 in Schwann cell migration
It is clear that ErbB2 phosphorylates and activates Dock7 in vitro; however, the question of whether Dock7 can be activated by an alternative mechanism remains. Besides the catalytic DHR-2 domain, Dock180-related GEFs contain another conserved domain, termed DHR-1 (also called City-zizimin homology domain 1). The putative phospholipid-binding C2 domain is found in the DHR-1 domain of Dock180. Dock180 binds to phosphatidylinositol-3,4,5-triphosphate, the product of phosphatidylinositol-3-kinase (Côté et al., 2005). Because ErbB2 can activate phosphatidylinositol-3-kinase (Garratt et al., 2000; Citri et al., 2003), it is possible that phosphatidylinositol-3,4,5-triphosphate modulates cellular functions of Dock7. Dock180 also has some protein–protein interactive domains. Dock180 interacts with the proline-rich region of engulfment and cell motility (ELMO) family proteins and the Src homology (SH) 3 domain of CrkII through the N-terminal SH3 domain and the C-terminal proline-rich sequence, respectively (Hasegawa et al., 1996; Brugnera et al., 2002). The ELMOs–CrkII–Dock180 complex is required for activating Rac1 along the periphery of a cell, leading to lamellipodial formation. The activation of Rac1 through Dock180 has an alternative mechanism when dealing with the Rho GTPase RhoG. Once activated, RhoG forms a ternary complex with ELMOs–CrkII–Dock180 (Katoh and Negishi, 2003). The RhoG-GTP–ELMOs–CrkII–Dock180 complex induces morphological changes at the cell periphery. However, Dock7 does not interact with ELMO1 and CrkII (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200709033/DC1) because it is unlikely that Dock7 contains either an SH3 domain or a proline-rich region. It will be of interest to examine the binding partners of Dock7 using yeast two-hybrid or affinity chromatography techniques and to analyze various regulatory mechanisms of Dock7 in Schwann cells. In addition, elucidation of the 3D structures of Dock7 and the other Dock180-related GEFs should provide valuable information concerning how the phosphorylation by ErbB2 activates Dock7 in Schwann cells.
The regulation of downstream signaling pathways involved in Schwann cell morphology and function
Rac1 and Cdc42 control the formation of membrane protrusions, including lamellipodia and filopodia, which are essential for the migration of many types of cells including neurons. Rac1 and Cdc42 regulate actin polymerization by activating the Arp2/3 complex through their effectors, the Wiskott-Aldrich syndrome protein (WASP) and the WASP family verprolin homologous protein families (Takenawa and Miki. 2001). In addition, the Rac1 and Cdc42 effector Pak family controls actin filament dynamics by phosphorylating myosin light chain kinases or LIM domain kinases (Zhao and Manser, 2005). The JNK signal is a key downstream effector of the Dock7-activated Rho GTPases in Schwann cell migration, but it is conceivable that effector proteins such as WASP, WASP family verprolin homologous protein, and Pak families can also influence migration by altering the actin cytoskeleton. Further studies may explain how signals through Rho GTPases are coordinately transduced with JNK to induce cell migration.
JNK has been originally identified as the kinase that phosphorylates the transcription factor c-Jun. Indeed, fibroblast migration likely requires c-Jun phosphorylation, but JNK has some key substrates that include cytoskeletal components (Huang et al., 2004). One particular JNK substrate candidate implicated in cell migration is the focal adhesion adaptor protein paxillin because JNK phosphorylates paxillin to regulate migration of bladder tumor epithelial NBT-II cells (Huang et al., 2003). Additionally, another candidate molecule may be the microtubule-associated proteins (MAPs). Mice deficient in JNK1, as well as pharmacological inhibition of JNK activity, exhibit a progressive morphological alteration associated with defective neuronal migration (Chang et al. 2003; Kawauchi et al., 2003). Hypophosphorylation of MAP2 and MAP1B is also observed with an increase in microtubule stability, although it is unclear whether JNK regulates microtubule dynamics by phosphorylating MAP2 and MAP1B. Because paxillin, MAP2, and MAP1B are widely expressed and control various cellular functions, they may act cooperatively as targets of JNK to assist migration of Schwann cells.
In the present study, we identify Dock7 as a downstream effector of ErbB2. This interaction mediates NRG1-induced migration of premyelinating Schwann cells. It is noteworthy to add that NRG1, acting through the ErbB2 and 3 heterodimer, enhances myelination by Schwann cells (Bunge, 1993; Garratt et al., 2000; Citri et al., 2003) as well as migration. Because migration precedes myelination, certain mechanisms may be preserved in both processes. Myelination by Schwann cells is mediated by the polarity protein Par-3, whose complex generally involves the Rac1 and Cdc42 effector Par-6 (Chan et al., 2006). Recently, Watabe-Uchida et al. (2006) reported that Dock7 regulates the polarity formation of axons and dendrites through Rac1 in hippocampal neurons. It is possible that the Dock7-mediated Rho GTPase activation may lead to the formation of a polarity complex that will ultimately trigger myelination. The chemical compound NSC23766 is a first generation Rac1-specific inhibitor identified by a structure-based in silico screening (Gao et al., 2004). It fits into a small GTPase binding groove on the Rac1-specific GEFs Tiam1 and Trio of the Dbl family. The development of chemical inhibitors specific for Dock7 and the in vivo application of siRNA oligonucleotides will help to elucidate the role of Dock7 in the myelination process both in vitro and in vivo, as well as in various pathological states originating from aberrant regulation of the ErbB receptors (Tsai et al., 1996; Citri et al., 2003).
Materials and methods
Antibodies and inhibitors
The following antibodies were purchased: anti-ErbB2, anti-ErbB3, anti-JNK1, anti-Tiam1, anti-Dbs, anti-HA, and anti-MBP (Santa Cruz Biotechnology, Inc.); anti-autophosphorylated (Tyr1112) ErbB2 (Invitrogen); anti-phosphorylated active (pThr183/pTyr185) JNK (Cell Signaling Technology); antiphosphorylated Tyr (pTyr) and anti-NGF (Millipore); anti-Rac1, anti-Cdc42, and anti-actin (BD Biosciences); anti-S100β (Dako); antineurofilament (Covance); anti-FLAG (Sigma-Aldrich); and anti-GFP (Medical & Biological Laboratories). The rabbit antiserum for Dock7 was generated against a KELFALHPSPDEEE peptide. The polyclonal anti-Dock7 antibody was affinity purified using a peptide-conjugated resin. The rabbit antiserum for phosphorylated (pTyr1118) Dock7 was generated against a phosphorylated peptide ETVPQLpYDFTET. The polyclonal anti-(pTyr1118)Dock7 antibody was affinity purified using a phosphorylated peptide-conjugated resin from nonadsorbed fractions of a nonphosphorylated peptide ETVPQLYDFTET-conjugated resin. Peroxidase and fluorescence-labeled secondary antibodies were purchased from GE Healthcare and Invitrogen, respectively. The following inhibitors were purchased: ErbB3-Fc, which possesses the extracellular domain of ErbB3 fused to the Fc region of an IgG (R&D Systems); and C. difficile Toxin B, Clostridium botulinum C3 exoenzyme, AG825, SP600125/JNK inhibitor II, and JNK inhibitor I (EMD).
Plasmids
The coding regions of three alternative splicing variants of Dock7 (available from GenBank/EMBL/DDBJ under accession nos. DQ118679, DQ118680, and DQ309763) were isolated by the method of 5′ and 3′ rapid amplification of cDNA from human brain (Marathon-Ready cDNA; Clontech Laboratories, Inc.), according to the manufacturer's protocol. The major variant DQ118679 was ligated into the mammalian FLAG- and GFP-tagged expression vectors pCMV-FLAG and pEGFP-C1. The cDNA fragments encoding DHR-1 (aa 1–691), middle region 1 (aa 692–1110), middle region 2 (aa 1111–1431), and DHR-2 (aa 1432–1992) of Dock7 were also inserted into pCMV-FLAG. The constructs of the full-length Dock7 harboring the Tyr-1118–to–Phe, Tyr-1138–to–Phe, Tyr-1225–to–Phe, Tyr-1233–to–Phe, Tyr-1375–to–Phe, or Tyr-1429–to–Phe mutation and the Y1118F, Y1138F, Y1225F, Y1233F, Y1375F, and Y1429F mutants of the middle region 2 were produced by the overlapping PCR method and ligated into pCMV-FLAG. The wild-type and Y1118, Y1138F, Y1225F, Y1233F, Y1375F, and Y1429F mutants of the full-length Dock7 were also inserted into pEGFP-C1. The wild-type and Y1118, Y1138F, Y1225F, Y1233F, Y1375F, and Y1429F constructs resistant to Dock7-1 siRNA were made by replacing 5′-AAGACGTTCAATGTCAATAGA-3′ with 5′-ACGTAGATCAATGAGTATTGA-3′ at nucleotides 207–227 without amino acid mutations (underlined nucleotides indicate replacements). The partial nucleotide sequences of mouse and rat Dock7 (Available from GenBank/EMBL/DDBJ under accession nos. DQ109674 and DQ124295) were isolated from cDNAs of mouse brain and rat Schwann cells, respectively. The regions encoding ErbB2 and ErbB3 (Available from GenBank/EMBL/DDBJ under accession no. AY686636) were isolated by 5′ and 3′ rapid amplification of cDNA from mouse brain Marathon-Ready cDNA and subcloned into pCMV. The coding region of ELMO1 was amplified from human brain cDNA and inserted into pCMV-HA. The mammalian expression plasmids pCMV–FLAG–constitutively active Dbs-DHPH and pCMV-MBP-CrkII and the Escherichia coli GST-tag expression plasmids pET42a–Rac1-GTP and Cdc42-GTP binding domain (Cdc42/Rac interactive binding domain [CRIB]) of Pak1, pET42a wild-type Rho GTPases (Rac1, Cdc42, and RhoA), and pET42a–guanine nucleotide–free Rho GTPases (Rac1G15A, Cdc42G15A, and RhoAG17A) were constructed as previously described (Yamauchi et al., 2005a,b). The mammalian expression plasmids encoding Dock180 were provided by M. Matsuda (Osaka University, Osaka, Japan) and K.S. Ravichandran (University of Virginia, Charlottesville, VA). All sequences were confirmed by automatic sequencers (Applied Biosystems).
Cell culture
Primary Schwann cells were prepared from sciatic nerves of Sprague-Dawley rats at postnatal day 1 (Yamauchi et al., 2004). Schwann cells were cultured on poly-lysine–coated dishes in DME containing 10% heat-inactivated FBS and 50 μg/ml gentamicin at 37°C and plated for experiments on collagen (type I)-coated dishes, unless otherwise indicated. Before the experiments were performed, Schwann cells were cultured in Sato medium containing 1 mg/ml BSA for 24 h. DRG neurons were dissociated from rat embryos at gestational day 15, purified, and cultured in DME-GlutaMax (Invitrogen) containing 10% FBS and 100 ng/ml NGF on collagen-coated dishes (Yamauchi et al., 2004). After 2–3 wk, DRG neurons were cultured in DME-GlutaMax containing 10% FBS and 20 ng/ml NGF. 293T and Cos-7 cells (Human Science Research Resource Bank) were cultured in tissue culture dishes in DME containing 10% FBS, 50 U/ml penicillin, and 50 μg/ml streptomycin, and Cos-7 cells were plated for experiments on collagen-coated dishes. Before the experiments, 293T and Cos-7 cells were cultured in DME containing 1% FBS and 1 mg/ml BSA for 24 h. Unless otherwise indicated, Schwann cells and Cos-7 cells were pretreated with or without 2 ng/ml C. difficile Toxin B for 24 h, 2 μg/ml C3 exoenzyme for 24 h, 10 μM AG825 for 45 min, 10 μM SP600125 for 45 min, or 20 μM JNK inhibitor I for 24 h before stimulation with 20 ng/ml NRG1 (R&D Systems), NT3 (PeproTech), or IGF-I (Invitrogen) for 0–360 min. To confirm cell viability under these experimental conditions, Schwann cells and Cos-7 cells were stained with 0.4% trypan blue. Trypan blue–incorporating cells were <1% in each experiment.
Boyden chamber migration assay
Cell migration was routinely measured using a 24-well Boyden chamber, as previously described (Yamauchi et al., 2004). In brief, in the case of assaying the effect of ErbB3-Fc on DRG neurons' conditioned medium, polyethylene terephthalate (8-μm pore size) filters were coated with axonal membranes from DRG neurons (Grimes et al., 1996). In other experiments, filters were essentially coated with collagen (type I), except for the comparison of extracellular matrix proteins collagen (type IV), fibronectin, and laminin. Cells (0.5 × 105 cells for Schwann cells or 5 × 105 cells for Cos-7 cells) in 500 μl of normal medium per well were loaded into the upper chambers, which were inserted into the tissue culture wells in 750 μl of conditioned medium containing 5 μg/ml anti-NGF antibody in the presence of 5 μg/ml of control IgG or ErbB3-Fc per well or in normal medium containing 50 ng/ml NRG1, NT3, or IGF-I per well. After incubation at 37°C for 6 h, the filters were stained with Giemsa solution or fixed with PFA to detect cells expressing GFP. No difference in cell number was observed at 6 h in the presence or absence of NRG1, NT3, or IGF-1. The number of stained or GFP-fluorescent migrating cells at the bottom surface of the filters was counted at four fields per filter in two to four independent experiments. In the presence of 5 μg/ml of control IgG or ErbB3-Fc for 8 h, trypan blue–incorporating cells were <0.5%.
Migration assay using reaggregated Schwann cells
To mimic physiological conditions, Schwann cell migration was also measured using DRG neurons and reaggregated Schwann cells essentially as described previously (Yamauchi et al., 2004). In brief, DRG neurons were plated onto the center of a collagen-coated dish and allowed to extend axons outwardly. Schwann cell reaggregates were formed by plating Schwann cells on Ultra Low Attachment dishes (Corning) for 4 h and on Petri dishes (Barloworld Scientific) for 20 h with gentle agitation every 3–4 h. In the case of analyzing the effect of 5 μg/ml ErbB3-Fc, 5 μg/ml anti-NGF antibody was added into culture medium of DRG neurons. For the other experiments, the medium was changed into normal medium containing vehicle or 20 ng/ml NRG1. Individual Schwann cells were allowed to migrate out of the reaggregates along the axons. After incubation at 37°C for 6 h, cells were fixed with PFA, blocked, and immunostained. The distance of migration was calculated by measuring the size of the reaggregates over time, subtracting the mean initial size of the reaggregates, and dividing the remaining distance in half. Experiments were performed by measuring eight reaggregates per dish in two independent experiments.
Fluorescence images
Cells on collagen-coated glass coverslips or filters of Boyden chamber and reaggregated Schwann cells were air-dried, fixed with 4% PFA, blocked with Immuno-Block (Dainippon Sumitomo Pharma) in phosphate-buffered saline–0.1% Tween-20, incubated with primary antibodies, and treated with fluorescence-labeled secondary antibodies. The coverslips and filters were mounted with Vectashield (Vector Laboratories) onto slides for observation by confocal and fluorescence microscopy. The confocal images were collected with a microscope (IX81; Olympus) with a laser-scanning FV500 system (Olympus) and analyzed with FluoView software (Olympus). The primary antibodies used for confocal images were anti-Dock7, anti-(pTyr1118)Dock7, anti-ErbB2, and anti-(pTyr1112)ErB2. The fluorescence images were captured with a microscope system (TE-300; Nikon) and analyzed with AxioVision software (Carl Zeiss, Inc.). The primary antibodies used for the fluorescence images were anti-S100β to identify Schwann cells when GFP constructs were not transfected and anti-neurofilament to identify DRG axons. The live imaging was performed using a microscope system (DMI4000B; Leica) equipped with an INUG2-ZILCS stage top incubator (Tokai Hit) and AF6000 software (Leica). The time frame was 60–300 min after putting Schwann cell reaggregates on DRG neurons, which were replaced with a fresh medium in the presence or absence of 20 ng/ml NRG1. To avoid fading of the GFP fluorescence, the intensity levels were fixed at less than position 2. Captured images were thus adjusted using the brightness switch on AF6000 software. Image sequence was recorded at one frame per 5 min and played at three frames per second.
Immunoprecipitation and immunoblotting
Cells were lysed in lysis buffer B (50 mM Hepes-NaOH, pH 7.5, 20 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethane sulfonylfluoride, 1 μg/ml leupeptin, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, and 0.5% NP-40) and the lysates were centrifuged. The supernatants were mixed with protein G resin that was preadsorbed with various antibodies. The immunoprecipitates or the proteins in the cell lysates were denatured and then subjected to SDS-PAGE. The electrophoretically separated proteins were transferred to PVDF membranes, blocked, and immunoblotted. The bound antibodies were detected using the ECL or ECL-Plus system (GE Healthcare). The band images were captured with a GT-7000U scanner (Epson) and analyzed with ImageJ software (National Institutes of Health; http://rsb.info.nih.gov/ij/).
In vitro tyrosine-phosphorylation reaction
250 ng of purified immobilized FLAG-Dock7 proteins were incubated with 20 μM of cold ATP in the presence or absence of 100 ng ErbB2 kinase in 30 μl of reaction buffer (20 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethane sulfonylfluoride, 1 μg/ml leupeptin, and 1 mM EDTA) at 30°C for 30 min and then chilled on ice. Tyrosine-phosphorylated FLAG-Dock7 proteins were washed with reaction buffer and used for guanine nucleotide releasing assays for Rac1 and Cdc42.
Guanine nucleotide exchange assays
Guanine nucleotide exchange assays were performed as previously described (Yamauchi et al., 2005a). In brief, for the guanine nucleotide binding assay, 125 ng of immobilized FLAG–Dock7–DHR-2 or the immunoprecipitates were incubated in 30 μl of reaction buffer containing 16 ng/μl each of GST-Rho GTPase, 33 ng/μl BSA, and 3 μM [3H]GDP (0.3 μCi/μl) at 30°C for 0–30 min. The reactions were stopped by adding 1 ml of ice-cold wash buffer (20 mM Hepes-NaOH, pH 7.5, and 10 mM MgCl2) and filtered through 0.45-μm nitrocellulose membranes. The membranes were immediately washed with ice-cold wash buffer and air dried. The radioactivity remaining on each membrane was measured using a LSC-6100 liquid scintillation counter (Aloka). For the guanine nucleotide–releasing assay, [3H]GDP-bound GST-Rho GTPases were obtained by incubation with reaction buffer containing 125 ng/μl each of Rho GTPase, 250 ng/μl BSA, 5 mM EDTA, and 0.3 μM [3H]GDP (0.3 μCi/μl) at 30°C for 90 min. The reaction was stopped by adding 5 mM MgCl2, and mixtures were immediately cooled on ice. 125 ng of immobilized FLAG-Dock7-DHR-2, 250 ng FLAG-Dock7 proteins, or the immunoprecipitates were incubated in 30 μl of reaction buffer containing 16 ng/μl GST-Rho GTPase–[3H]GDP, 33 ng/μl BSA, and 3 μM of cold GDP at 30°C for 0–30 min. The reaction was stopped and filtered. The radioactivity remaining on each membrane was measured. 3–10 separate experiments were performed.
RNA preparation and RT-PCR analysis
Total RNA was isolated by Trizol reagent (Invitrogen). The cDNA were prepared from 1 μg of total RNA with Superscript II (Invitrogen), according to the manufacturer's instructions. PCR amplification (Takara Bio Inc.) was performed at 30 cycles, each cycle consisting of denaturation at 94°C for 1 min, annealing at 56.5°C for 1 min, and extension at 72°C for 1 min. The primers used were the following: 5′-CCTTCATTCCTTCGGGGAAAAGTG-3′ (sense) and 5′-GGACCTGGAGTGCATCTTCTTC-3′ (antisense) for Dock180; 5′-CTTTTGAGCCAGTTCCCCAACG-3′ (sense) and 5′-CTCCTCCACTTTGGGAGTCTTG-3′ (antisense) for Dock2; 5′-TTCTTCCTTCGGAATAAAGAGTATGTGTG-3′ (sense) and 5′-GTCACCCATCATCATCATGTTTCC-3′ (antisense) for Dock3; 5′-CCATTTTTCTTAAGAAATAAGAAGTTTGTATGTCGAG-3′ (sense) and 5′-TGATTGCCCTGTAATATTGCTTACTTCAGC-3′ (antisense) for Dock4; 5′-GTGGGATACTACGGGCAG-3′ (sense) and 5′-CAGGATGGAACCATCAGATCC-3′ (antisense) for Dock5; 5′-ATCCTTGAAGCCCACCGAGAC-3′ (sense) and 5′-TGACCGCCGGCCCAAGCTC-3′ (antisense) for Dock6; 5′-GAACACCAGGAGGATCCTGAAATGTTG-3′ (sense) and 5′-GAGATCCATTTTGCGAAGGCTCATTCG-3′ (antisense) for Dock7; 5′-ACCTCTACTCACGACAAGCTGC-3′ (sense) and 5′-TCAGCTGCCCTGTGACAACTG-3′ (antisense) for Dock8; 5′-ATCCTGACAGTTAGTGTTCCCAC-3′ (sense) and 5′-TCAGCAGCAAGTCATGGAGAG-3′ (antisense) for Dock9; 5′-GACTTCAAAAAATTATCGGACCTCTATTATG-3′ (sense) and 5′-GAGATTCTGGCTGCCACTTTTG-3′ (antisense) for Dock10; 5′-GAGAAACTGACTCAAGTCTATAGAACTC-3′ (sense) and 5′-TCATACTTCAGAGTATCTTGGGGAAC-3′ (antisense) for Dock11; and 5′-ATGGATGACGATATCGCTGCGCTC-3′ (sense) and 5′-CTAGAAGCATTTGCGGTGCACGATG-3′ (antisense) for β-actin.
Recombinant proteins
Unless otherwise indicated, all steps were performed at 4°C as previously described (Yamauchi et al., 2005a,b; Chan et al., 2006). FLAG-tagged DHR-2, wild-type, and Y1118F proteins of Dock7 were purified from serum-starved 293T cells transiently transfected with pCMV–FLAG–Dock7–DHR-2, pCMV-FLAG-Dock7, and pCMV-FLAG-Dock7Y1118F, respectively, using the CalPhos transfection reagent (Takara Bio Inc.) according to the manufacturer's protocol. In brief, cells were lysed in lysis buffer A (50 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 3 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethane sulfonylfluoride, 1 μg/ml leupeptin, 1 mM EDTA, and 0.5% NP-40) and centrifuged. The supernatants were mixed with protein G resin (GE Healthcare) that was preadsorbed with an anti-FLAG antibody. Bound FLAG-Dock7 proteins were extensively washed with lysis buffer A containing 500 mM NaCl and subsequently with lysis buffer A containing 500 mM NaCl and 50 mM EDTA and eluted with lysis buffer A containing 20 mM FLAG peptide (Sigma-Aldrich), according to the manufacturer's protocol. The buffer contained in elution fractions was exchanged with reaction buffer (20 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethane sulfonylfluoride, 1 μg/ml leupeptin, and 1 mM EDTA). The aliquots were stored at −80°C until use. The intracellular kinase domain of ErbB2 (HTScan ErbB2 kinase) was purchased from Cell Signaling Technology. GST-tagged Pak1-CRIB, wild-type Rho GTPases, and guanine nucleotide–free Rho GTPases were purified from E. coli BL21 (DE3) pLysS cells transformed with pET42a-Pak1-CRIB, pET42a-wild-type Rho GTPases, and pET42a–guanine nucleotide–free Rho GTPases, respectively. In brief, cells were treated with 0.4 mM isopropyl-1-thio-β-d-galactopyranoside at 37°C for 1.5 h and harvested by centrifugation. A cell-free extract was made by the addition of 500 μg/ml lysozyme and 100 μg/ml DNase I in extraction buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethane sulfonylfluoride, 1 μg/ml leupeptin, 1 mM EDTA, and 0.5% NP-40). The lysates were centrifuged, and the supernatants were mixed with glutathione resin (GE Healthcare). Bound proteins were washed with extraction buffer and eluted with extraction buffer containing 20 mM glutathione. The buffer contained in elution fractions was dialyzed against reaction buffer for GST-Pak1-CRIB or against reaction buffer containing 0.1 μM GDP for GST-Rho GTPases. The aliquots were stored at −80°C until use. The Coomassie brilliant blue staining was performed by using the Rapid Coomassie or One Step Coomassie kit (Nakalai), according to the manufacturer's protocol.
siRNA transfection
The siRNAs were transfected into primary Schwann cells using the Oligofectamine or Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. The medium was replaced at 24 h after transfection. The efficiencies of protein depletion were 95 ± 3.1% for Dock7-1 siRNA, 83 ± 5.7% for Dock7-2 siRNA, 87 ± 6.7% for ErbB2 siRNA, 93 ± 5.5% for ErbB3 siRNA, 92 ± 5.7% for Tiam1 siRNA, 91 ± 7.9% for Dbs siRNA, 98 ± 0.33% for Rac1 siRNA, 81 ± 2.9% for Cdc42-1 siRNA, and 98 ± 1.8% for Cdc42-2 siRNA at 48 h after transfection.
siRNA preparation
The 21-nt siRNA duplexes were synthesized by Nippon EGT. The target nucleotide sequences for the first Dock7 (Dock7-1) siRNA (5′- AAGACGTTCGATGTCAATAGA-3′), the second, nonoverlapping Dock7 (Dock7-2) siRNA (5′-AAGTCTTAATTTTGCCAACCG-3′), ErbB2 siRNA (5′-AAGTCTCACAGAGATCCTGAA-3′), ErbB3 siRNA (5′-AAGTTCACTCAGCTAACAGAG-3′), Tiam1 siRNA (5′-AAGAACATTTAACAAGCAACG-3′; Yamauchi et al., 2005b), Dbs siRNA (5′-AAGGCTAAAGTGAAGGAGGAT-3′; Yamauchi et al., 2005a), Rac1 siRNA (5′-AAGATTATGACAGACTGCGTC-3′), the first Cdc42 (Cdc42-1) siRNA (5′-AACTATGCAGTCACAGTTATG-3′), and the second, nonoverlapping Cdc42 (Cdc42-2) siRNA (5′-AAACCGTTAAGTTATCCACAG-3′) were designed according to an online software, siRNA Sequence Selector (Clontech Laboratories, Inc.; http://bioinfo.clontech.com/rnaidesigner/). The target sequence of the control Photinus pyralis luciferase siRNA was 5′-AAGCCATTCTATCCTCTAGAG-3′, which does not have significant homology to any mammalian gene sequences. To confirm cell viability under these experimental conditions, Schwann cells were stained with trypan blue. Trypan blue–positive cells in tissue culture dishes numbered <1% at 48 h after siRNA transfection (<0.5% for control luciferase siRNA, 0.7 ± 0.02% for Dock7-1 siRNA, 0.5 ± 0.03% for Dock7-2 siRNA, 0.5 ± 0.03% for ErbB2 siRNA, 0.7 ± 0.06% for ErbB3 siRNA, 0.5 ± 0.09% for Tiam1 siRNA, 0.7 ± 0.07% for Dbs siRNA, 0.7 ± 0.05% for Rac1 siRNA, 0.5 ± 0.02% for Cdc42-1 siRNA, and 0.8 ± 0.03% for Cdc42-2 siRNA).
Plasmid transfection
For primary Schwann cells, pEGFP, pEGFP-Dock7-1 siRNA-resistant wild-type, Y1118F, Y1138F, Y1225F, Y1233F, Y1375F, or Y1429F Dock7 was cotransfected with control, Dock7-1, Rac1, or Cdc42-2 siRNA by using the Lipofectamine 2000 reagent or Nucleofector II (Amaxa Biosystems) with the Basic Neuron Nucleofector Transfection kit (Amaxa Biosystems), according to the manufacturer's protocol. Transfection efficiency was 15–20% using GFP-expressing plasmid as the control. The medium was replaced at 24 h after transfection. To perform the Boyden chamber migration assay, Schwann cells were cultured in Sato medium containing 1 mg/ml BSA for another 24 h. To assay the migration of reaggregated Schwann cells, cells were allowed to form reaggregates in DME containing 10% FBS for another 24 h. For 293T and Cos-7 cells, pCMV-FLAG-Dock7, pCMV-FLAG-Dock7Y1118F, pCMV-FLAG-Dock7Y1138F, pCMV-FLAG-Dock7Y1225F, pCMV-FLAG-Dock7Y1233F, pCMV-FLAG-Dock7Y1375F, pCMV-FLAG-Dock7Y1429F, pCMV–FLAG-Dock7–DHR-1, pCMV–FLAG–Dock7–middle region 1, pCMV–FLAG-Dock7–middle region 2 or the Tyr-to-Phe mutant, or pCMV–FLAG–Dock7–DHR-2 was transfected with or without pCMV-ErbB2 and pCMV-ErbB3 using the CalPhos transfection reagent. Transfection efficiency typically exceeded 95% using GFP-expressing plasmid as the control.
Affinity precipitation of GEFs
Dock7 proteins or Dbs-DHPH in the cell lysates was affinity precipitated with 20 μg GST-Rac1G15A, GST-Cdc42G15A, or GST-RhoAG17A, which are guanine nucleotide–free Rho GTPases. A Gly-to-Ala mutation of residue 15 in Rac1 and Cdc42 or residue 17 in RhoA decreases their nucleotide binding (Arthur et al., 2002). Active GEFs preferentially interact with guanine nucleotide–free forms of the small GTPases (Arthur et al., 2002; Schmidt and Hall, 2002; Rossman et al., 2005). The affinity precipitation was also performed using 20 μg GST wild-type Rho GTPase (Rac1, Cdc42, or RhoA). Affinity-precipitated GEFs were detected by immunoblotting (Yamauchi et al., 2005a,b).
Detection of active Rho GTPases
To detect active GTP-bound Rac1 and Cdc42 in the cell lysates, we performed affinity precipitation by using 20 μg GST-Pak1-CRIB, which binds to their GTP-bound forms. To compare the total amount of GTPase, immunoblotting was also performed with an anti-Rac1 or Cdc42 antibody. Two to five separate experiments were performed. The band intensity in the immunoblot was quantified, and the levels of Rac1-GTP and Cdc42-GTP were normalized to the amount of each total GTPase (Yamauchi et al., 2005a,b).
JNK assay
The cell lysates were immunoblotted with an anti-(pThr183/pTyr185)JNK antibody that recognizes the active form. To compare the total amount of JNK, immunoblotting was also performed with an anti-JNK antibody. Three to five separate experiments were performed. The band intensity in the immunoblot was quantified, and the levels of the phosphorylated forms were normalized to the amount of total kinase.
Statistical analysis
Values shown represent the mean ±SD from separate experiments. Analysis of variance (ANOVA) was followed by Fisher's protected least significant difference post hoc comparisons (*, P < 0.01; **, P < 0.015; ***, P < 0.02).
Online supplemental material
Fig. S1 demonstrates that Schwann cell migration requires the activation of the Rho family of small GTPases but is not dependent on Tiam1 or Dbs of the Dbl family GEFs. Fig. S2 characterizes the anti-Dock7 and anti-(pTyr1118)Dock7 antibodies. Fig. S3 shows the purification of the DHR-2, wild-type, and Y1118F mutant proteins of Dock7. Fig. S4 illustrates that the middle region 2 of Dock7 inhibits NRG1-induced migration and activation of Rac1 and Cdc42 in Cos-7 cells cotransfected with ErbB2 and ErbB3. Fig. S5 demonstrates that Dock7 does not interact with ELMO1 and CrkII. Videos 1 and 2 illustrate time-lapse imaging of vehicle-stimulated migration from reaggregates of control siRNA-transfected Schwann cells. Videos 3 and 4 demonstrate NRG1-stimulated migration from reaggregates of control siRNA-transfected Schwann cells. Video 5 demonstrates vehicle-stimulated migration from reaggregates of Rac1 siRNA-transfected Schwann cells. Video 6 illustrates NRG1-stimulated migration from reaggregates of Rac1 siRNA-transfected Schwann cells. Video 7 illustrates vehicle-stimulated migration from reaggregates of Cdc42 siRNA-transfected Schwann cells. Video 8 shows NRG1-stimulated migration from reaggregates of Cdc42 siRNA-transfected Schwann cells. Video 9 shows vehicle-stimulated migration from reaggregates of Dock7 siRNA-transfected Schwann cells. Video 10 represents NRG1-stimulated migration from reaggregates of Dock7 siRNA-transfected Schwann cells.
Supplemental Material
Acknowledgments
We thank Drs. E.M. Shooter and Y. Kaziro for their participation in insightful discussions and for providing encouragement. We thank Drs. S. Kusakawa and S. Takashima for their participation in helpful discussions, M. Matsuda and K.S. Ravichandran for providing plasmids, and H. Aizawa and M. Yamamoto for assistance in the time-lapse imaging studies.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Japan Society for the Promotion of Science, and the Ministry of Human Health and Welfare of Japan and was supported partially by research grants from the Astellas Metabolic Disease Foundation, the Human Science Foundation of Japan, the Kampou Medical Foundation, the Kato Bioscience Foundation, the Kowa Life Science Foundation, the Nakajima Foundation, the Samuro Kakiuchi Memorial Foundation, and the Takeda Science Foundation.
Abbreviations used in this paper: ANOVA, analysis of variance; CRIB, Cdc42/Rac interactive binding domain; DH, Dbl homology; DHR, Dock homology region; DRG, dorsal root ganglion; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; IGF, insulin-like growth factor; MAP, microtubule-associated protein; NRG1, neuregulin-1; NT3, neurotrophin-3; PH, pleckstrin homology; SH, Src homology; WASP, Wiskott-Aldrich syndrome protein.
References
- Arthur, W.T., S.M. Ellerbroek, C.J. Der, K. Burridge, and K. Wennerberg. 2002. XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J. Biol. Chem. 277:42964–42972. [DOI] [PubMed] [Google Scholar]
- Benninger, Y., T. Thurnherr, J.A. Pereira, S. Krause, X. Wu, A. Chrostek-Grashoff, D. Herzog, K.A. Nave, R.J. Franklin, D. Meijer, et al. 2007. Essential and distinct roles for cdc42 and rac1 in the regulation of Schwann cell biology during peripheral nervous system development. J. Cell Biol. 177:1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnera, E., L. Haney, C. Grimsley, M. Lu, S.F. Walk, A.C. Tosello-Trampont, I.G. Macara, H. Madhani, G.R. Fink, and K.S. Ravichandran. 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 4:574–582. [DOI] [PubMed] [Google Scholar]
- Bunge, R.P. 1993. Expanding roles for the Schwann cell: ensheathment, myelination, trophism and regeneration. Curr. Opin. Neurobiol. 3:805–809. [DOI] [PubMed] [Google Scholar]
- Chan, J.R., C. Jolicoeur, J. Yamauchi, J. Elliott, J.P. Fawcett, B.K. Ng, and M. Cayouette. 2006. The polarity protein Par-3 directly interacts with the p75 neurotrophin receptor to regulate myelination. Science. 314:832–836. [DOI] [PubMed] [Google Scholar]
- Chang, L., Y. Jones, M.H. Ellisman, L.S. Goldstein, and M. Karin. 2003. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell. 4:521–533. [DOI] [PubMed] [Google Scholar]
- Cheng, H.L., M.L. Steinway, J.W. Russell, and E.L. Feldman. 2000. GTPases and phosphatidylinositol 3-kinase are critical for insulin-like growth factor-I-mediated Schwann cell motility. J. Biol. Chem. 275:27197–27204. [DOI] [PubMed] [Google Scholar]
- Citri, A., K.B. Skaria, and Y. Yarden. 2003. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp. Cell Res. 284:54–65. [DOI] [PubMed] [Google Scholar]
- Côté, J.F., and K. Vuori. 2002. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115:4901–4913. [DOI] [PubMed] [Google Scholar]
- Côté, J.F., A.B. Motoyama, J.A. Bush, and K. Vuori. 2005. A novel and evolutionarily conserved PtdIns(3, 4, 5)P3-binding domain is necessary for DOCK180 signaling. Nat. Cell Biol. 7:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., J.B. Dickerson, F. Guo, J. Zheng, and Y. Zheng. 2004. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA. 101:7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garratt, A.N., S. Britsch, and C. Birchmeier. 2000. Neuregulin, a factor with many functions in the life of a Schwann cell. Bioessays. 22:987–996. [DOI] [PubMed] [Google Scholar]
- Grimes, M.L., J. Zhou, E.C. Beattie, E.C. Yuen, D.E. Hall, J.S. Valletta, K.S. Topp, J.H. LaVail, N.W. Bunnett, and W.C. Mobley. 1996. Endocytosis of activated TrkA: evidence that nerve growth factor induces formation of signaling endosomes. J. Neurosci. 16:7950–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, H., E. Kiyokawa, S. Tanaka, K. Nagashima, N. Gotoh, M. Shibuya, T. Kurata, and M. Matsuda. 1996. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell. Biol. 16:1770–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, Y.S., S.K. Kim, C.I. Seo, Y.K. Kim, B.J. Sung, H.S. Lee, J.I. Lee, S.Y. Park, J.H. Kim, K.Y. Hwang, et al. 2004. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 23:2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, M., T. Ishizaki, N. Watanabe, M. Uehata, O. Kranenburg, W.H. Moolenaar, F. Matsumura, M. Maekawa, H. Bito, and S. Narumiya. 1998. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J. Cell Biol. 141:1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Z. Rajfur, C. Borchers, M.D. Schaller, and K. Jacobson. 2003. JNK phosphorylates paxillin and regulates cell migration. Nature. 424:219–223. [DOI] [PubMed] [Google Scholar]
- Huang, C., K. Jacobson, and M.D. Schaller. 2004. MAP kinases and cell migration. J. Cell Sci. 117:4619–4628. [DOI] [PubMed] [Google Scholar]
- Just, I., J. Selzer, M. Wilm, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 375:500–503. [DOI] [PubMed] [Google Scholar]
- Katoh, H., and M. Negishi. 2003. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 424:461–464. [DOI] [PubMed] [Google Scholar]
- Kawauchi, T., K. Chihama, Y. Nabeshima, and M. Hoshino. 2003. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 22:4190–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanthappa, N.K., E.S. Anton, and W.D. Matthew. 1996. Glial growth factor 2, a soluble neuregulin, directly increases Schwann cell motility and indirectly promotes neurite outgrowth. J. Neurosci. 16:4673–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, N., M. Irani-Tehrani, W.B. Kiosses, M.A. Del Pozo, and M.A. Schwartz. 2002. Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat. Cell Biol. 4:639–647. [DOI] [PubMed] [Google Scholar]
- Nave, K.A., and J.L. Salzer. 2006. Axonal regulation of myelination by neuregulin 1. Curr. Opin. Neurobiol. 16:492–500. [DOI] [PubMed] [Google Scholar]
- Nodari, A., D. Zambroni, A. Quattrini, F.A. Court, A. D'Urso, A. Recchia, V.L. Tybulewicz, L. Wrabetz, and M.L. Feltri. 2007. β1 integrin activates Rac1 in Schwann cells to generate radial lamellae during axonal sorting and myelination. J. Cell Biol. 177:1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetto, C., A. Bardelli, F. Maina, P. Longati, G. Panayotou, R. Dhand, M.D. Waterfield, and P.M. Comoglio. 1993. A novel recognition motif for phosphatidylinositol 3-kinase binding mediates its association with the hepatocyte growth factor/scatter factor receptor. Mol. Cell. Biol. 13:4600–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman, K.L., C.J. Der, and J. Sondek. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180. [DOI] [PubMed] [Google Scholar]
- Rushton, E., R. Drysdale, S.M. Abmayr, A.M. Michelson, and M. Bate. 1995. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 121:1979–1988. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., and A. Hall. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587–1609. [DOI] [PubMed] [Google Scholar]
- Takenawa, T., and H. Miki. 2001. WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 114:1801–1809. [DOI] [PubMed] [Google Scholar]
- Tsai, C.M., A. Levitzki, L.H. Wu, K.T. Chang, C.C. Cheng, A. Gazit, and R.P. Perng. 1996. Enhancement of chemosensitivity by tyrphostin AG825 in high-p185(neu) expressing non-small cell lung cancer cells. Cancer Res. 56:1068–1074. [PubMed] [Google Scholar]
- Watabe-Uchida, M., K.A. John, J.A. Janas, S.E. Newey, and L. Van Aelst. 2006. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 51:727–739. [DOI] [PubMed] [Google Scholar]
- Wu, Y.C., and H.R. Horvitz. 1998. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 392:501–504. [DOI] [PubMed] [Google Scholar]
- Yamauchi, J., J.R. Chan, and E.M. Shooter. 2004. Neurotrophins regulate Schwann cell migration by activating divergent signaling pathways dependent on Rho GTPases. Proc. Natl. Acad. Sci. USA. 101:8774–8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, J., J.R. Chan, Y. Miyamoto, G. Tsujimoto, and E.M. Shooter. 2005. a. The neurotrophin-3 receptor TrkC directly phosphorylates and activates the nucleotide exchange factor Dbs to enhance Schwann cell migration. Proc. Natl. Acad. Sci. USA. 102:5198–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, J., Y. Miyamoto, A. Tanoue, E.M. Shooter, and J.R. Chan. 2005. b. Ras activation of a Rac1 exchange factor, Tiam1, mediates neurotrophin-3-induced Schwann cell migration. Proc. Natl. Acad. Sci. USA. 102:14889–14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z.S., and E. Manser. 2005. PAK and other Rho-associated kinases-effectors with surprisingly diverse mechanisms of regulation. Biochem. J. 386:201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.