Abstract
Diversification of cone pigment spectral sensitivities during evolution is a prerequisite for the development of color vision. Previous studies have identified two naturally occurring mechanisms that produce variation among vertebrate pigments by red-shifting visual pigment absorbance: addition of hydroxyl groups to the putative chromophore binding pocket and binding of chloride to a putative extracellular loop. In this paper we describe the use of two blue-shifting mechanisms during the evolution of rodent long-wave cone pigments. The mouse green pigment belongs to the long-wave subfamily of cone pigments, but its absorption maximum is 508 nm, similar to that of the rhodopsin subfamily of visual pigments, but blue-shifted 44 nm relative to the human red pigment, its closest homologue. We show that acquisition of a hydroxyl group near the retinylidene Schiff base and loss of the chloride binding site mentioned above fully account for the observed blue shift. These data indicate that the chloride binding site is not a universal attribute of long-wave cone pigments as generally supposed, and that, depending upon location, hydroxyl groups can alter the environment of the chromophore to produce either red or blue shifts.
Visual pigments differ between species and between different photoreceptor cells within the same species in their wavelengths of maximal absorption. This diversity of visual pigment absorption spectra is determined by interactions between the 11-cis-retinal chromophore and the particular opsin to which it binds, or in rare instances by the alternate use of an 11-cis-dehydroretinal chromophore (1). Among vertebrate visual pigments that use 11-cis-retinal, absorption maxima have been identified throughout the wavelength interval from 360 nm (2, 3) to 570 nm (4). Sequencing of visual pigments from diverse species shows a limited range of wavelengths of maximal absorption among members of each of the three major branches of the visual pigment family: the short-wave subfamily absorbs maximally at wavelengths less than 500 nm, the rhodopsin subfamily absorbs maximally at wavelengths near 500 nm, and the long-wave subfamily absorbs at wavelengths greater than 500 nm (5–7).
Experiments with retinal and its derivatives in solution have led to a number of plausible mechanisms for spectral tuning in visual pigments (8–11). The two photochemical properties of retinal that are most relevant are the shift in absorption maximum from 360 nm to 440 nm upon formation of a protonated Schiff base (12, 13) and the increase in π-electron delocalization and the concomitant change in dipole moment that accompanies photoexcitation (14, 15). Therefore, amino acids from the apoprotein that alter the polarity, polarizability, or charge density around either the conjugated π-electron system or the retinylidene Schiff base would be expected to shift the absorption spectrum of the pigment. In keeping with these models, sequence comparisons and spectroscopic studies of site-directed mutants have identified a glutamate counterion that is present in all vertebrate photoreceptor pigments sequenced to date and is required for protonation of the retinylidene Schiff base (16–18), and three positions where increases in side-chain polarity lead to red shifts of 5–14 nm that together account for most of the variation in absorption maxima among previously characterized members of the long-wave subfamily (19–22). A third red-shifting mechanism involves an interaction between chloride and a histidine in the second putative extracellular loop in members of the long-wave pigment subfamily (23–28); the mechanistic basis of the chloride effect is unclear.
Like other nonprimate mammals, rodents have only two types of cone pigments, one resembling the human blue pigment (ref. 29; the short-wave subfamily) and the other presumed to resemble the human green and red pigments (the long-wave subfamily). The spectral sensitivities of the two corresponding cone types in the mouse retina show maxima at 360 nm and 510 nm as determined by electroretinography (3). Similar maxima are observed in gerbils, gophers, and rats. An absorption maximum of 510 nm makes the mouse green pigment the most blue-shifted member of the long-wave subfamily described to date. In this paper we report the amino acid sequence of the mouse green pigment and show by site-directed mutagenesis experiments that two spectral tuning interactions account for the difference in absorption maximum that distinguishes it from other members of the long-wave subfamily.
MATERIALS AND METHODS
cDNA Cloning.
cDNA clones encoding the mouse green pigment were isolated by low-stringency hybridization of a Lambda ZAP (Stratagene) mouse eye cDNA library (A. Lanahan, H.S., and J.N., unpublished results) using a human red pigment cDNA as probe (5).
In Situ Hybridization.
Digoxigenin in situ hybridization was performed as described in ref. 30. The probes consisted of a full-length mouse green pigment cDNA or a segment of macaque rhodopsin cDNA as described in ref. 31.
Creation of Chimeric Genes and Site-Directed Mutagenesis.
Chimeric genes were created using an in vivo recombination method (32). In brief, a plasmid containing the two visual pigment coding regions in a head-to-tail tandem arrangement was propagated in a strain of Escherichia coli that is proficient in homologous recombination. Incubation with a restriction enzyme that cleaves only at a site between the two coding regions followed by transformation of the resulting DNA molecules enriches for those recombinants that have eliminated the restriction site by homologous recombination between the tandem coding regions. Chimeric junctions were determined by PCR using gene-specific primers and by DNA sequencing. Site-directed mutagenesis was performed using PCR. All DNA segments that were subjected to mutagenesis were sequenced to rule out spurious mutations.
Reconstitution of Visual Pigments.
Production of visual pigments from transfected 293 cells was performed as described in ref. 33 except that the cells were harvested 36 hr after transfection and the membrane pellets were solubilized in 1% rather than 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS). Spectra were obtained from membranes harvested from 20 10-cm plates of transfected 293 cells. To study the effect of iodide on the absorption spectra, 140 mM NaI was used instead of 140 mM NaCl in the 4% BSA solution through which the membranes were centrifuged to remove free retinal and in the CHAPS solution used to solubilize the final membrane pellet.
Absorption Spectra.
Measurement of photobleaching difference absorption spectra, data analysis, and determination of absorption maxima were performed as described (33) except that for photobleaching the sample was exposed for 5 min using a 550-nm narrow band-pass filter. The rhodopsin spectra were determined in the presence of 50 mM hydroxylamine. To improve the signal-to-noise ratio, photobleaching difference spectra were calculated from averages of four prebleach and four postbleach spectra. The absorption maximum for each spectrum was determined by calculating the best-fitting fifth-order polynomial, and it is accurate to within 1–2 nm.
RESULTS
To investigate the molecular basis for the extreme blue shift in the mouse green pigment, we determined the amino acid sequence of this pigment by analyzing cloned cDNA. The identity of the corresponding mRNA was confirmed by localizing it to mouse photoreceptors by in situ hybridization (Fig. 1c) and by functional expression (Fig. 2a). As expected for cone pigment hybridization in a rod-dominant retina, the in situ hybridization pattern shows that a subset of photoreceptor cell bodies contains the mouse green pigment transcript. In contrast, uniform labeling is seen with a rhodopsin probe. When expressed in transfected cells and reconstituted in vitro with 11-cis-retinal, the mouse green pigment reveals a photobleaching difference spectrum with an absorption maximum at 508 nm, in good agreement with the value of 510 nm determined by electroretinography (3).
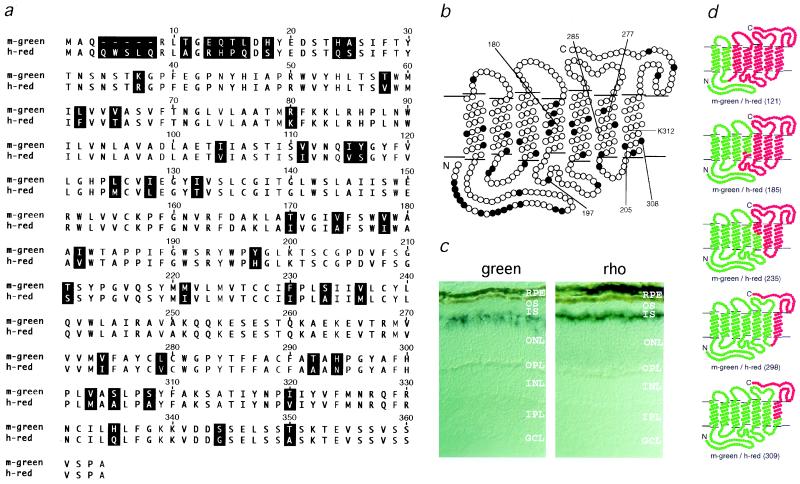
Figure 1.
(a) Sequence comparison of the mouse green pigment and the human red (A180) pigment. Residues that differ are shaded in black. The numbers above the sequence correspond to the human red pigment numbering system, and are used in this paper to refer to both mouse green and human red pigment positions. (b) Topographical model showing the amino acid differences (filled circles) between the mouse green and the human red (A180) pigments. The positions of residues referred to in the text are noted. (c) In situ hybridization comparing green pigment transcripts and rhodopsin transcripts in the mouse retina. Green pigment transcripts (Left) are present in a subset of photoreceptor cells that are located at various positions in the outer third of outer nuclear layer, while rhodopsin transcripts (Right) are localized to rod inner segments. RPE, retinal pigment epithelium; OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. (d) Structure of chimeras constructed between the mouse green pigment (in green) and the human red (A180) pigment (in red). The amino acid at which each crossover occurs is indicated in parentheses.
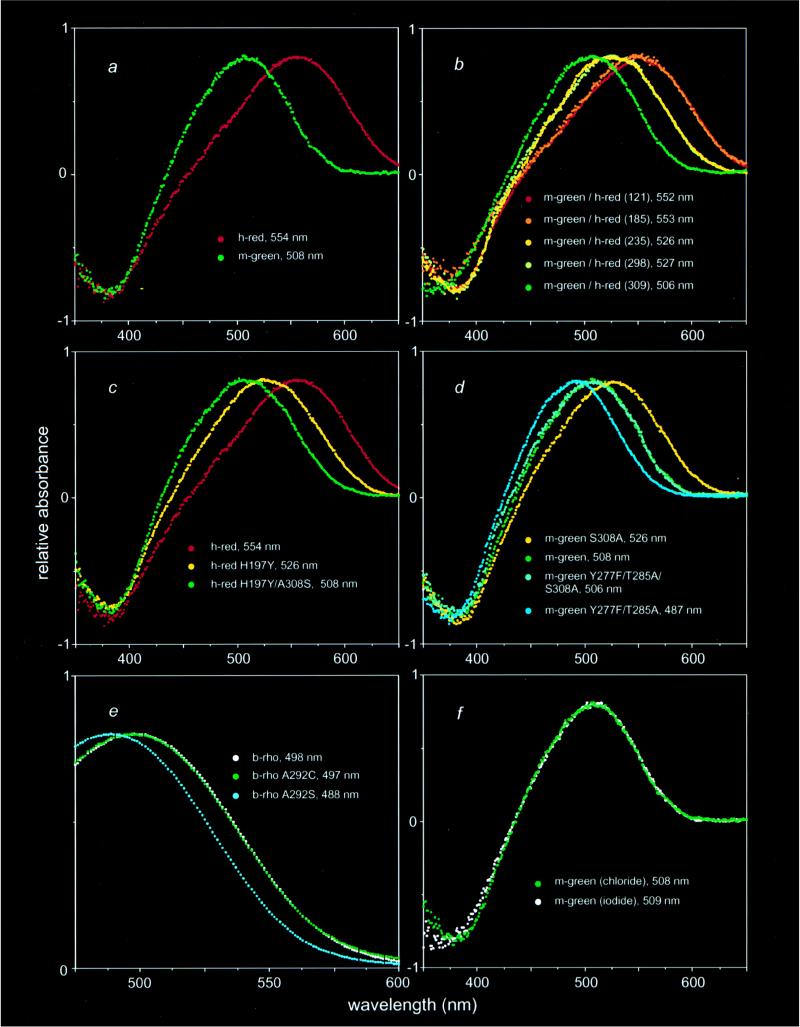
Figure 2.
Visual pigment photobleaching difference absorption spectra obtained by subtracting a spectrum measured after photobleaching from a spectrum measured before light exposure. The amplitudes of maximal absorption ranged from 0.007 to 0.034 optical density units for cone pigments, and from 0.072 to 0.179 optical density units for rhodopsin. For the purposes of this figure the spectra were scaled to the same height. (a) Absorption spectra of the mouse green pigment and the human red (A180) pigment. (b) Absorption spectra of chimeras with mouse green pigment sequences at the amino terminus and human red (A180) pigment sequences at the carboxyl terminus. (c) Amino acid substitutions at positions 197 and 308 in the human red (A180) pigment together produce a blue-shifted pigment with the same absorption spectrum as the mouse green pigment. (d) In the mouse green pigment, the blue shift produced by S308 and the red shifts produced by Y277 and T285 are independent and additive. (e) Serine but not cysteine at position 292 in bovine rhodopsin produces a blue shift. (f) The absorption spectrum of the mouse green pigment is unaffected by the substitution of NaI for NaCl. Note that to compare absorption maxima in terms of photoexcitation energy differences, one should convert these values to a frequency scale (i.e., wavelength−1); for the range of absorption maxima considered here the comparison of wavelengths approximates the energy difference to within 10%.
Surprisingly, the amino acid sequence of the mouse green pigment (Fig. 1 a and b) more closely resembles the human red pigment (absorption maximum 552 nm or 557 nm for the A180 or S180 polymorphic variants, respectively) than the human green pigment (absorption maximum 530 nm; refs. 5, 19, 20, 22). In particular, the mouse green pigment shares with the human red pigment Y277 and T285, residues that differ between human red and green pigments and that together account for 21 nm of the red shift that distinguishes the human red and green pigments (19–22). At position 180, the third position that plays a major role in human red and green pigment spectral differences, the mouse green pigment has an alanine, as does the human green pigment and the 552-nm variant of the red pigment. Thus, the mouse green pigment is blue-shifted 44 nm relative to the human red (A180) pigment, with which it shares the greatest similarity.
To identify the residues that account for the spectral sensitivity of the mouse green pigment, we constructed chimeras between it and the human red (A180) pigment (Fig. 1d). Each of the five chimeras in which mouse sequences were amino-terminal to human sequences produced a photolabile pigment upon incubation with 11-cis-retinal, whereas four of five chimeras with the converse arrangement failed to produce a pigment. The failure of chimeric or mutant pigments to form a photolabile pigment is presumed to reflect protein instability or an inability to stably bind 11-cis-retinal; for simplicity we shall refer to these as unstable pigments. Comparisons of the absorption spectra of the parental and chimeric pigments point to two small regions as the sole determinants of the 44-nm difference in the absorption maxima of the mouse green and human red (A180) pigments (Fig. 2). [Calculations in this paper use the published absorption maximum of the human red (A180) pigment, 552.4 ± 1.1, i.e. 552 nm (ref. 20); the example shown in Fig. 2 has a measured absorption maximum of 554 nm, within the 1–2-nm accuracy of this method.] Chimeras m-green/h-red(121) [i.e., a chimeric protein with mouse green pigment sequences amino-terminal to position 121 and human red (A180) pigment sequences carboxyl-terminal to position 121] and m-green/h-red(185) have absorption maxima indistinguishable from that of the human red (A180) pigment; chimera m-green/h-red(309) has an absorption maximum indistinguishable from that of the mouse green pigment; but chimeras m-green/h-red(235) and m-green/h-red(298) have identical absorption spectra at a position intermediate between those of the mouse green and human red (A180) pigments. The sequences responsible for spectral tuning therefore must reside in two small regions of the protein, one region encompassing the second extracellular loop and a second region in the seventh transmembrane segment (Fig. 1b). These regions are responsible for spectral shifts of approximately 26 nm and 20 nm, respectively.
Comparison of the mouse green pigment amino acid sequence with the sequences of other long-wave pigments suggests that a loss of chloride sensitivity (23–27) could account for the blue shift produced by sequence differences in the second extracellular loop. The mouse green pigment is the only member of the long-wave subfamily of visual pigments sequenced to date that does not have a histidine at position 197 in the second extracellular loop, a residue which has been shown by site-directed mutagenesis to mediate a chloride-dependent red shift of 30 nm in the human long-wave family (28). This residue is presumed to account for the 10- to 30-nm chloride-dependent red shift observed in long-wave pigments from other animals. In the photoreceptor cell and under standard in vitro conditions, the pigment is exposed to saturating concentrations of chloride, leading to the full red shift. Fig. 2f shows that the mouse green pigment differs from other pigments in the long-wave subfamily in that its spectrum is not changed upon replacement of chloride by iodide, an anion that does not produce a chloride-type red shift (23–27). To determine whether this difference in spectral behavior could be attributed solely to a difference at residue 197, we examined the properties of single amino acid substitution mutations at this position. When Y197 in the mouse green pigment was replaced by histidine (Y197H), the corresponding amino acid in the human red (A180) pigment, the resulting pigment was unstable. However, the converse mutation, H197Y in the human red (A180) pigment, generated a functional pigment with a 28-nm blue shift (Fig. 2c), a spectral shift that can fully account for the difference in absorption maxima between chimeras m-green/h-red(185) and m-green/h-red(235), and consistent with the earlier work of Wang and colleagues (28).
Within the region defined by chimeras m-green/h-red(298) and m-green/h-red(309), positions 305 and 308 are highly conserved among long-wave pigments but differ in the mouse green pigment (Fig. 1; refs. 5–7). Mutation of these residues in the human red (A180) pigment to the corresponding residues from the mouse green pigment shows that A305S does not produce a spectral shift (data not shown) and that A308S causes the pigment to be unstable. However, when the A308S and H197Y mutations were introduced together into the human red (A180) pigment, the double mutant formed a functional pigment with an absorption maximum indistinguishable from that of the mouse green pigment (Fig. 2c). Thus, sequence differences at positions 197 and 308 can fully account for the 44-nm spectral difference between the mouse green pigment and the human red (A180) pigment. In agreement with this conclusion, mutation S308A in the mouse green pigment produced an 18-nm red shift (Fig. 2d), which corresponds to the difference in absorption maxima between chimeras m-green/h-red(298) and m-green/h-red(309).
With respect to pigment instability, an interesting pattern emerged from testing the four possible single- and double-substitution mutants at positions 197 and 308 in both the mouse green and human red (A180) pigments (i.e., H197/A308; H197/S308; Y197/A308; and Y197/S308). Functional pigments are observed whenever position 197 is occupied by tyrosine. However, when position 197 is occupied by histidine, only the red (A180) pigment with alanine at position 308 is stable. Although we cannot offer a structural explanation for this pattern of pigment instability, it suggests an intimate relationship between the chloride binding site at position 197 and the spectral tuning residue at position 308.
The 18-nm blue shift produced by introduction of serine at position 308 appears to be independent of and additive with spectral shifts produced by substitutions at positions 277 and 285, the two most significant positions for producing the spectral separation between human red and green pigments. This was demonstrated for the mouse green pigment by introducing mutations Y277F and T285A, which together produce a blue shift of 21 nm in the human red pigment (19, 21, 22). As seen in Fig. 2d, the Y277F/T285A double mutation produces blue shifts of 20 or 21 nm in mouse green pigments that carry A308 or S308, respectively. The Y277F/T285A mutant of the mouse green pigment, with a maximal absorption at 487 nm, is the most blue-shifted member of the long-wave subfamily produced to date.
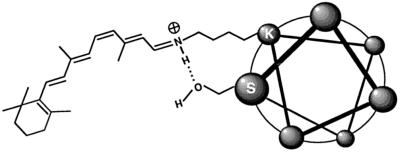
Position 308 is located one helical turn from K312, the site of the retinylidene Schiff base, suggesting that the hydroxyl group of S308 may stabilize the protonated Schiff base, thereby decreasing π-electron delocalization in the attached 11-cis-retinal and producing a higher energy gap between the ground and photoexcited states—i.e., a blue shift (14, 15). This stabilization could occur through a hydrogen bond between S308 and the protonated retinylidene Schiff base, as depicted in Fig. 3, or by stabilization of the interaction between the retinylidene Schiff base and its counterion, E129 (equivalent to E113 in bovine rhodopsin; refs. 16–18). Both models are consistent with earlier work on retinal derivatives in solution (8–10).
Figure 3.
Schematic diagram of one model of spectral tuning by means of S308 in which a hydrogen bond between S308 and the protonated retinylidene Schiff base at K312 stabilizes the positive charge on the Schiff base nitrogen. The Schiff base counterion E129 is not shown. The helical wheel representation of an α-helical seventh transmembrane segment of opsin is shown, looking from the extracellular face.
The sulfhydryl group of cysteine has a weaker dipole moment and is a weaker hydrogen bond acceptor than is the hydroxyl group of serine. Therefore, one prediction of the charge stabilization model described above is that the introduction of cysteine at position 308 should produce little or no spectral shift, despite its similarity in size to serine. The most direct test of this prediction could not be performed because the C308 mutants of the human red (A180) and mouse green pigments were found to be unstable (data not shown). However, we found that the corresponding mutation could be studied using the more stable rod pigment, rhodopsin (Fig. 2e). Bovine rhodopsin has alanine at position 292 (34), which corresponds to position 308 in the long-wave pigments. In bovine rhodopsin, substitution A292S produces a blue shift of 10 nm, whereas substitution A292C produces no spectral shift. This result implies that the blue shift produced by S308 is related to its side-chain polarity or hydrogen bonding potential rather than its size, consistent with the model described above.
DISCUSSION
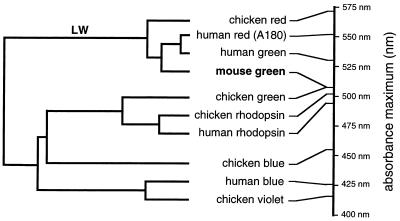
The experiments reported here define an evolutionary pathway for spectral tuning in which the present-day mouse green pigment appears to have evolved from an ancestral red pigment by a two-step mutational pathway involving substitutions H197Y and A308S. In this evolutionary pathway, the loss of a chloride binding site by substitution H197Y can be regarded as a loss-of-function mutation, whereas the acquisition of a blue-shifting hydroxyl group by substitution A308S can be regarded as a gain-of-function mutation. An alternative view in which the mouse green pigment is considered an “intermediate” species that has yet to acquire a chloride binding site seems unlikely in view of the widespread existence of the chloride binding site in vertebrate long-wave pigments, including those of birds, fish, and amphibia (23–27), and the corresponding presence of histidine at position 197 in these pigments (5–7, 35). The proposed tuning mechanisms, together with those previously defined in the human red and green pigments, extend the range of spectral sensitivities that can be accommodated within the long-wave subfamily to include absorption maxima as low as 487 nm (the mouse green pigment carrying the Y277F and T285A mutations), a range that largely overlaps that of the rhodopsin subfamily (Fig. 4). At the long-wave end of the spectrum, retinal-based pigments in this subfamily have been documented with absorption maxima as high as 570 nm (chicken iodopsin; ref. 4), and the use by some organisms of dehydroretinal produces long-wave pigments with absorption maxima as high as 620 nm (1). Thus, pigments within the long-wave subfamily can exhibit a range of at least 130 nm in absorption maxima, permitting an evolutionary plasticity that has been exploited in different ways among vertebrates (37, 38).
Figure 4.
Dendrogram of a representative sample of vertebrate visual pigments based on amino acid sequence homology (calculated using Geneworks software). Human and chicken visual pigments and the mouse green pigment are included. The wavelengths of maximal absorption are shown to the right (refs. 4, 20, and 36, and this study). The mouse green pigment (in boldface type) is in the long-wave pigment branch, indicated by “LW”; all other members of this branch show a chloride-dependent red shift.
Acknowledgments
We thank Dr. Sriram Subramaniam for helpful comments on the manuscript. This work was supported by the Howard Hughes Medical Institute.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank data base (accession no. AF011389).
References
- 1.Wald G. Nature (London) 1968;219:800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 2.Harosi F I, Hashimoto Y. Science. 1983;222:1021–1023. doi: 10.1126/science.6648514. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs G H, Neitz J, Deegan J F. Nature (London) 1991;353:655–656. doi: 10.1038/353655a0. [DOI] [PubMed] [Google Scholar]
- 4.Okano T, Fukada Y, Artamonov I D, Yoshizawa T. Biochemistry. 1989;28:8848–8856. doi: 10.1021/bi00448a025. [DOI] [PubMed] [Google Scholar]
- 5.Nathans J, Thomas D, Hogness D S. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 6.Okano T, Kojima D, Fukada Y, Shichida Y, Yoshizawa T. Proc Natl Acad Sci USA. 1992;89:5932–5936. doi: 10.1073/pnas.89.13.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson R L, Grant K B, Zankel T C, Boehm M F, Merbs S L, Nathans J, Nakanishi K. Biochemistry. 1993;32:208–214. doi: 10.1021/bi00052a027. [DOI] [PubMed] [Google Scholar]
- 8.Blatz P E, Mohler J H, Navangul H V. Biochemistry. 1972;11:848–855. doi: 10.1021/bi00755a026. [DOI] [PubMed] [Google Scholar]
- 9.Blatz P E, Mohler J H. Biochemistry. 1975;14:2304–2309. doi: 10.1021/bi00682a005. [DOI] [PubMed] [Google Scholar]
- 10.Baasov T, Sheves M. Biochemistry. 1986;25:5249–5258. [Google Scholar]
- 11.Koutalos Y, Ebrey T G, Tsuda M, Odashima K, Lien T, Park M H, Shimuzu N, Derguini F, Nakanishi K, Gilson H R, Honig B. Biochemistry. 1989;28:2732–2739. doi: 10.1021/bi00432a055. [DOI] [PubMed] [Google Scholar]
- 12.Ball S, Collins F D, Dalvi P D, Morton R A. Biochem J. 1949;45:304–307. doi: 10.1042/bj0450304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morton R A, Pitt G A J. Biochem J. 1955;59:128–134. doi: 10.1042/bj0590128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kropf A, Hubbard R. Ann NY Acad Sci. 1958;74:266–280. doi: 10.1111/j.1749-6632.1958.tb39550.x. [DOI] [PubMed] [Google Scholar]
- 15.Mathies R, Stryer L. Proc Natl Acad Sci USA. 1976;73:2169–2173. doi: 10.1073/pnas.73.7.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhukovsky E A, Oprian D D. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 17.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathans J. Biochemistry. 1990;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- 19.Neitz M, Neitz J, Jacobs G H. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- 20.Merbs S L, Nathans J. Nature (London) 1992;356:433–435. doi: 10.1038/356433a0. [DOI] [PubMed] [Google Scholar]
- 21.Merbs S L, Nathans J. Photochem Photobiol. 1993;58:706–710. doi: 10.1111/j.1751-1097.1993.tb04956.x. [DOI] [PubMed] [Google Scholar]
- 22.Asenjo A B, Rim J, Oprian D D. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 23.Crescitelli F. Science. 1977;195:187–188. doi: 10.1126/science.831267. [DOI] [PubMed] [Google Scholar]
- 24.Fager L Y, Fager R S. Exp Eye Res. 1979;29:401–408. doi: 10.1016/0014-4835(79)90056-3. [DOI] [PubMed] [Google Scholar]
- 25.Shichida Y, Kato T, Sasayama S, Fukada Y, Yoshizawa T. Biochemistry. 1990;29:5843–5848. doi: 10.1021/bi00476a028. [DOI] [PubMed] [Google Scholar]
- 26.Crescitelli F, Karlavy B. Vision Res. 1991;31:945–950. doi: 10.1016/0042-6989(91)90202-g. [DOI] [PubMed] [Google Scholar]
- 27.Kleinschmidt J, Harosi F I. Proc Natl Acad Sci USA. 1992;89:9181–9185. doi: 10.1073/pnas.89.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Asenjo A, Oprian D D. Biochemistry. 1993;32:2125–2130. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- 29.Chiu M I, Zack D J, Wang Y, Nathans J. Genomics. 1994;21:440–443. doi: 10.1006/geno.1994.1292. [DOI] [PubMed] [Google Scholar]
- 30.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 31.Allikmets R, Singh N, Sun H, Shroyer N F, Hutchinson A, Chidambaram A, Gerrard B, Baird l, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson K L, Lewis R A, Nathans J, Leppert M, Dean M, Lupski J R. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 32.Levin L R, Reed R R. J Biol Chem. 1995;270:7573–7579. doi: 10.1074/jbc.270.13.7573. [DOI] [PubMed] [Google Scholar]
- 33.Merbs S L, Nathans J. Photochem Photobiol. 1992;56:869–881. doi: 10.1111/j.1751-1097.1992.tb09708.x. [DOI] [PubMed] [Google Scholar]
- 34.Nathans J, Hogness D S. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 35.Kojima D, Okano T, Fukada Y, Shichida Y, Yoshizawa T, Ebrey T G. Proc Natl Acad Sci USA. 1992;89:6841–6845. doi: 10.1073/pnas.89.15.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wald G, Brown P K. Science. 1958;127:222–226. doi: 10.1126/science.127.3292.222. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs G H. Comparative Color Vision. New York: Academic; 1981. [Google Scholar]
- 38.Lythgoe J N. The Ecology of Vision. Oxford: Clarendon; 1979. [Google Scholar]