Abstract
Saponins are glycosylated plant secondary metabolites found in many major food crops [Price, K. R., Johnson, I. T. & Fenwick, G. R. (1987) CRC Crit. Rev. Food Sci. Nutr. 26, 27–133]. Because many saponins have potent antifungal properties and are present in healthy plants in high concentrations, these molecules may act as preformed chemical barriers to fungal attack. The isolation of plant mutants defective in saponin biosynthesis represents a powerful strategy for evaluating the importance of these compounds in plant defense. The oat root saponin avenacin A-1 fluoresces under ultraviolet illumination [Crombie, L., Crombie, W. M. L. & Whiting, D. A. (1986) J. Chem. Soc. Perkins 1, 1917–1922], a property that is extremely rare among saponins. Here we have exploited this fluorescence to isolate saponin-deficient (sad) mutants of a diploid oat species, Avena strigosa. These sad mutants are compromised in their resistance to a variety of fungal pathogens, and a number of lines of evidence suggest that this compromised disease resistance is a direct consequence of saponin deficiency. Because saponins are widespread throughout the plant kingdom, this group of secondary metabolites may have general significance as antimicrobial phytoprotectants.
Plants produce a myriad of secondary metabolites, many of which can inhibit the growth of microbes in vitro. These inhibitory compounds may be synthesized during normal growth and development (preformed antimicrobial compounds, or “phytoanticipins”) (1–3). Alternatively, they may be absent from healthy plants, accumulating only in response to pathogen attack or stress (phytoalexins) (4, 5). Collectively, antimicrobial compounds of plant origin encompass a diverse array of different classes of compounds, including saponins, phenolics, cyclic hydroxamic acids, cyanogenic glycosides, isoflavonoids, sesquiterpenes, sulfur-containing indole derivatives, and many others. The issue of whether these compounds protect plants against pathogen attack has intrigued plant pathologists since the early part of this century.
Investigations of the contribution of antimicrobial compounds to plant defense have tended to focus on phytoalexins, because these molecules are actively synthesized as part of the battery of induced defense responses associated with disease resistance. There are some indications that phytoalexins can act as antimicrobial phytoprotectants. For example, the ability of pea- and chickpea-infecting isolates of the fungus Nectria haematococca to detoxify host-plant phytoalexins has been shown to be important for full virulence (6, 7). Also, experiments in which levels of phytoalexins in plants have been altered, either by the generation of mutants or by transformation-mediated manipulation of gene expression, have provided evidence to link phytoalexins with disease resistance (8–11). Preformed antimicrobial compounds have attracted less attention, although they are likely to represent the first chemical barriers to infection. Nevertheless, there is increasing evidence that such compounds do contribute to plant defense, as demonstrated by the enhanced disease susceptibility of maize mutants defective in the ability to synthesize 2,4-dihydroxy-1,4-benzoxazin-3-one (DIBOA) (12, 13) and of transgenic tobacco lines in which the accumulation of preformed phenylpropanoids has been suppressed (14).
This study investigates the role of saponins, an important group of preformed plant secondary metabolites, in protecting plants against fungal attack. Saponins are glycosylated triterpenoid, steroid, or steroidal alkaloid molecules that occur constitutively in many plant species (1, 15–17). Because many saponins have potent antifungal activity and are often present at high levels in healthy plants, these molecules have been implicated as antimicrobial phytoprotectants (1, 3, 15–17). Direct genetic evidence for this is, however, lacking. This work involves a family of four structurally related triterpenoid saponins, avenacins A-1, B-1, A-2, and B-2, found in the roots of oat (Avena spp.) (18, 19) (Fig. 1). These saponins have been implicated as determinants of the resistance of oats to the root-infecting fungus Gaeumannomyces graminis var. tritici (20), which causes “take-all” disease of wheat and barley but is unable to infect oats. Indirect evidence for a role for avenacins in plant protection has come from the demonstration that an oat-attacking variant of G. graminis (G. graminis var. avenae) requires the saponin-detoxifying enzyme avenacinase for successful infection of oat (21). A single diploid oat species, Avena longiglumis, has been identified that lacks avenacins. Significantly, A. longiglumis is susceptible to G. graminis var. tritici (22). However, genetic analysis of the association between avenacins and disease resistance has not been carried out because A. longiglumis does not hybridize readily with other avenacin-producing oat species. Here, we have generated avenacin-deficient mutants of the diploid oat species Avena strigosa to assess the contribution of these saponins to the resistance of oats to G. graminis var. tritici and other fungal pathogens.
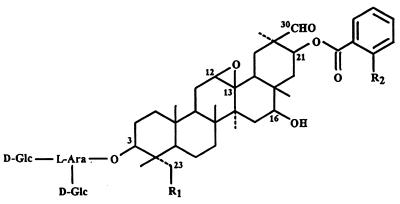
Figure 1.
The structures of the four avenacins. Avenacin A-1: R1 = OH, R2 = NHCH3; avenacin A-2: R1 = OH, R2 = H; avenacin B-1: R1 = H, R2 =NHCH3; avenacin B-2: R1 = H, R2 = H; d-Glc = d-glucose; l-Ara = l-arabinose.
Methods
Oat Mutagenesis and Genetic Analysis.
Seed of the diploid oat species A. strigosa (from the Institute of Grasslands and Environmental Research, Aberystwyth, Wales, United Kingdom; accession no. S75) was mutagenized with 10 mM sodium azide in 0.1 M sodium phosphate buffer (pH 3.2) as described (23). M2 seed from individual M1 plants was kept separate to avoid the isolation of siblings. The efficiency of the mutagenesis was assessed by monitoring the frequency of chlorophyll-deficient seedlings, which was ≈4.6% [consistent with data previously reported for sodium azide mutagenesis of A. strigosa (23)]. Amplified fragment length polymorphism (AFLP) analysis was used to confirm that the saponin-deficient mutants identified in this study originated from the seed used for the mutagenesis. For genetic analysis, F1 progeny generated by crossing and F2 plants generated by selfing were grown to maturity in the greenhouse.
Analysis of Root Extracts.
M3 seeds were germinated on moist filter paper for 2 days, and terminal 0.5-cm sections of the roots from 20 seedlings per line were harvested and extracted in methanol. For HPLC analysis, crude methanolic root extracts from M3 seedlings were prepared in triplicate, and 100–μl aliquots were analyzed directly on a Hichrom Nucleosil 5 C18 reverse phase column (4.5 × 250 mm) under isocratic conditions in 75% methanol (flow rate 1 ml/min) with detection at 225 nm, following published methods (18, 19). The four avenacins were quantified by comparison of peak areas with those of standards of known concentration [prepared as described (18, 19)]. Extracts for TLC analysis were dried, resuspended in 1 ml of water, and applied to SepPak C18 reverse-phase cartridges (Waters) that had been preconditioned with 10 ml of methanol followed by 10 ml of distilled water. After elution with 75% methanol, samples were dried, resuspended in 15 μl of 100% methanol, and analyzed by TLC (13:6:1 vol/vol/vol, chloroform/methanol/water) (24). Avenacins A-1 and B-1 and other fluorescent components were visualized under UV illumination (302 nm). The TLC plate was then sprayed with p-anisaldehyde/sulfuric acid/acetic acid (1:1:48, vol/vol/vol) and baked at 130°C for 5 min to detect all four saponins (16). Root extracts derived from either M3 or F3 seedlings were compared on at least seven occasions with essentially the same outcome. For bioassay experiments to assess the presence of inhibitory substances in root extracts, crude methanolic root extracts were prepared and fractionated by TLC as described above. The TLC plate was then sprayed with homogenized mycelium of G. graminis var. tritici isolate T5 and assessed for zones of inhibition of fungal growth after incubation (22). Bioassay experiments were carried out at least three times for all oat lines.
Determination of Avenacoside Content in Leaves.
Avenacosides A and B were extracted from 8-day-old oat seedlings and analyzed by TLC as described (25). The experiment was carried out three times for all oat lines.
Pathogenicity Tests.
Pathogenicity tests to assess root infection were carried out as described (21, 24). Fungi included G. graminis var. tritici, G. graminis var. avenae, F. culmorum, and F. avenaceum (isolates T5, A3, F712, and F616 respectively). Cross sections of roots infected with G. graminis var. tritici were viewed under bright-field illumination with a Zeiss Axioskop microscope after staining with trypan blue/lactophenol (10 ml of lactic acid, 10 g of phenol, 10 ml of glycerol, 10 ml of water, 10 mg of trypan blue, mixed 1:1 with ethanol). Inoculation of detached leaf sections with Stagonospora nodorum and Stagonospora avenae (isolates S212/80 and WAC1293) followed the procedure of Wubben et al. (25).
Statistical Analysis.
Generalized linear modeling of a three-way contingency table involving the different mutant lines, the two generations, and the plant response (resistant or susceptible) was used to test whether the proportions of diseased seedlings occurring in homozygous F3 mutant lines and in those F2 progeny derived from wild-type × mutant crosses with reduced root fluorescence were significantly different. The analysis was done using the statistical package genstat for Windows, fourth edition (26).
Results
Isolation and Biochemical Characterization of Saponin-Deficient Mutants of A. strigosa.
The major oat-root saponin avenacin A-1 contains N-methyl anthranilic acid (Fig. 1) and so is fluorescent under UV illumination. Avenacin A-1 is primarily responsible for the bright blue fluorescence of young oat roots (22), enabling root fluorescence to be used as a preliminary screen to identify saponin-deficient (sad) oat mutants. Seed of the diploid oat species A. strigosa was mutagenized with the chemical mutagen sodium azide. Seed of individual M2 families was germinated and assessed for root fluorescence, and 10 independent mutants with reduced fluorescence were identified after screening seedlings representing 1,289 M2 families.
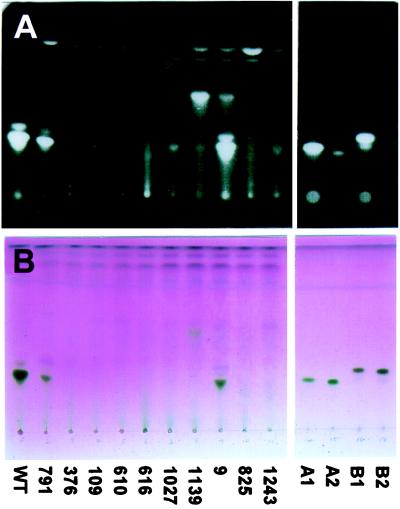
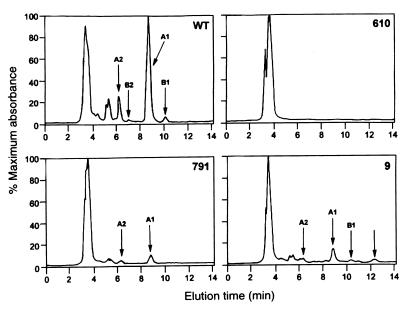
The avenacin content of methanolic root extracts of homozygous M3 seedlings of the putative mutants was analyzed by TLC and HPLC. Extracts from eight of the ten mutants either lacked or contained only trace amounts of avenacins as assessed by TLC, while those from mutants 791 and 9 contained reduced but readily detectable amounts of the saponins (Fig. 2). HPLC analysis of crude root extracts (Fig. 3) confirmed the absence of all four avenacins in the first eight mutants, while extracts from 791 and 9 both contained reduced levels of avenacins (with approximately 15% and 30% of wild-type avenacin A-1 levels, respectively). Fluorescent compounds that were less polar than the avenacins and that reacted with the chromogenic detection reagent for saponins and related compounds were visible in extracts of mutants 1,139 and 9 after TLC (Fig. 2). Both compounds had a retention time of ≈12 min on HPLC (data for mutant 9 is given in Fig. 3). The behavior of these compounds on TLC and HPLC was identical to that of monodeglucosyl avenacin A-1 (data not shown). When crude methanolic root extracts were separated by TLC and the plate was sprayed with the avenacin-sensitive fungus G. graminis var. tritici, clear zones of inhibition of fungal growth were visible corresponding to the avenacins in the wild-type extract and in extracts from 791 and 9, but were not visible for root extracts from any of the other mutants (data not shown). Mutants 1,139 and 9 were the only mutants that were obviously affected in growth. Seedlings of both mutants had stunted roots, whereas 9 also had reduced leaf growth.
Figure 2.
TLC analysis of partially purified root extracts from wild-type and mutant oats. The fluorescent avenacins, A-1 and B-1, were visualized under UV illumination (Upper), and all four saponins were detected using a chromogenic reagent (Lower). Preparations enriched for each of the four avenacins (3 μg per lane) are on the Right, and extract from wild-type (WT) oat roots is on the Left. The major fluorescent compound present in the wild-type root extract is avenacin A-1, and the minor one with a higher Rf value is avenacin B-1.
Figure 3.
HPLC analysis of crude extracts from wild-type and mutant oat roots. The peaks corresponding to the four avenacins are indicated for extracts from wild-type (WT) roots (Upper Left); the peaks eluting at around 5 min derive from micelles of the four avenacins, which are resolved into the separate peaks on re-running. Extracts from 376, 109, 610, 616, 1,027, 1,139, 825, and 1,243 did not contain detectable levels of any of the four avenacins (an example of an HPLC trace for 610 is shown), whereas those from 791 and 9 contained reduced levels (Lower). Extracts from 9 and 1,139 both had an additional peak with a retention time of 12 min, which is likely to correspond to monodeglucosyl avenacin A-1 (indicated for mutant 9 by the unlabeled arrow Lower Right). The full-scale deflection was 4.1 mV for the wild type and 5.5–8.5 mV for the mutants.
Whereas the avenacins are restricted to the roots of oat and are the only saponins present in this part of the plant, the leaves contain another family of saponins, the steroidal avenacosides, A and B (27, 28). Both the triterpenoid avenacins and the steroidal avenacosides are predicted to be synthesized from squalene-2,3-epoxide (16), but their biosynthetic pathways diverge after this point. We therefore analyzed the avenacoside content of leaves of the wild-type oat line and all 10 sad mutants by TLC to determine whether the mutants were affected in the synthesis of these foliar saponins. The avenacoside content of the mutants was normal with the exception of mutant 9, which contained reduced levels of avenacoside B (data not shown).
Genetic Analyses of sad Mutants.
Test crosses were performed between the sad mutants and the wild-type A. strigosa parent to determine whether the saponin-deficient phenotype of each mutant was likely to be due to a single mutation. The ratio of the numbers of seedlings with wild-type root fluorescence to those with reduced fluorescence in the F2 progeny was generally consistent with a 3:1 segregation (Table 1), suggesting that the mutations are recessive alleles of single genes. Seed was not obtained for the cross between the wild-type and 1,243. The growth defects observed for mutants 1,139 and 9 cosegregated with reduced root fluorescence in populations of 321 and 197 F2 progeny, respectively, indicating that the effects on growth are either a consequence of defects in saponin biosynthesis or are due to closely linked mutations. With the exception of mutant 1,243, all of the biochemical analyses and pathogenicity tests described here have been confirmed using homozygous sad lines derived from crosses to the wild-type parent.
Table 1.
Segregation ratios for the F2 generations from crosses of sad mutants with the wild-type parent
| Mutant | Fluorescence
|
χ2 Wild-type: reduced = 3:1* | |
|---|---|---|---|
| Wild-type | Reduced | ||
| 109 | 40 | 19 | 1.6 |
| 610 | 101 | 32 | 0.06 |
| 791 | 74 | 27 | 0.16 |
| 1,027 | 72 | 20 | 0.52 |
| 1,139 | 251 | 70 | 1.3 |
| 616 | 84 | 21 | 1.4 |
| 9 | 150 | 47 | 0.13 |
| 376 | 58 | 18 | 0.07 |
| 825 | 71 | 33 | 2.5 |
*Segregation analysis is consistent with a 3:1 ratio in all cases (P > 0.05).
Analysis of the F2 generations from intermutant crosses identified at least four complementation groups in the mutant collection (Table 2). We have designated the respective loci Sad1 through Sad4. All F2 progeny from crosses of mutants 109 with 610, and of 1,027 with 791, showed reduced fluorescence, indicating mutant alleles of single loci (designated Sad1 and Sad2, respectively). All other intermutant crosses gave a proportion of progeny with wild-type root fluorescence. Two more complementation groups are defined by mutants 1,139 and 616, corresponding to Sad3 and Sad4, respectively. Mutants 9, 376, 825, and 1,243 are yet to be assigned to complementation groups.
Table 2.
Complementation tests
| Cross | Fluorescence
|
|
|---|---|---|
| Wild-type | Reduced | |
| 109 × 610 | 0 | 80 |
| 109 × 376 | 26 | 34 |
| 109 × 791 | 13 | 17 |
| 610 × 1,139 | 14 | 22 |
| 610 × 616 | 150 | 110 |
| 610 × 1,027 | 28 | 28 |
| 610 × 791 | 37 | 43 |
| 1,027 × 791 | 0 | 268 |
| 1,027 × 616 | 30 | 27 |
| 1,027 × 1,139 | 107 | 108 |
| 1,027 × 825 | 9 | 9 |
| 1,027 × 9 | 80 | 70 |
| 791 × 9 | 19 | 24 |
| 791 × 1,139 | 54 | 51 |
| 791 × 616 | 38 | 29 |
| 1,139 × 616 | 25 | 26 |
| 1,139 × 9 | 67 | 51 |
| 1,139 × 376 | 31 | 40 |
| 9 × 616 | 85 | 75 |
| 616 × 825 | 35 | 27 |
The combined data from a number of independent crosses between M3 plants is presented for each mutant. The segregation ratios for different crosses involving the same mutant pairs were all similar. Mutants 9, 376, 825, and 1,243 have not yet been assigned to complementation groups.
sad Mutants Are Compromised in Disease Resistance.
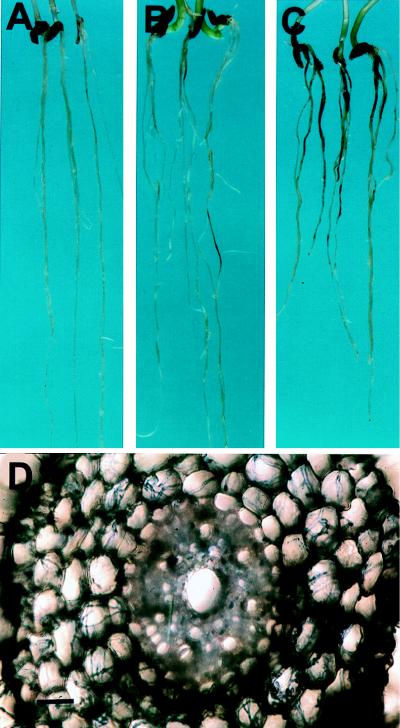
The homozygous mutant A. strigosa lines and the wild-type line were inoculated with fungal pathogens to assess the effects of the sad mutations on plant disease resistance. A. strigosa normally fails to give any discernible signs of disease when inoculated with the wheat pathogen G. graminis var. tritici (22) (Fig. 4A). In contrast, seedlings of all 10 sad mutants developed lesions on their roots to a greater or lesser extent (Table 2; Fig. 4 B and C). In three separate experiments, black lesions were seen on the roots of seedlings of all nine sad mutants tested but never on the wild-type. Mutants 9 and 1,139 tended to give less extensive lesions than the other mutants, but these mutants both have shorter roots and so may avoid infection to some extent. The proportion of seedlings showing disease symptoms and the relative severity of the lesions for the remaining eight mutants varied from experiment to experiment, but 791 (which has ≈15% of wild-type avenacin levels) consistently showed only limited lesions (Table 2; Fig. 4B). Microscopic analysis of the infected oat lines revealed that fungal hyphae were clearly visible ramifying through the root cortex, and in some cases the stele, of the sad mutants (e.g., Fig. 4D), whereas hyphal growth was never seen inside the roots of the wild-type. In seedling tests with homozygous M3 seed of mutants 610 and 1,243, both of these mutants also showed increased susceptibility to Fusarium culmorum and Fusarium avenaceum (shown for mutant 610 in Fig. 5). All 10 sad mutants were also consistently more susceptible to G. graminis var. avenae, which is a successful pathogen of oats (data not shown). No differences were observed between the wild type and mutants when the leaves were inoculated with the leaf-infecting fungi Stagonospora nodorum (a non-pathogen of oat) or Stagonospora avenae (a pathogen of oat) (data not shown).
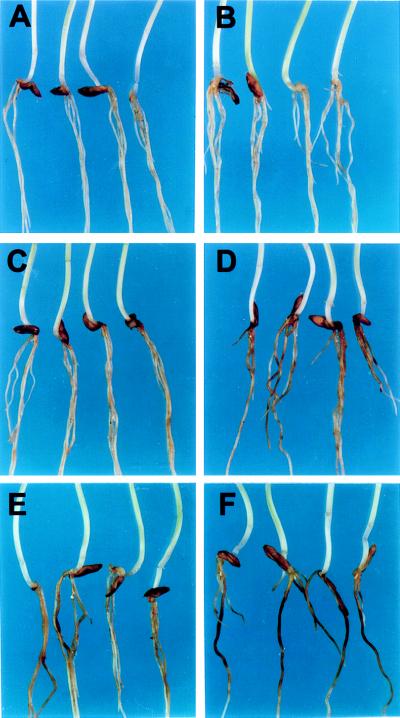
Figure 4.
Infection of sad mutants by Gaemannomyces graminis var. tritici. (A) A. strigosa wild type; (B and C) sad2 mutants 791 and 1027, respectively (21 days after inoculation); (D) cross-section of an infected root of mutant 1027 viewed under bright field illumination after staining with trypan blue/lactophenol. (Bar = 6 μm.)
Figure 5.
Susceptibility of the wild-type A. strigosa line and mutant 610 to Fusarium spp. (A, C, and E) Wild-type seedlings; (B, D, and F) sad mutant 610. (A and B) Mock-inoculated; (C and D) inoculated with Fusarium culmorum isolate F712; (E and F) inoculated with Fusarium avenaceum isolate F616 (assessed after 15 days).
Avenacin Content and Disease Resistance Are Correlated in Segregating Progeny.
The association between the sad mutations and increased disease susceptibility was tested in segregating F2 progeny from wild-type × mutant crosses (with between 53 and 89 seedlings per cross) for mutants 610, 616, 1,027, 1,139, 376, 825, and 791. Seed of F2 progeny was germinated on moist filter paper for 2 days, and the roots of the seedlings were scored as having either wild-type or reduced fluorescence as described above. Individual seedlings were then inoculated with G. graminis var. tritici and assessed for disease symptoms after incubation. An example of the outcome of these experiments is given for mutant 610 (Table 4), where it can be seen that after challenge with G. graminis var. tritici, all F2 seedlings identified as having wild-type root fluorescence remained healthy, whereas 16 of the 17 seedlings with reduced root fluorescence were clearly diseased. Similarly, in tests with the other mutants, those F2 seedlings with wild-type root fluorescence all failed to show signs of disease. For mutants 610, 616, 1,027, 1,139, 376, and 825, the majority of seedlings (between 70 and 95%) with reduced fluorescence gave clear disease symptoms. Although incubation conditions were optimized to maximize the chances of infection, inevitably some seedlings escaped infection in some experiments. This proportion ranged from 0 to around 20%, as assessed by the proportion of homozygous mutant seedlings included as controls in the same experiment that failed to show disease. For wild type × 791 crosses, ≈20% of F2 progeny with reduced fluorescence gave limited disease symptoms, whereas the remainder appeared healthy. This is consistent with the intermediate-susceptibility phenotype of this mutant (Table 2; Fig. 4). The proportions of seedlings showing disease symptoms in the F2 progeny with reduced fluorescence and in the homozygous mutant control lines were not significantly different as assessed using generalized linear modeling (χ2 = 56.6, 6 df, P ≪10−3), and this was consistent for all seven mutants tested (χ2 = 10.74, 6 df, P > 0.1).
Table 4.
Disease scores for F2 progeny with wild-type or reduced root fluorescence derived from a cross between the wild-type oat line and sad mutant 610 after inoculation with G. graminis var. tritici
| No.
of seedlings with disease ratings of:
|
||||
|---|---|---|---|---|
| 0 | + | ++ | +++ | |
| Homozygous parent lines | ||||
| Wild type | 9 | 0 | 0 | 0 |
| 610 | 2 | 1 | 3 | 4 |
| F2 progeny | ||||
| Wild-type fluorescence | 47 | 0 | 0 | 0 |
| Reduced fluorescence | 1 | 2 | 5 | 9 |
Seed of segregating F2 progeny from a cross between the wild-type oat line and sad mutant 610 was germinated, and the seedlings were scored for root fluorescence and assigned to one of two categories (wild type or reduced). The seedlings were then inoculated with G. graminis var. tritici (isolate T5) and assessed for disease after incubation. The wild-type line S75 and a homozygous mutant M3 oat line were included as controls. Disease was scored 21 days after inoculation on an arbitrary scale ranging from 0 to +++ (0, no disease; +, ++, and +++, increasing severity of lesions on the roots).
Discussion
Little is known about the contribution of preformed antimicrobial compounds to plant defense. Here, we have addressed this question for the triterpenoid oat root avenacin saponins by generating mutants of the diploid oat species A. strigosa that are defective in saponin biosynthesis (saponin-deficient, or sad, mutants). A number of lines of evidence indicate that avenacins are likely to act as determinants of disease resistance in oats. First, all 10 independent sad mutants were clearly more susceptible to fungal infection than was the wild-type A. strigosa line. These mutants represent at least four different complementation groups. Second, mutant 791, which shows a partial reduction in avenacin content (to ≈15% of wild-type avenacin levels), gave only limited disease symptoms when inoculated with G. graminis var. tritici. In contrast, sad mutants that lacked detectable levels of avenacins were substantially more susceptible to disease than mutant 791 (including mutant 1,027, which is in the same complementation group). This result provides a further link between avenacins and disease resistance. Third, the association between saponin deficiency (as assessed by reduced root fluorescence) and increased disease susceptibility was supported in segregating F2 progeny from crosses of the mutants with the wild type. It is possible that some sad mutants may harbor additional genetically linked mutations that are unrelated to saponin deficiency but that affect disease resistance. It also is possible that some sad mutations may lead to pleiotropic defects in defense responses in addition to effects on avenacin content. However, despite these caveats, all of the evidence considered together provides a compelling argument to indicate that saponin deficiency and compromised disease resistance are causally related.
The first committed step in avenacin biosynthesis is likely to be the cyclization of squalene-2,3-epoxide to β-amyrin. The steroidal avenacoside saponins are also predicted to be synthesized from squalene-2,3-epoxide but by cyclization of squalene-2,3-epoxide to cycloartenol as part of the normal sterol biosynthetic pathway (16). The avenacoside content of nine of the sad mutants was indistinguishable from that of the wild-type oat line, indicating that this pathway was unaffected. The exception, mutant 9, contained normal levels of avenacoside A but reduced levels of avenacoside B. Mutants 9 and 1,139, which are in different complementation groups, both accumulate monodeglucosyl avenacin A-1. Because avenacoside B differs from avenacoside A only in having an additional d-glucosyl residue (27, 28), it appears that mutant 9 is defective in the ability to glucosylate both avenacin A-1 and avenacoside A. The reduced avenacoside B levels had no obvious effect on the interactions of mutant 9 with the leaf-infecting Stagonospora spp. included in these experiments.
Very little is known about saponin biosynthesis in plants in general. The intermediates in the biosynthetic pathway between β-amyrin and the avenacins have not been characterized, nor have any of the cognate genes been isolated. The ability to synthesize avenacins appears to be restricted to the genus Avena (19, 22), and saponins of any kind do not appear to be well represented in cereals. This may be because the Gramineae are generally lacking in these secondary metabolites or because saponins have been bred out of most cultivated cereals. In maize, the complete biosynthetic pathway for the antimicrobial secondary metabolite DIBOA is encoded by five clustered genes. Preliminary genetic analysis suggests that some of the Sad genes identified in this study may also be linked. It is clear that the conversion of β-amyrin to avenacins will be a multistep process involving many biosynthetic enzymes, and the elucidation of this pathway will present an exciting challenge for the future. The mutants described here will be valuable in the dissection of the avenacin biosynthetic pathway and its control.
Table 3.
Disease scores for infection of A. strigosa lines with Gaeumannomyces graminis var. tritici
| Oat line | Disease rating, % seedlings
|
|||
|---|---|---|---|---|
| 0 | + | ++ | +++ | |
| Wild type | 100 | 0 | 0 | 0 |
| 610 | 0 | 25 | 12 | 63 |
| 109 | 28 | 28 | 28 | 16 |
| 791* | 62 | 38 | 0 | 0 |
| 1,027 | 31 | 44 | 6 | 19 |
| 616 | 12 | 71 | 17 | 0 |
| 376 | 27 | 40 | 33 | 0 |
| 825 | 6 | 44 | 50 | 0 |
| 9*† | 74 | 21 | 5 | 0 |
| 1,139† | 56 | 38 | 6 | 0 |
Homozygous mutant oat lines and the wild-type line S75 were tested for resistance to G. graminis var. tritici (isolate T5) by using a seedling assay. The mutant lines in this experiment were homozygous F3 progeny derived from crosses to the wild type. Between 15 and 19 seedlings were tested per line. Disease was scored 21 days after inoculation on an arbitrary scale ranging from 0 to +++ (0, no disease; +, ++, and +++, increasing severity of lesions on the roots). Line 1,243 was not included in this experiment because F3 seed was not available. However, in pathogenicity tests using M3 seedlings, this mutant was clearly susceptible to infection by G. graminis var. tritici.
*Mutants containing reduced levels of avanacins.
†Mutants with growth defects.
Acknowledgments
We thank Andreas Freialdenhoven for amplified fragment length polymorphism (AFLP) analysis of the oat lines, Paul Nicholson for providing isolates of F. culmorum and F. avenaceum, James Brown for statistical analysis, and all those colleagues who helped us in the sowing and harvesting of oat plants. We would also like to thank Shauna Somerville, Paul Schulze-Lefert and Mike Ambrose for their advice and comments. The Sainsbury Laboratory is supported by the Gatsby Charitable Foundation, and K. Papadopoulou is funded by a European Community Marie Curie Research Training Grant.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schönbeck F, Schlösser E. In: Physiological Plant Pathology. Heitefuss R, Williams P H, editors. Berlin: Springer; 1976. pp. 653–678. [Google Scholar]
- 2.VanEtten H D, Mansfield J W, Bailey J A, Farmer E E. Plant Cell. 1994;6:1191–1192. doi: 10.1105/tpc.6.9.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osbourn A E. Plant Cell. 1996;8:1821–1831. doi: 10.1105/tpc.8.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller K O, Börger H. Arb Biol Reichsasnstalt Landw Forstw Berlin. 1940;23:189–223. [Google Scholar]
- 5.Paxton J D. Phytopathol Z. 1981;101:106–109. [Google Scholar]
- 6.Wasmann C C, VanEtten H D. Mol Plant–Microbe Interact. 1996;9:793–803. doi: 10.1094/mpmi-9-0483. [DOI] [PubMed] [Google Scholar]
- 7.Enkerli J, Bhatt G, Covert S F. Mol Plant–Microbe Interact. 1998;11:317–326. doi: 10.1094/MPMI.1997.10.6.742. [DOI] [PubMed] [Google Scholar]
- 8.Hain R, Reif H J, Krause E, Langebartels R, Kindl H, Vornam B, Wiese W, Schmelzer E, Schreier P H, Stocker R H, Stenzel K. Nature (London) 1993;361:153–156. doi: 10.1038/361153a0. [DOI] [PubMed] [Google Scholar]
- 9.Glazebrook J, Ausubel F M. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glazebrook J, Zook M, Mert F, Kagan I, Rogers E E, Crute I R, Holub E B, Hammerschmidt R, Ausubel F M. Genetics. 1997;146:381–392. doi: 10.1093/genetics/146.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomma B P H J, Eggermont K, Penninckx I A M A, Mauch-Mani B, Vogelsang R, Cammue B P A, Broekaert W F. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton R H. Weeds. 1964;12:27–30. [Google Scholar]
- 13.Frey M, Chomet P, Glawischnig E, Stettnet C, Grün S, Winklmair A, Eisenreich W, Bacher A, Meeley R B, Briggs S P, et al. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- 14.Maher E A, Bate N J, Ni W, Elkind Y, Dixon R A, Lamb C J. Proc Natl Acad Sci USA. 1994;91:7802–7806. doi: 10.1073/pnas.91.16.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price K R, Johnson I T, Fenwick G R. CRC Crit Rev Food Sci Nutr. 1987;26:27–133. doi: 10.1080/10408398709527461. [DOI] [PubMed] [Google Scholar]
- 16.Hostettman K A, Marston A. Saponins. Cambridge, U.K.: Cambridge Univ. Press; 1991. [Google Scholar]
- 17.Osbourn A E. Trends Plant Sci. 1996;1:4–9. [Google Scholar]
- 18.Crombie L, Crombie W M L, Whiting D A. J. Chem. Soc. Perkins Trans. 1. 1986. , 1917–1922. [Google Scholar]
- 19.Crombie W M L, Crombie L. Phytochemistry. 1986;25:2069–2073. [Google Scholar]
- 20.Turner E M. J Exp Bot. 1953;4:264–271. [Google Scholar]
- 21.Bowyer P, Clarke B R, Lunness P, Daniels M J, Osbourn A E. Science. 1995;267:371–374. doi: 10.1126/science.7824933. [DOI] [PubMed] [Google Scholar]
- 22.Osbourn A E, Clarke B R, Lunness P, Scott P R, Daniels M J. Physiol Mol Plant Pathol. 1994;45:457–467. [Google Scholar]
- 23.Rines H W. Env Exp Bot. 1985;25:7–16. [Google Scholar]
- 24.Osbourn A E, Clarke B R, Dow J M, Daniels M J. Physiol Mol Plant Pathol. 1991;38:301–312. [Google Scholar]
- 25.Wubben J P, Price K R, Daniels M J, Osbourn A E. Phytopathology. 1996;86:986–992. [Google Scholar]
- 26.Genstat 5 Committee. Genstat 5 Release 41 Reference Manual Supplement. Oxford, U.K.: Numerical Algorithms Group; 1997. [Google Scholar]
- 27.Tschesche R, Tauscher M, Fehlhaber H W, Wulff G. Chem Ber. 1969;102:2072–2082. [Google Scholar]
- 28.Tschesche R, Lauven P. Chem Ber. 1971;104:3549–3555. [Google Scholar]