Abstract
The release of cytotoxic granule contents by cytotoxic T lymphocytes triggers apoptotic target cell death. Cytotoxic granules contain a pore-forming protein, perforin, and a group of serine proteases called granzymes. We expressed human granzyme A in bacteria as a proenzyme capable of in vitro activation by enterokinase. The recombinant activated enzyme has catalytic activity against substrates with Arg, preferably, or Lys at the P1 position, comparable to trypsin. An enzymatically inactive recombinant granzyme A, with the active site Ser mutated to Ala, was produced and used with affinity chromatography to identify potential substrates. Two granzyme A-binding cytoplasmic proteins of molecular mass 33 and 44 kDa were isolated and identified by tryptic fragment sequencing as PHAP I and II, ubiquitous putative HLA-associated proteins, previously coisolated by binding to an HLA class II peptide. PHAP II forms an SDS-stable complex with recombinant mutant granzyme A and coprecipitates with it from cytoplasmic extracts. PHAP II, either purified or in cell lysates, is cleaved by the recombinant enzyme at nanomolar concentrations to a 25-kDa fragment. PHAP II begins to be degraded within minutes of initiation of cytotoxic T lymphocyte attack. PHAP I and II are candidate participants in the granzyme A pathway of cell-mediated cytotoxicity.
When cytotoxic T lymphocytes (CTLs) release their cytotoxic granule contents into the intercellular space formed when the CTL binds to a target cell, apoptotic target cell death is triggered (1). CTL granules contain a pore-forming protein, perforin, and a group of serine proteases, termed granzymes, in a proteoglycan matrix (2–7). Perforin is thought to poke holes in the target cell, allowing granzymes to enter and initiate cell death. Recently, the molecular pathways involved in the induction of cell death by granzyme B have been identified. Granzyme B cleaves members of the ICE/ced-3 family of cysteine proteases, thereby activating a ubiquitous apoptotic cascade (8–12). Little is known about the mechanism of action of the other granzymes. In the mouse, only granzymes A and B are proteolytically active and expressed in significant amounts after CTL activation (13).
Noncytotoxic rat basophilic leukemia cells transfected with perforin and either granzyme A or B can trigger DNA degradation of target cells (14). Moreover, purified rat granzymes A and B synergistically enhance apoptosis of perforin-treated target cells (15). Perforin-deficient mice, generated by homologous recombination technology, have impaired immunity to lymphocytic choriomeningitis virus, Listeria monocytogenes infection, or syngeneic tumors (16). The granzyme A-deficient mouse is not deficient in its immune response to these challenges, but is unusually susceptible to infection with the mousepox virus, ectromelia (17, 18). This is noteworthy because pox viruses express cytokine response modifier proteins that inactivate granzyme B and other caspase proteases implicated in the granzyme B pathway (19). These results suggest that the granzyme A and B pathways are redundant, each sufficient to trigger apoptosis, but likely to act by different mechanisms.
Granzyme A, the most abundant protease in human CTL granules, is a tryptic protease which cleaves synthetic substrates with Lys or preferably Arg at the P1 position. It is the only granzyme that forms a disulfide-linked homodimer. It is produced in CTLs as a proenzyme, which can be activated by the dipeptidyl exopeptidase cathepsin C (20). Granzyme A has been shown to cleave nucleolin, interleukin 1β, fibronectin, type IV collagen, thrombin receptor, and pro-urokinase-type plasminogen activator in vitro, but the biological significance of these findings remains uncertain (21–25).
Previous attempts to produce enzymatically active granzyme A in bacteria resulted in insoluble and inactive enzyme (26). However, progranzyme A, capable of in vitro activation, has been produced in yeast (G. Duke Virca, personal communication) and from a vaccinia recombinant virus (20). In this paper we describe the production in Escherichia coli of a soluble granzyme A precursor, cleaved in vitro with enterokinase to an active protease (rGranA). RGranA cleaves known granzyme A synthetic substrates and is inhibited by known inhibitors of native granzyme A. We also produced an inactive enzyme by mutating the active site Ser to Ala and used it to identify two cytoplasmic proteins, one of which forms an SDS-soluble complex with mutant rGranA and is a substrate of the active enzyme. This protein begins to be degraded within minutes of CTL attack. These proteins have properties that support their role in a cell death pathway.
EXPERIMENTAL PROCEDURES
Production of Active rGranA.
Human granzyme A cDNA was PCR amplified with Vent polymerase (New England Biolabs) from reverse transcribed mRNA isolated from mitogen-activated peripheral blood cells (27). Primers were constructed from the human granzyme A sequence to contain BamHI and EcoRI restriction sites. The PCR product was directionally ligated into pGBT9 (CLONTECH). The granzyme insertion was excised and modified by PCR amplification with primers encoding an enterokinase site 5′ of the predicted first amino acid of the active enzyme and BamHI and XhoI restriction sites for insertion into pet26b (Novagen). (Fig. 1A) The sequence was verified by dideoxynucleotide sequencing (Sequenase; United States Biochemical/Amersham). Plasmid expression of transfected colonies of BL21-DE3 (Novagen) was induced with 1 mM isopropyl β-dthiogalactoside (IPTG). Bacterial pellets, resuspended in nickel column binding buffer (5 mM imidazole/500 mM NaCl/20 mM Tris⋅HCl, pH 7.9) with 0.1% Nonidet P-40 (Sigma), were sonicated and centrifuged for 20 min at 40,000 × g. rGranA was eluted with 125 mM imidazole elution buffer from a Novagen nickel column by using a BioLogic chromatography system (Bio-Rad). Fractions containing granzyme A were pooled and desalted over a HITRAP column (Pharmacia) into enterokinase buffer (20 mM Tris⋅HCl/50 mM NaCl/2 mM CaCl2) and digested with 10 units of porcine enterokinase (Sigma) at 21°C for 12 hr, desalted into nickel binding buffer, and repurified over a nickel column. The final material was stored in 500 mM NaCl/125 mM imidazole/50 mM Tris⋅HCl, pH 7.9, at −20°C. Protein concentration of granzyme A was determined by BCA assay (Pierce).
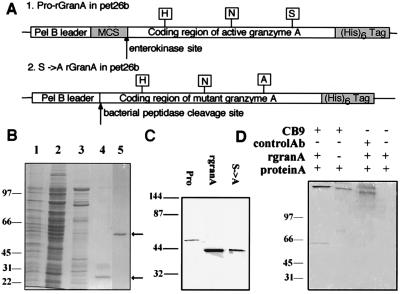
Figure 1.
rGranA and its Ser → Ala mutant S→ArGranA produced in E. coli yield a 42-kDa homodimer that reacts with mAb to native human granzyme A. (A) The plasmid for rGranA encodes an enterokinase site for in vitro activation within a multiple cloning site (MCS), but the S→ArGranA insert encodes a mutant enzyme whose active coding sequence begins after the bacterial peptidase cleavage site. (B) Pro-rGranA (arrows) purified by nickel chromatography from E. coli lysate and visualized after SDS/PAGE by Coomassie blue staining. Lanes: 1, bacterial lysate; 2, Ni column flow-through; 3, 60 mM imidazole wash; 4, eluate, reduced; and 5, eluate, nonreduced. (C) After enterokinase treatment, the 52-kDa proenzyme is cleaved to a 42-kDa active enzyme. The S→ArGranA protein comigrates with rGranA. (D) CB9 mAb to native human granzyme A specifically precipitates pro-rGranA.
Production of Inactive Mutated Granzyme A.
Mutation of the catalytic site Ser-184 to Ala was carried out by means of PCR mutagenesis. The mutated sequence was reamplified using a 5′ primer containing the bacterial pel B leader sequence 5′ of the coding sequence and the previously used 3′ primer. The sequence was confirmed by dideoxynucleotide sequencing. The mutant protein (S→ArGranA) was expressed and purified as above without enterokinase cleavage.
Production of mAb and Polyclonal Antiserum Against Human Granzyme A.
BALB/c mice were immunized with human granule proteins, extracted from human CTL clones by N2 cavitation and Percoll density gradient centrifugation as described (2). The primary immunization [60 μg of protein in complete Freund’s adjuvant (CFA) in the footpads] was followed at 4-week intervals with i.p. injection of 75–100 μg of granule protein in incomplete Freund’s adjuvant (IFA). Three days after the second boost, splenocytes from an immunized mouse were fused to X63.653 myeloma cells for hybridoma production as previously described (28). Culture supernatants and ascites fluid from hybridoma CB9 immunoprecipitated from human granules a 48-kDa protein on nonreducing gels and a 28-kDa protein on reducing gels, corresponding to the molecular mass of native human granzyme A (data not shown). Polyclonal antiserum was produced by immunizing mice with 10 μg of rGranA in CFA and boosting 3 times with rGranA in IFA.
Determination of Substrate and Inhibition Kinetics.
The active site concentration of rGranA was determined by titration with p-nitrophenyl p-guanidinobenzoate (NPGB) by measuring the change in absorption at 412 nm for the initial acylation step production of p-nitrophenol (ɛ = 12,600 M−1⋅cm−1). The number of catalytic sites was divided by 2 to give moles of granzyme. Granzyme A activity was determined by measuring the hydrolysis of thioester substrates in 0.1 M Hepes/0.01 M CaCl2, pH 7.5, buffer containing 5% dimethyl sulfoxide at 25°C in the presence of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (29). Substrate stock solutions were prepared in dimethyl sulfoxide and stored at −20°C. Initial rates were measured at 412 nm (ɛ412 = 14,150 M−1⋅cm−1) in a Beckman DU-650 spectrophotometer when 10 μl of an enzyme stock solution was added to a cuvette containing 1.0 ml of buffer, 0.236 mM DTNB, and 25 μl of substrate stock solution. After subtraction for background hydrolysis, duplicate measurements for each concentration were graphed on Lineweaver–Burk plots to determine the Km, kcat, and kcat/Km for each substrate. Correlation coefficients for all plots were >0.99. Specific inhibitors (25 μl in dimethyl sulfoxide) were added to 76 μM rGranA in 500 μl of 0.1 M Hepes/0.01 M CaCl2, pH 7.5, at 25°C. Residual enzymatic activity was measured by assaying cleavage of 200 μM Z-Arg-SBzl substrate (Z, benzyloxycarbonyl; SBzl, thiobenzyl ester) by 10-μl aliquots removed at various times. First-order inactivation rate constants (kobs) were obtained by plotting ln v0/vt vs. time. Correlation coefficients were ≥ 0.98.
Affinity Chromatography with S→ArGranA.
Purified S→ArGranA (4 mg) was coupled to 1 ml of Affi-Gel 10 (Bio-Rad) to produce an affinity column. Cytoplasmic extracts prepared from 1 × 109 K562 cells treated with Nonidet P-40 lysis buffer [0.5% Nonidet P-40/25 mM KCl/5 mM MgCl2/1 mM phenylmethanesulfonyl fluoride (PMSF)/10 mM Tris⋅HCl, pH 7.6] were applied to the column and eluted sequentially with 200 mM and 1 M NaCl in 50 mM Tris⋅HCl, pH 7.6. Protein-containing fractions were analyzed by SDS/PAGE. Fractions 13–15 and 21 were further purified by ion-exchange chromatography on a Bio-Scale Q2 column (Bio-Rad), and protein bands at 33 and 44 kDa were subjected to tryptic digestion and peptide sequencing by the Harvard Microchemistry Facility.
Granzyme A Cleavage Assay.
Pooled putative HLA-associated protein (PHAP) fractions 13–15 from the S→ArGranA column (250 ng of protein) or K562 cell lysates were incubated for 1–4 hr at 30°C with 125–400 nM rGranA, pro-rGranA, S→ArGranA, or rGranA pretreated with 1 mM PMSF in 20 μl of 1 mM CaCl2/1 mM MgCl2/50 mM Tris⋅HCl, pH 7.5. Reaction products were separated by SDS/PAGE on an 18% polyacrylamide gel and transferred to nitrocellulose for immunoblotting.
Immunoprecitation, Coimmunoprecipitation, and Immunoblotting.
For immunoprecipitation, purified rGranA was added to 10 μl of CB9 ascites fluid and 20 μl of a 70% slurry of Sepharose-protein A (Pharmacia) in 300 μl of PBS and incubated for 30 min at 4°C. After washing, samples were boiled in SDS sample buffer and analyzed by SDS/PAGE and stained with Coomassie blue. For coimmunoprecipitation, 5 μg of rGranA, S→ArGranA, or buffer was added to K562 cell lysate (1 × 106 cell equivalents) in 1 ml of Nonidet P-40 lysis buffer with 1% Triton X-100, previously cleared twice with preimmune rabbit serum and protein A-Sepharose. After incubation with 2.5 μl of anti-PHAP II rabbit antiserum and protein A-Sepharose for 2 hr at 4°C, the beads were washed extensively with RIPA buffer (150 mM NaCl/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/50 mM Tris⋅ HCl, pH 7.6) and boiled in SDS/PAGE sample buffer for electrophoresis. The coprecipitation with nickel resin was performed similarly, except that cells were lysed in 0.1% Nonidet P-40/50 mM Tris⋅HCl, pH 8, the preclearing was omitted, nickel resin was substituted for antiserum and protein A-Sepharose, and washes were in nickel binding buffer. For immunoblotting, electrophoresed samples were transferred to nitrocellulose. After blocking with 5% nonfat dry milk/0.05% Tween in Tris-buffered saline (TBS), the blot was incubated for 1 hr with 1/500 dilution of polyclonal mouse anti-granzyme A or anti-PHAP II peptide polyclonal rabbit antiserum [a kind gift of T. Copeland, National Cancer Institute (29)], washed, and incubated for 1 hr with 1/5000 donkey anti-rabbit immunoglobulin conjugated to horseradish peroxidase (Amersham) and visualized by chemiluminescence with Luminol/Enhancer solution (Pierce).
CTL Assay.
Effector CTLs [1 × 105 per condition of human lymphokine-activated killer cell lines generated by exposure to 1000 international units/ml interleukin 2 (Cetus Oncology) for 14 days] were incubated in 200 μl of RPMI medium 1640 supplemented with 10% FCS and 1 mM EDTA, with or without the addition of 250 μM Ph-NHCONH-CiTEtOIC (see Table 2), a granzyme A-specific inhibitor, in Eppendorf tubes for 1 hr at 37°C. The effector cells were then mixed with an equal number of K562 target cells and pelleted. After addition of 5 mM CaCl2, cells were harvested at various times and lysed at room temperature directly in SDS/PAGE sample buffer to which was added a mixture of inhibitors [0.5 mg/ml EDTA, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 1 μg/ml aprotinin, 1 mg/ml Pefabloc, 10 μg/ml E64 (Boehringer Mannheim), and 250 μM Ph-NHCONH-CiTEtOIC]. Samples were boiled for 5 min prior to electrophoresis and blotting. Control samples without added Ca2+ were also analyzed.
Table 2.
Specific tryptic protease inhibitors inhibit rGranA activity
| Inhibitor* | [I], μM |
kobs/[I],† M−1⋅s−1
|
|
|---|---|---|---|
| rGran A‡ | Human rGrz A§ | ||
| DCI | 400 | 15 | 50¶ |
| Ph-NHCONH-CiTEtOIC | 0.42 | 57,000 | 94,000 |
| GuaC6H4COOC6H4-pCN | 7.8 | 2,400 | 3,400 |
| C6H4CH2SO2-Gly-Pro(4-AmPhGly)P(OPh)2 | 5 | 4,000 | 4,000 |
Structural formulas:
Inhibition rate constants were measured in 0.1 M Hepes/0.01 M CaCl2, pH 7.5, and 5% (vol/vol) dimethyl sulfoxide at 21°C by the incubation method.
Z-Arg-SBzl (200 μM) was used as the substrate to determine the residual enzyme activity of rGranA.
Data were obtained with recombinant granzyme A produced in yeast (S. Odake and J.C.P., unpublished results).
This value is for native human granzyme A.
RESULTS
Production of Active Recombinant Bacterial Granzyme A.
A granzyme A bacterial expression plasmid was constructed which contains a pel B leader sequence for periplasmic export, an enterokinase site immediately 5′ of the active coding sequence to enable in vitro enzyme activation, and a C-terminal (His)6 tag for purification (Fig. 1A). rGranA was purified from bacterial lysates by nickel chelation chromatography to yield a single 52-kDa protein, which forms a 28-kDa monomer under reducing conditions (Fig. 1B). After enterokinase cleavage, the homodimer migrates with an apparent molecular mass 10 kDa less than the proenzyme (Fig. 1C). This difference is greater than the 3 kDa anticipated from cleavage of each monomer’s 15 amino acid propeptide containing the multiple cloning and enterokinase sites. This difference is not due to failure to cleave the pel B sequence; N-terminal sequencing of the proenzyme confirmed the anticipated cleavage site by the bacterial signal peptidase.
A recombinant inactive granzyme A (S→ArGranA) was generated by mutation of the active-site Ser to Ala. The mutant enzyme was designed so that the bacterial signal peptidase cleavage produced a protein whose N terminus coincides with that of the active enzyme. The mutant enzyme migrated with the same mobility as the enterokinase-cleaved rGranA (Fig. 1C). The final yield of purified rGranA from 4 liters of bacterial culture was 100 μg after enterokinase cleavage and purification; the yield of the mutant protein was higher (approximately 1 mg/4 liters).
To confirm the similarity of rGranA and S→ArGranA to native granzyme, mAb CB9, produced toward native human granzyme, was used to immunoprecipitate rGranA. CB9 recognizes a granzyme A conformational determinant, since it does not react with native granzyme A in immunoblotting. Pro-rGranA, rGranA, and S→ArGranA are specifically precipitated with CB9 (Fig. 1D and data not shown). Because the intact and mutated proteins adopt a common conformation which mimics that of native enzyme, these recombinant proteins could be used to identify potential granzyme A substrates.
Substrate Recognition and Enzyme Kinetics of rGranA.
Michaelis–Menten parameters were determined for known synthetic thioester substrates of native granzyme A (Table 1). After quantitation of active sites, rGranA was tested for cleavage of the synthetic thioester substrates Z-Arg-SBzl, Z-Lys-SBzl, Boc-Ala-Ala-Arg-SBzl and Boc-Trp-Arg-SBzl. The kcat/Km for cleavage of Z-Arg-SBzl by rGranA of 2.86 × 106 is more than 10-fold greater than that reported for purified granzyme A from human or mouse granules and is comparable to the activity of trypsin. For short substrates Arg was preferred to Lys at the P1 site. The kcat/Km for the Arg substrate was approximately 6-fold higher than for the comparable Lys substrate. For the longer Arg substrates Boc-Ala-Ala-Arg-SBzl and Boc-Trp-Arg-SBzl, the kcat/Km values were 9.4 × 104 μM and 3.8 × 105 μM, respectively. Typical substrates for Aspase and Metase activity were not hydrolyzed. The S → A mutant and the proenzyme had no detectable Tryptase activity against Z-Arg-SBzl.
Table 1.
Kinetic constants for the hydrolysis of thioester substrates by mouse granzyme A and human rGranA
| Substrate | Kinetic constant* | Value
|
|
|---|---|---|---|
| rGranA | Mouse granzyme A† | ||
| Z-Arg-SBzl | kcat | 506 | 83 |
| Km | 177 | 315 | |
| kcat/Km | 2,860,000 | 260,000 | |
| Z-Lys-SBzl | kcat | 369 | 22 |
| Km | 767 | 130 | |
| kcat/Km | 481,000 | 170,000 | |
| Boc-Trp-Arg-SBzl | kcat | 66 | |
| Km | 170 | ||
| kcat/Km | 388,000 | ||
| Boc-Ala-Ala-Arg-SBzl | kcat | 83 | 45 |
| Km | 887 | 140 | |
| kcat/Km | 94,000 | 320,000 | |
| Boc-Ala-Ala-Asp-SBzl | NH | NH | |
| Boc-Ala-Ala-Met-SBzl | NH | NH | |
Boc, t-butyloxycarbonyl. NH, no hydrolysis. The rGran A concentrations were 0.007–0.42 nM.
The units of kcat, Km, and kcat/Km are s−1, μM, and M−1⋅s−1, respectively.
Data from ref. 30.
Inhibition Kinetics.
Four known inhibitors of granzyme A were used to inhibit rGranA (Table 2). 3,4-Dichloroisocoumarin (DCI), a general serine protease inhibitor (31), inhibits rGranA weakly with kobs/[I] of 15 M−1⋅s−1. DCI also inhibits native human and mouse granzyme A weakly with kobs/[I] of 50 M−1⋅s−1 (30). The other three inhibitors (an isocoumarin, a guanidinobenzoate, and a phosphonate) contain basic functional groups (isothioureido, guanidino, and amidino) and are more specific for trypsin-like enzymes. They inhibit rGran A much more potently, with kobs/[I] values of 103 to 104 M−1⋅s−1. Among these compounds, the isocoumarin derivative, PhNHCONH-CiTEtOIC, is the best inhibitor. All three compounds have been found to be potent irreversible inhibitors for human recombinant granzyme A produced in Saccharomyces cerevisiae (S. Odake and J.C.P., unpublished results). The inhibition constants of rGran A are comparable to those for native and yeast recombinant human granzyme A.
Isolation of Candidate Substrates for Granzyme A by Affinity Chromatography.
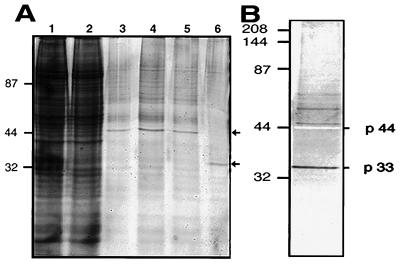
Since rGranA and its mutant variant had the expected enzymatic and immunological properties, S→ArGranA was coupled to a matrix for affinity chromatography to isolate candidate substrates for granzyme A. Two predominant proteins were eluted with 200 mM and 1 M NaCl. The 200 mM NaCl eluate contains a 44-kDa protein, and the 1 M NaCl eluate contains a 33-kDa protein, visible by SYPRO Orange staining (Fig. 2). The 33-kDa protein was probably present in the 200 mM NaCl fractions at lower levels, since a band of this size was visualized by silver staining. After ion exchange chromatography purification, the 33- and 44-kDa proteins (p33 and p44, respectively) were subjected to tryptic digestion and peptide sequencing. The sequenced peptides show nearly 100% homology to two previously coisolated proteins, PHAP I and PHAP II, isolated by binding to an HLA class II cytoplasmic peptide (32). The p33 tryptic fragment sequences IPNLTHLNSGKN and DLSTIEPLK were identical to amino acids 87–98 and 102–110 of PHAP I, except that the initial I was a C in the PHAP I sequence. The p44 sequences SGYRIDFYFDENPYFEN and EFHLNESGDPSSK were identical to amino acids 120–136 and 142–154 of cloned PHAP II.
Figure 2.
Two predominant proteins in K562 cytoplasmic extracts bind to S→ArGranA and are eluted sequentially with 200 mM and 1 M NaCl. (A) SDS/PAGE analysis of the following: lane 1, cell lysate; lane 2, flow-through; lanes 3–5, 200 mM NaCl eluate; and lane 6, 1 M NaCl eluate, all visualized with SYPRO Orange (Molecular Probes). (B) Silver stain of 200 mM NaCl eluate reveals presence of both p33 and p44. Protein sequencing of tryptic fragments shows homology to the putative HLA-binding proteins, PHAP I and PHAP II.
The PHAP (putative HLA-associated) proteins are ubiquitously expressed. They both have acidic-residue-rich C-terminal domains and inhibit the serine phosphatase PP2A (33, 34). PHAP I (p33; also known as I1PP2A) has two N-terminal leucine/isoleucine-rich regions and a canonical nuclear localization signal. It is also homologous to two small nuclear ribonucleoproteins (snRNPs), implicated in RNA splicing (35, 36). PHAP II (p44, also known as set, TAF-1, or I2PP2A) can be phosphorylated at Ser-9 and Ser-24 and is homologous to a yeast nucleosome assembly protein (29, 32, 34, 37, 38). PHAP II has been associated with the putative protooncogene can in a t(9;9) translocation or inversion in acute undifferentiated leukemia (39–42). Although the PHAPs were isolated initially and by us from cytoplasmic lysates, PHAP I has a nuclear localization signal, and the PHAPs have been identified by immunohistochemistry as nuclear proteins (32, 37). The PHAPs are likely to be transported from the cytoplasm to the nucleus.
PHAP II Is a Substrate for rGranA and Coprecipitates from Cell Lysates with Inactive rGranA.
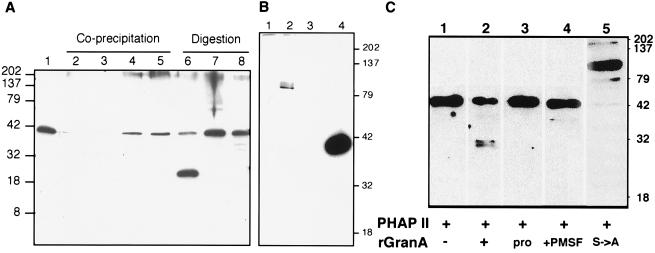
Because granzyme A is a basic protein and both PHAPs have acidic-residue-rich domains, we were concerned that the PHAP isolation from the S→ArGranA column might be a weak electrostatic interaction. To verify that the PHAP isolation from the column represents a physiologically significant interaction and to determine whether the PHAPs might be substrates for granzyme A, we treated cell lysates for 1 hr at 30°C with 400 nM rGranA, S→ArGranA, pro-rGranA, or buffer and analyzed the reaction mixture by immunoblotting, probing with antiserum to an N-terminal PHAP II peptide (29) (Fig. 3A). The 44-kDa PHAP II band was diminished and a new band of 25 kDa was seen in lysates treated with active enzyme but not in samples treated with inactive variants. When the treated cell lysates were incubated with nickel resin to isolate His-tagged granzyme A and probed with the PHAP II antiserum, PHAP II coprecipitated with the inactive mutant and proenzyme but not with the active rGranA. PHAP II is cleaved by active rGranA in cell lysates. PHAP II forms a stable complex with inactive variants of rGranA, but the active enzyme–substrate interaction is transitory and therefore not visualized by coprecipitation.
Figure 3.
PHAP II is coprecipitated from K562 cell lysates with S→ArGranA and pro-rGranA and is cleaved by rGranA. (A) Cell lysates treated with rGranA or inactive variants and blotted with antiserum to a PHAP II N-terminal peptide. Either no granzyme was added (lanes 1 and 2) or lysates were treated with rGranA (lanes 3 and 6), S→ArGranA (lanes 4 and 7), or pro-rGranA (lane 8); lanes 1 and 6–8 were analyzed directly and lanes 2–5 were analyzed after nickel resin precipitation. (B) Cell lysates incubated with rGranA (lane 1), S→ArGranA (lane 2), or buffer (lane 3) were immunoprecipitated with PHAP II antisera and blotted with granzyme A antisera. In lane 4, S→ArGranA was electrophoresed and immunoblotted as a positive control. (C) Immunoblot with antisera to three PHAP II peptides (39) identifies PHAP II as a substrate for rGranA. PHAP fractions 13–15 were incubated with no enzyme, active rGranA, or inactive granzyme A as the proenzyme, the S→A mutant, or after PMSF treatment. The active enzyme cleaves PHAP II into 25- and 27-kDa fragments. The 100-kDa SDS-stable complex of S→ArGranA and PHAP II is seen in B and C.
Similar results were found when cell lysates treated with rGranA, S→ArGranA, or buffer were immunoprecipitated with PHAP II antiserum, separated by SDS/PAGE, and immunoblotted with polyclonal anti-granzyme A antiserum (Fig. 3B). No rGranA coimmunoprecipitated with PHAP II in cell lysates as before, but a higher molecular mass complex of approximately 100 kDa was observed when S→ArGranA was added to the cell lysates. This suggests that the PHAP II–mutant granzyme interaction is strong enough to withstand SDS denaturing conditions. Although higher molecular mass material immunoreactive with PHAP II antiserum was present in the different buffer conditions of Fig. 3A, the 100-kDa complex was not seen. Its formation and stability may depend on ionic concentration and pH.
The cell lysate experiments were confirmed when pooled PHAP fractions 13–15 were incubated for 4 hr with 100 nM rGranA or inactive pro-rGranA, PMSF-treated rGranA, or S→ArGranA and analyzed by SDS/PAGE. The p33 PHAP I band after rGranA treatment was unchanged, suggesting it is not a substrate for granzyme A (data not shown). After immunoblotting with PHAP II antisera against N-terminal peptide (29), a cleavage product of 25 kDa is visualized after treatment with active rGranA. After incubation with S→ArGranA, the PHAP II band disappears and a new PHAP II-containing band at approximately 100 kDa is again observed, confirming that PHAP II forms an SDS-stable complex with S→ArGranA.
PHAP II Is Degraded After CTL Attack.
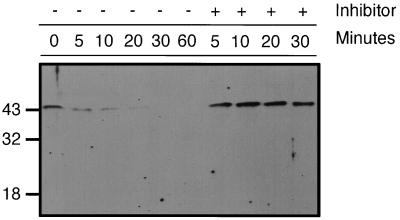
To verify the physiological significance of PHAP II in cell-mediated lysis, we analyzed the effects of CTL attack on cellular PHAP II (Fig. 4). Effector cells were allowed to bind to targets in the presence of EGTA to inhibit Ca-dependent granule-mediated lysis. Cell extracts were analyzed by SDS/PAGE and Western blotting with PHAP II antisera at sequential times after addition of CaCl2. The relative intensities of the PHAP II signal in separated LAK cells and K562 are comparable (data not shown). Within 5 min of CTL attack, the PHAP II signal of the cell mixture is reduced, and it is undetectable after 30 min. Comparable results are found when other human or murine CTLs are incubated with specific target cells, although the exact kinetics and extent of PHAP II degradation depends on the cytolytic activity of the effector cells (data not shown). The degradation depends on mixing the CTLs and target together in the presence of Ca, since the PHAP II band does not degrade when CTLs or targets in Ca-containing medium are lysed separately (data not shown). However, the 25-kDa product of PHAP II degradation by granzyme A is not seen. This suggests that the 25-kDa cleavage product is probably unstable within cells. When CTLs are preincubated with the potent and specific granzyme A inhibitor Ph-NHCONH-CiTEtOIC, there is no degradation of PHAP II, demonstrating that PHAP II is likely to be a physiological substrate of granzyme A.
Figure 4.
PHAP II is degraded within minutes of CTL attack. Cell lysates extracted before and at 5–60 min after addition of CaCl2 to initiate CTL lysis were analyzed by SDS/PAGE and Western blotting with PHAP II antiserum. No degradation was found in control samples which contained LAK cells pretreated for 1 hr with a potent granzyme A inhibitor, PhNHCONH-CiTEtOIC. When the blot was stripped and reprobed with polyclonal rabbit serum to the cytoplasmic protein moesin (a gift of E. Remold-O’Donnell, Center for Blood Research) as a control, there was no change in the moesin signal (data not shown).
DISCUSSION
The key to the prokaryotic production of granzyme A in a properly folded enzymatically active form was the synthesis of an inactive proenzyme, designed to be exported to the bacterial periplasm. When granzyme A was expressed without a periplasmic leader, it formed enzymatically inactive aggregates (data not shown). After enterokinase cleavage, an unexpectedly large shift in apparent molecular mass of the dimer from 53 kDa to 43 kDa coincided with enzyme activation. This may reflect a conformational shift in the enzyme from an extended form of the zymogen. We initially attempted to design a construct encoding the active enzyme directly following the periplasmic leader sequence, but protein expressed by this plasmid had much lower specific enzymatic activity. However, the inactive S→ArGranA, cleaved by the bacterial signal peptidase, appeared to assume a conformation similar to that of the active enzyme, based on its migration in SDS/PAGE and its reactivity to CB9 mAb.
Substrate specificity and inhibition of rGranA is comparable to that of purified native human granzyme. The Km values for rGranA are at most 2- to 5-fold different from the Km values previously reported for native granzyme A. Large differences in the kcat are potentially due to impurities from other granule constituents in previous partially purified native granzyme preparations. Three classes of irreversible inhibitors were evaluated as inhibitors of rGran A. As expected, the general serine protease inhibitor DCI inhibits rGranA weakly, as it does native granzyme A. The isocoumarin PhNHCONH-CiTEtOIC, containing a basic group, inhibits rGran A about 6,000-fold more potently than DCI, which indicates the importance of the interaction between the basic side chain of the inhibitor and Asp in the S1 subsite of rGran A. The amidine-containing peptide phosphonate (41) C6H4-CH2-SO2-Gly-Pro-(4-AmPhGly)P(OPh)2 inhibits rGranA quite potently, although its inhibition constant is one order of magnitude lower than that for PhNHCONH-CiTEtOIC. Guanidinobenzoates are also potent inhibitors of trypsin-like enzymes and form stable acyl enzyme derivatives upon reaction with Ser-184. FUT-175 (6-amidino-2-naphthyl-4-guanidinobenzoate) has previously been shown to inhibit human granzyme A very potently with a kobs/[I] value of 105 M−1⋅s−1 (43). Our inhibitor, p-cyanophenyl p-guanidinobenzoate, has a lower inhibition rate constant than FUT-175 by 2 orders of magnitude. FUT-175 either has a much better leaving group than p-cyanophenyl p′-guanidinobenzoate or has significant additional binding interactions with granzyme A.
For some serine proteases, it is known that crystal structure is preserved after active site mutagenesis (44). In this work we have successfully used a mutated inactive enzyme as bait for identifying candidate serine protease substrates. An affinity column produced with the S → A mutant was used to isolate two cytoplasmic proteins, which are candidates for participation in a cell death pathway induced by granzyme A. The interaction of one of these proteins, PHAP II, with granzyme A was substantiated by the formation of an SDS-stable complex with the mutant rGranA and by its cleavage by active recombinant enzyme. Although PHAP I does not appear to be a substrate for granzyme A, it may exist as a complex with PHAP II or regulate its activity. When pooled PHAP fractions 13–15 are analyzed on a 200-kDa-cutoff gel-filtration column, the PHAP II-containing material is found within the void volume (data not shown), suggesting that it may exist in a high molecular weight complex. Moreover, the coisolation of the two PHAP proteins by independent methods (this report, ref. 32) is consistent with this. Although PHAP II is eluted predominantly in fractions 13–15 and PHAP I in fraction 21, there appears to be some of each protein in both fractions (Fig. 2B). Although PHAP I requires higher salt concentrations than PHAP II to elute from the S→ArGranA column, we have not excluded the possibility that PHAP I binds indirectly to rGranA through an interaction with PHAP II. PP2A inhibition by PHAP I and/or PHAP II may be involved in regulating the activity of PHAP II, known to be phosphorylated on serines 9 and 24 (29). Further study of the role of PHAP I will be aided by developing PHAP I antisera.
The properties of the PHAPs make them attractive candidates for participation in a cell death pathway that leads to DNA degradation. Although isolated from the cytoplasm, where they may first encounter granzyme A, PHAP I has a nuclear localization signal (32), and PHAP II has been localized to the nucleus by immunohistochemistry (37). They both have extended acidic-residue-rich domains, common to factors which bind histones and other DNA-associated proteins (45, 46). The leucine-rich motif of PHAP I is common to proteins that inhibit protein phosphatases involved in cell cycle regulation (47). PHAP I is homologous to snRNPs involved in RNA splicing, and PHAP II has homology to a yeast nucleosome assembly protein. It is possible that granzyme A cleaves the N-terminal fragment of PHAP II with PP2A inhibitory activity (33) from the acidic C-terminal domain likely to be involved in chromatin binding (38, 48, 49). Nucleosome disassembly could be the first step in the DNA degradation seen with granzyme A-induced cell death.
We have taken the first step in suggesting a role for the PHAP proteins in the granzyme A pathway. PHAP II disappearance within minutes of CTL attack is completely inhibited by preincubating CTL with a potent specific granzyme A inhibitor. This suggests that granzyme A is the protease responsible for PHAP II degradation during CTL attack. The complete disappearance of PHAP II signal indicates that PHAP II is degraded in both the CTL and its target during cell-mediated lysis. The puzzle remains—if PHAP II is also degraded in CTLs, why are CTLs resistant to the lytic effects of their granule components (50)? The role of cleaved PHAP II in the myriad events that occur during cell-mediated lysis remains to be defined.
Acknowledgments
We thank T. Copeland for his gift of PHAP II antisera, E. Remold for rabbit antiserum to moesin, M. Yoon and J. Hirsch for rGranA preparation, and K. Blackwell, H. Eisen, E. Remold, and D. Tkachuk for helpful discussions. This work was supported by National Institutes of Health Grant CA01449 and a Pew Scholar Award in the Biomedical Sciences (J.L.) and National Institutes of Health Grant GM54401 (J.C.P.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: Am, amidino; Boc, t-butyloxycarbonyl; CTL, cytotoxic T lymphocyte; DCI, 3,4-dichloroisocoumarin; PHAP, putative HLA-associated protein; PMSF, phenylmethanesulfonyl fluoride; rGranA, recombinant granzyme A; S → ArGranA, Ser → Ala mutant rGranA; SBzl, thiobenzyl; Z, benzyloxycarbonyl.
References
- 1.Russell J H. Immunol Rev. 1983;72:97–118. doi: 10.1111/j.1600-065x.1983.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 2.Henkart P A, Millard P J, Reynolds C W, Henkart M P. J Exp Med. 1984;160:75–93. doi: 10.1084/jem.160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Podack E R, Koenigsberg P J. J Exp Med. 1984;160:695–710. doi: 10.1084/jem.160.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasternack M S, Eisen H N. Nature (London) 1985;314:743–745. doi: 10.1038/314743a0. [DOI] [PubMed] [Google Scholar]
- 5.Gershenfeld H K, Weissman I L. Science. 1986;232:854–858. doi: 10.1126/science.2422755. [DOI] [PubMed] [Google Scholar]
- 6.Bleackley R C, Lobe C G, Duggan B, Ehrman N, Fregeau C, Meier M, Letellier M, Havele C, Shaw J, Paetkau V. Immunol Rev. 1988;103:5–20. doi: 10.1111/j.1600-065x.1988.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 7.Jenne D E, Tschopp J. Immunol Rev. 1988;103:53–71. doi: 10.1111/j.1600-065x.1988.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 8.Darmon A J, Nicholson D W, Bleackley R C. Nature (London) 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 9.Quan L T, Tewari M O, Rourke K, Dixit V, Snipas S J, Poirier G G, Ray C, Pickup D J, Salvesen G S. Proc Natl Acad Sci USA. 1996;93:1972–1976. doi: 10.1073/pnas.93.5.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan H, Orth K, Chinnaiyan A M, Poirier G G, Froelich C J, He W W, Dixit V M. J Biol Chem. 1996;271:16720–16724. doi: 10.1074/jbc.271.28.16720. [DOI] [PubMed] [Google Scholar]
- 11.Orth K, Chinnaiyan A M, Garg M, Froelich C J, Dixit V M. J Biol Chem. 1996;271:16443–16446. [PubMed] [Google Scholar]
- 12.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Sanz J A, MacDonald H R, Jenne D E, Tschopp J, Nabholz M. J Immunol. 1990;145:3111–3118. [PubMed] [Google Scholar]
- 14.Nakajima H, Park H L, Henkart P A. J Exp Med. 1995;181:1037–1046. doi: 10.1084/jem.181.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi L, Kam C M, Powers J C, Aebersold R, Greenberg A H. J Exp Med. 1992;176:1521–1529. doi: 10.1084/jem.176.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kagi D, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Nature (London) 1994;369:31–37. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 17.Ebnet K, Hausmann M, Lehmann-Grube F, Mullbacher A, Kopf M, Lamers M, Simon M M. EMBO J. 1995;14:4230–4239. doi: 10.1002/j.1460-2075.1995.tb00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullbacher A, Ebnet K, Blanden R V, Hla R T, Stehle T, Museteanu C, Simon M M. Proc Natl Acad Sci USA. 1996;93:5783–5787. doi: 10.1073/pnas.93.12.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quan L T, Caputo A, Bleackley R C, Pickup D J, Salvesen G S. J Biol Chem. 1995;270:10377–10379. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
- 20.Kummer J A, Kamp A M, Citarella F, Horrevoets A J, Hack C E. J Biol Chem. 1996;271:9281–9286. doi: 10.1074/jbc.271.16.9281. [DOI] [PubMed] [Google Scholar]
- 21.Pasternack M S, Bleier K J, McInerney T N. J Biol Chem. 1991;266:14703–14708. [PubMed] [Google Scholar]
- 22.Simon M M, Ebnet K, Kramer M D. In: Cytotoxic Cells: Recognition, Effector Functions, Generation, Methods. Sitkowsky M V, Henkart P A, editors. Boston: Birkhaeuser; 1993. pp. 278–294. [Google Scholar]
- 23.Irmler M, Hertig S, MacDonald H R, Sadoul R, Becherer J D, Proudfoot A, Solari R, Tschopp J. J Exp Med. 1995;181:1917–1922. doi: 10.1084/jem.181.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanguri P, Lee E, Henkart P, Shin M L. J Immunol. 1993;150:2431–2439. [PubMed] [Google Scholar]
- 25.Sower L E, Froelich C J, Allegretto N, Rose P M, Hanna W D, Klimpel G R. J Immunol. 1996;156:2585–2590. [PubMed] [Google Scholar]
- 26.Kummer J A, Kamp A M, van Katwijk M, Brakenhoff J P, Radosevic K, van Leeuwen A M, Borst J, Verweij C L, Hack C E. J Immunol Methods. 1993;163:77–83. doi: 10.1016/0022-1759(93)90241-x. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Köhler G, Milstein C. Nature (London) 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 29.Adachi Y, Pavlakis G N, Copeland T D. FEBS Lett. 1994;340:231–235. doi: 10.1016/0014-5793(94)80144-4. [DOI] [PubMed] [Google Scholar]
- 30.Odake S, Kam C M, Narasimhan L, Poe M, Blake J T, Krahenbuhl O, Tschopp J, Powers J C. Biochemistry. 1991;30:2217–2227. doi: 10.1021/bi00222a027. [DOI] [PubMed] [Google Scholar]
- 31.Harper J W, Hemmi K, Powers J C. Biochemistry. 1985;24:1831–1841. doi: 10.1021/bi00329a005. [DOI] [PubMed] [Google Scholar]
- 32.Vaesen M, Barnikol-Watanabe S, Gotz H, Awni L A, Cole T, Zimmermann B, Kratzin H D, Hilschmann N. Biol Chem Hoppe-Seyler. 1994;375:113–126. doi: 10.1515/bchm3.1994.375.2.113. [DOI] [PubMed] [Google Scholar]
- 33.Li M, Makkinje A, Damuni Z. Biochemistry. 1996;35:6998–7002. doi: 10.1021/bi960581y. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Makkinje A, Damuni Z. J Biol Chem. 1996;271:11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 35.Matsuoka K, Taoka M, Satozawa N, Nakayama H, Ichimura T, Takahashi N, Yamakuni T, Song S Y, Isobe T. Proc Natl Acad Sci USA. 1994;91:9670–9674. doi: 10.1073/pnas.91.21.9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sillekens P T, Beijer R P, Habets W J, van Verooij W J. Nucleic Acids Res. 1989;17:1893–1906. doi: 10.1093/nar/17.5.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K. Proc Natl Acad Sci USA. 1995;92:4279–4283. doi: 10.1073/pnas.92.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adachi Y, Pavlakis G N, Copeland T D. J Biol Chem. 1994;269:2258–2262. [PubMed] [Google Scholar]
- 40.Fink T M, Vaesen M, Kratzin H D, Lichter P, Zimmer M. Genomics. 1995;29:309–310. doi: 10.1006/geno.1995.1257. [DOI] [PubMed] [Google Scholar]
- 41.Fornerod M, Boer J, van Baal S, Jaegle M, von Lindern M, Murti K G, Davis D, Bonten J, Buijs A, Grosveld G. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- 42.Oleksyszyn J, Boduszek B, Kam C-M, Powers J C. J Med Chem. 1994;37:226–231. doi: 10.1021/jm00028a004. [DOI] [PubMed] [Google Scholar]
- 43.Poe M, Blake J T, Boulton D A, Gammon M, Sigal N H, Wu J K, Zweerink H J. J Biol Chem. 1991;266:98–103. [PubMed] [Google Scholar]
- 44.Sprang S R, Fletterick R J, Graf L, Rutter W J, Craik C S. Crit Rev Biotech. 1988;8:225–236. doi: 10.3109/07388558809147559. [DOI] [PubMed] [Google Scholar]
- 45.Kuehl L, Childers T J, McCauley R M. Arch Biochem Biophys. 1986;248:272–281. doi: 10.1016/0003-9861(86)90424-8. [DOI] [PubMed] [Google Scholar]
- 46.Earnshaw W C. J Cell Biol. 1987;105:1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohkura H, Yanagida M. Cell. 1991;64:149–157. doi: 10.1016/0092-8674(91)90216-l. [DOI] [PubMed] [Google Scholar]
- 48.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 49.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Nature (London) 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 50.Kranz D M, Eisen H N. Proc Natl Acad Sci USA. 1987;84:3375–3379. doi: 10.1073/pnas.84.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]