Abstract
Vegetable oils that contain fatty acids with conjugated double bonds, such as tung oil, are valuable drying agents in paints, varnishes, and inks. Although several reaction mechanisms have been proposed, little is known of the biosynthetic origin of conjugated double bonds in plant fatty acids. An expressed sequence tag (EST) approach was undertaken to characterize the enzymatic basis for the formation of the conjugated double bonds of α-eleostearic (18:3Δ9cis,11trans,13trans) and α-parinaric (18:4Δ9cis,11trans,13trans,15cis) acids. Approximately 3,000 ESTs were generated from cDNA libraries prepared from developing seeds of Momordica charantia and Impatiens balsamina, tissues that accumulate large amounts of α-eleostearic and α-parinaric acids, respectively. From ESTs of both species, a class of cDNAs encoding a diverged form of the Δ12-oleic acid desaturase was identified. Expression of full-length cDNAs for the Momordica (MomoFadX) and Impatiens (ImpFadX) enzymes in somatic soybean embryos resulted in the accumulation of α-eleostearic and α-parinaric acids, neither of which is present in untransformed soybean embryos. α-Eleostearic and α-parinaric acids together accounted for as much as 17% (wt/wt) of the total fatty acids of embryos expressing MomoFadX. These results demonstrate the ability to produce fatty acid components of high-value drying oils in transgenic plants. These findings also demonstrate a previously uncharacterized activity for Δ12-oleic acid desaturase-type enzymes that we have termed “conjugase.”
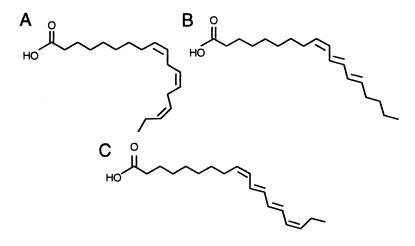
Hundreds of unusual fatty acid structures are known to occur in the seed oils of various plant species (1). The biosynthetic pathways of many of these fatty acids are unknown or have not been well characterized. One such class consists of fatty acids with double bonds that are conjugated. This structural configuration is in contrast to that of linoleic (18:2Δ9cis,12cis) and α-linolenic (18:3Δ9cis,12cis,15cis) acids, the typical polyunsaturated fatty acids of plant seed oils, which contain double bonds that are separated by methylene (−CH2−) groups (Fig. 1). Among the fatty acids with conjugated double bonds that occur in the plant kingdom are α-eleostearic acid (18:3Δ9cis,11cis,13trans) and α-parinaric acid (18:4Δ9cis,11trans,13trans,15cis) (Fig. 1). α-Eleostearic acid accounts for >65% (wt/wt) of the total fatty acids of tung oil, a high-value drying oil obtained from seeds of Aleurites fordii (2). Other sources of this fatty acid include the seed oil of Momordica charantia, which contains approximately 65% (wt/wt) α-eleostearic acid (2). In addition, α-parinaric acid composes 30 to 65% (wt/wt) of the seed oils of plants such as Parinarium laurinum and Impatiens species (1, 3, 4).
Figure 1.
Comparison of the structures of fatty acids with methylene interrupted double bonds (A) vs. those that contain conjugated double bonds (B and C). Shown are α-linolenic acid (18:3Δ9cis,12cis,15cis) (A), α-eleostearic acid (18:3Δ9cis,11trans,13trans) (B), and α-parinaric acid (18:4Δ9cis,11trans,13trans,15cis) (C).
The presence of conjugated double bonds in fatty acids markedly increases their rate of oxidation relative to polyunsaturated fatty acids with methylene-interrupted double bonds (5). This property makes seed oils, such as tung oil, that are enriched in fatty acids with conjugated double bonds well suited for use as drying agents in paints, varnishes, and inks because they require less oxygen for the polymerization reactions that occur during the drying process (5). As a result, coating materials that contain oils with conjugated unsaturation dry more quickly than those with methylene-interrupted unsaturation, and the resulting films are more resistant to water and alkali (6).
It has been demonstrated that conjugated double bonds arise from the modification of an existing cis-double bond. Liu et al. (7), for example, reported that linoleic acid bound to phosphatidylcholine serves as the precursor of α-eleostearic acid in developing seeds of M. charantia. In this reaction, the cis-Δ12 double bond of linoleic acid is converted into the two trans-Δ11 and Δ13 double bonds found in α-eleostearic acid. The mechanism involved in this reaction has yet to be determined, although a number of biosynthetic scenarios have been proposed. The possible origins of conjugated double bonds include reactions similar to those catalyzed by lipoxygenases (8), because oxidation of the Δ12 double bond of linoleic acid by 13-lipoxygenases results in conjugated Δ9cis and Δ11trans double bonds. However, this reaction also yields a hydroperoxy group at the Δ13-carbon atom rather than a third conjugated double bond, such as that found in α-eleostearic acid. In addition, Gunstone (9) proposed a reaction scheme involving sequential hydroxylation and reduction of the two methylene groups that flank the carbon atoms at an existing double bond.
As a step toward clarifying the biosynthetic origin of conjugated double bonds, we undertook an expressed sequence tag (EST) approach. As described here, cDNAs for variant forms of the Δ12-oleic acid desaturase (Fad2) were identified by random sequencing of libraries prepared from developing seeds of M. charantia and Impatiens balsamina. Expression of these cDNAs in transgenic somatic soybean embryos resulted in the accumulation of fatty acids with conjugated double bonds. These findings demonstrate a catalytic activity not previously described for fatty acid desaturase-related enzymes or other diiron-oxo proteins.
Materials and Methods
cDNA Library Construction.
Developing seeds of M. charantia were obtained from fresh fruits purchased at Asian produce markets in Philadelphia. Immature cotyledons were dissected from seed coats for use in RNA isolation. Developing I. balsamina seeds were harvested from seed pods collected from plants grown on the grounds of the DuPont Experimental Station.
Total RNA was extracted from developing seeds of M. charantia by using Trizol reagent (GIBCO-BRL) according to the manufacturer’s protocol and from developing seeds of I. balsamina as described by Jones et al. (10). PolyA+ RNA was subsequently enriched by passing total RNA from each tissue twice over oligo-dT cellulose columns by using reagents supplied in the QuickPrep mRNA purification kit (Pharmacia Biotech). cDNA libraries in plasmid form were then synthesized from 7.5 μg of polyA+ RNA from Momordica and Impatiens seeds by using a Uni-ZAP XR cDNA synthesis kit (Stratagene) with several modifications of the manufacturer’s protocol. These modifications included the use of Superscript II RNase H− reverse transcriptase (GIBCO-BRL) for synthesis of first-strand cDNA. In addition, selection of cDNA inserts that contained >500 bp was performed by using cDNA Size Fractionation Columns (GIBCO-BRL). The resulting cDNA inserts, which contained flanking 5′ EcoRI and 3′ XhoI sites, were ligated into the corresponding sites of pBluescript SK(−) and then transformed into Escherichia coli ElectroMAX DH10B cells (GIBCO-BRL) by electroporation. Bacterial cells harboring the libraries in plasmid form were maintained as glycerol stocks at −80°C. The libraries derived from developing seeds of Momordica and Impatiens represented >500,000 independent cDNAs.
Generation of ESTs and Identification of Diverged Fad2 cDNAs.
Plasmids for EST analysis were prepared from randomly picked colonies from the Momordica and Impatiens cDNA libraries by using the Qiagen R.E.A.L. Prep 96 System according to the manufacturer’s protocol. Nucleotide sequence was obtained from the T3 priming site of cDNA clones in pBluescript SK (−) by dye-terminator cycle sequencing by using an ABI 377 DNA fluorescence sequencer. To assign putative identities to ESTs, the resulting sequence information was compared with translated sequences in the public databases by using the National Center for Biotechnology Information blastx program (11).
Among the pools of ESTs from the Momordica and Impatiens libraries, full-length cDNAs for diverged forms of the Fad2 were identified. (The polypeptides encoded by the Momordica and Impatiens cDNAs were designated MomoFadX and ImpFadX, respectively.) Nucleotide sequences were determined from both strands of the MomoFadX and ImpFadX cDNAs in pBluescript SK(−) by dye-terminator sequencing by using the instrumentation described above.
Expression of ImpFadX in Saccharomyces cerevisiae.
A full-length ImpFadX cDNA in pBluescript SK(−) (obtained from the EST analysis described above) was recovered by digestion with EcoRI and XhoI. A full-length cDNA for MomoFadX was also isolated from pBluescript SK(−) as a NotI/XhoI fragment. The purified cDNA inserts were ligated into the corresponding sites of the S. cerevisiae expression vector pYES2 (Invitrogen). The resulting plasmids containing the ImpFadX or MomoFadX cDNA behind the GAL1 promoter were introduced into S. cerevisiae INVSc1 cells (Invitrogen) by lithium acetate-mediated transformation (12). Transformed cells were selected for their ability to grow on media lacking uracil. Individual colonies of transformed cells were then grown for 2 days at 30°C in growth media lacking uracil [0.17% (wt/vol) yeast nitrogen base without amino acids (Difco)/0.5% (wt/vol) ammonium sulfate/0.18% SC-URA (Bio101)] supplemented with glycerol and glucose to a final concentration of 5% (vol/vol) and 0.5% (wt/vol), respectively. Cells were then washed twice in the growth media described above, which was supplemented instead with galactose to a final concentration of 2% (wt/vol). The washed cells were then diluted to OD600 ≈0.2 in the galactose growth media that also contained Tergitol Nonidet P-40 (Sigma) at a concentration of 0.2% (wt/vol). Aliquots of these cells were grown without exogenous fatty acids or with the addition of either linoleic acid (18:2Δ9cis,12cis) or α-linolenic acid (18:3Δ9cis,12cis,15cis) at a final concentration of 2 mM. After 4 days of growth at 16°C, the S. cerevisiae cells were harvested and examined for the accumulation of fatty acids containing conjugated double bonds as described below.
Expression of cDNAs in Somatic Soybean Embryos.
The vector pKS67 was used for expression of cDNAs for MomoFadX and ImpFadX in soybean (Glycine max) somatic embryos. This vector contains a unique NotI site for cloning of transgenes. This site is flanked by the promoter of the gene for the α′-subunit of β-conglycinin (for seed-specific expression of transgenes) (13) and phaseolin termination sequence (14). Bacterial selection with this vector is conferred by a hygromycin B phosphotransferase gene (15) under control of the T7 RNA polymerase promoter, and plant selection is conferred by a second hygromycin B phosphotransferase gene under control of the cauliflower mosaic virus 35S promoter.
Full-length MomoFadX and ImpFadX cDNAs were amplified by PCR to generate flanking NotI sites for subcloning into the pKS67 vector. PCR reactions were conducted by using Pfu polymerase (Stratagene) and plasmids containing the Impatiens and Momordica cDNAs as templates. For amplification of the MomoFadX, the following oligonucleotide primer combination was used: 5′-aaggaaaaaagcggccgcATGGGGGGCAGAGGAGCTATT-3′ (sense) and 5′-aaggaaaaaagcggccgcTCAGAGCTTGTTGTGGTACCA-3′ (antisense). The ImpFadX cDNA was amplified by using the primer combination: 5′-aaggaaaaaagcggccgcATGGGAGAAGTGGGACCCACA-3′ (sense) and 5′- aaggaaaaaagcggccgcTCAAATGTCGTATTGTACCA-3′. (Note: The sequences shown in lower case contain an added NotI site along with additional bases to facilitate restriction enzyme digestion.) Products from PCR reactions were cloned into the intermediate vector pGEM-T (Promega). Before ligation, a single adenine base was added to the 3′ end of each strand by incubation of purified PCR products for 20 min at 72°C with 200 μM dATP/2.5 units Amplitaq DNA polymerase (Perkin–Elmer)/1.5 mM MgCl2 in a final volume of 100 μL. The subcloned PCR products were then digested with NotI and introduced into the corresponding site of the plant expression vector pKS67.
Gene fusions of the ImpFadX and MomoFadX cDNAs with the β-conglycinin promoter and phaseolin termination sequences in pKS67 were introduced into soybean embryos by using the particle bombardment method of transformation (16). To induce somatic embryos, cotyledons (3–5 mm in length) were dissected from surface sterilized immature soybean seeds of soybean cultivar A2872, unless otherwise indicated. Secondary somatic embryos to be used for bombardment were excised and maintained essentially as described in ref. 16. Seven to eight weeks after bombardment, green transformed tissue growing from untransformed necrotic embryogenic clusters was removed and inoculated into individual flasks to generate new clonally propagated transformed embryogenic suspension cultures. Selection of transformants was achieved by the addition of hygromycin to the media (50 μg/ml final concentration). Each new line was treated as an independent transformation event. These suspensions were then subcultured and maintained as clusters of immature embryos until analyzed for fatty acid composition.
Fatty Acid Analysis of Transgenic Soybean Embryos.
Fatty acid methyl esters were prepared from transgenic soybean embryos or from S. cerevisiae cultures by transesterification in 1% (wt/vol) sodium methoxide in methanol. Single soybean embryos were homogenized with a glass stirring rod in 0.5 ml of the sodium methoxide solution and incubated at room temperature for 20 min. At the end of this period, 0.5 ml of 1 M sodium chloride was added, and fatty acid methyl esters were extracted with 0.5 ml of heptane. In the case of S. cerevisiae, cells from 3 ml cultures were collected by centrifugation, dried under vacuum, and then resuspended in 0.4 ml of sodium methoxide solution. After 20-min incubation in the transesterification reagent, fatty acid methyl esters were extracted as described for soybean embryos. Fatty acid methyl esters in the soybean embryo and yeast extracts were resolved and quantified by using a Hewlett Packard 6890 gas chromatograph fitted with a 30 m x 0.32 mm (inner diameter) Omegawax column (Supelco). The oven temperature was programmed from 220°C (2-min hold) to 240°C at a rate of 20°C/min, and carrier gas was supplied by a Whatman hydrogen generator. Eluted compounds were detected by flame ionization. Fatty acid methyl esters were also analyzed by GC-MS by using a Hewlett Packard 6890 gas chromatograph interfaced with a Hewlett Packard 5973 mass selective detector. Samples were separated with a 30-m x 0.25-mm (inner diameter) INNOWax column (Hewlett Packard). The oven temperature was programmed from 185°C (3.5-min hold) to 215°C (5-min hold) at a rate of 2°C/min and then to 230°C at a rate of 5°C/min. The mass spectra of methyl α-eleostearic and α-parinaric acids were characterized by abundant molecular ions of 292 m/z and 290 m/z, respectively. Compounds in transgenic soybean embryo and S. cerevisiae extracts identified as methyl α-eleostearic acid or α-parinaric acid had retention times and mass spectra that were identical to the corresponding fatty acid methyl esters in extracts of M. charantia and I. balsamina seeds. Although positions and cis-trans orientations of double bonds were not determined, small differences in these structural features have a measurable effect on the retention times of isomers of methyl α-eleostearic and α-parinaric acids [see supplemental Figs. 5 and 6 for additional gas chromatographic and mass spectral data (www.pnas.org)].
Northern Blot Analysis.
Total RNA was extracted from leaves and developing seeds of M. charantia and from leaves and seed pods (containing developing seeds) of I. balsamina by using methods described above. Total RNA (10 μg) from each tissue and RNA standards were electrophoresed in a 1% agarose gel. Formaldehyde was included in the sample loading buffer at a final concentration of 6.5% (wt/vol). After electrophoresis, RNA was transferred from the gel to Hybond N+ (Amersham Pharmacia) using 20 X SSC (17). The RNA was fixed to the membrane by UV crosslinking. The membrane was rinsed with 2 X SSC and then hybridized with 32P-labeled probes for 18 h at 65°C in 5 X SSC/5 X Denhardt’s reagent/0.5% (wt/vol) SDS/5% (wt/vol) dextran sulfate/100 μg/ml salmon sperm DNA. Probes were prepared from the full-length cDNAs for MomoFadX and ImpFadX and were labeled by using random hexamer priming (17). Blots were washed twice with 2 X SSC/0.1% SDS at room temperature and twice at 65°C with 0.5 X SSC/0.1% SDS. Radioactivity on filters was detected by phosphorimaging.
Results
Identification of cDNAs for Diverged Forms of Fad2 from Seeds of Momordica and Impatiens.
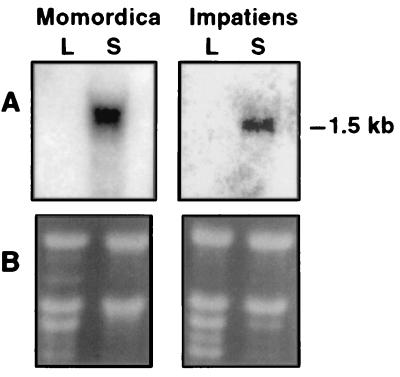
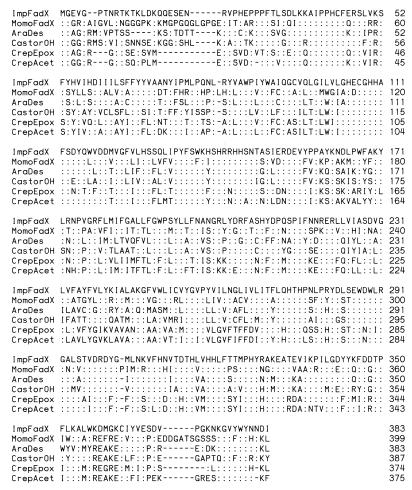
To identify cDNAs for enzymes involved in the synthesis of conjugated double bonds of fatty acids, ESTs were generated by random sequencing of cDNAs from libraries prepared from developing seeds of M. charantia and I. balsamina. Seeds of both plants are rich sources of fatty acids with conjugated double bonds. In this regard, α-eleostearic acid (18:3Δ9cis,11trans,13trans) composes ≈65% of the seed oil of M. charantia (7), and α-parinaric (18:4Δ9cis,11trans,13trans,15cis) and α-eleostearic acids compose ≈30% and ≈2%, respectively, of the seed oil of Impatiens (ref. 4; supplemental Fig. 5). These fatty acids are absent from leaves of both species (data not shown). Previous reports provided few definitive clues as to the biosynthetic origin of conjugated double bonds in fatty acids (7–9, 18). As such, our analyses of ESTs from each library focused broadly on variant forms of fatty acid modification enzymes, including desaturases and lipoxygenases. Homology searches of ESTs revealed the occurrence of full-length cDNAs for a diverged form of the microsomal Fad2 in both the Momordica and Impatiens libraries. This class of cDNAs was found to occur at a frequency of 12 per 2,792 ESTs in the Impatiens library and three per 3,096 ESTs in the Momordica library. Results from Northern analysis indicated that transcripts for these cDNAs were present in developing seeds of both species but absent in leaves (Fig. 2). This expression profile is consistent with the seed-specific occurrence of fatty acids with conjugated double bonds in Momordica and Impatiens (unpublished observation). The polypeptides deduced from the Fad2-like cDNAs from Momordica and Impatiens were designated MomoFadX and ImpFadX, respectively. These polypeptides share 53% amino acid sequence identity and contain histidine motifs that are characteristic of membrane-bound diiron-oxo proteins (19). In addition, MomoFadX and ImpFadX share from 50 to 60% amino acid sequence identity with the Arabidopsis Fad2 (20) and diverged forms of Fad2 including the castor oleic acid hydroxylase (21) and the Crepis acetylenase and epoxygenase (22) (Fig. 3).
Figure 2.
Northern analysis of the expression of genes for a diverged Δ12-oleic acid desaturase or Fad2 from Momordica (MomoFadX) and Impatiens (ImpFadX). 32P-labeled probes derived from the MomoFadX or ImpFadX cDNAs were hybridized to 10 μg of total RNA from leaves (L) and developing seeds (S) of M. charantia or from leaves (L) and seed pods (that contain developing seeds) (S) of I. balsamina as indicated in A. The ethidium bromide stained gel corresponding to the Northern blot is shown in B.
Figure 3.
Comparison of amino acid sequences of diverged forms of the Fad2 from Impatiens (ImpFadX) and Momordica (MomoFadX) with known Fad2 and Fad2-related polypeptides. The alignment includes the amino acid sequences of the Arabidopsis Fad2 (AraDes), the castor oleic acid hydroxylase (CastorOH), and the Crepis epoxygenase (CrepEpox) and acetylenase (CrepAcet). Colons indicate residues that are identical to those found in the ImpFadX sequence. Gaps in alignments are maintained by dashes (−).
In addition to cDNAs for MomoFadX and ImpFadX, cDNAs encoding less diverged forms of Fad2 were detected in both the Impatiens and Momordica libraries by random sequencing or by PCR using degenerate Fad2 oligonucleotides (data not shown). The polypeptides corresponding to these cDNAs shared >70% amino acid sequence identity with Arabidopsis Fad2 and other Δ12-oleic acid desaturases. Given their closer relation to the Arabidopsis Fad2, we hypothesized that these enzymes, unlike MomoFadX and ImpFadX, function as Δ12 desaturases in the synthesis of linoleic acid.
Expression of Diverged Fad2s in S. cerevisiae.
As a preliminary analysis of the function of diverged Fad2s from Momordica and Impatiens, a full-length cDNA for ImpFadX was expressed in S. cerevisiae behind a GAL1 promoter. Cultures were supplemented with linoleic acid (18:2Δ9cis,12cis) or α-linolenic acid (18:3Δ9cis,12cis,15cis), which were considered possible substrates for ImpFadX. In extracts of cells grown in media containing linoleic acid, α-eleostearic acid was detected in amounts of up to 0.6% (wt/wt) of the total fatty acids (results not shown). No α-parinaric acid was detected in extracts of these cultures. Conversely, in extracts of cultures grown in media containing α-linolenic acid, α-parinaric acid accumulated to amounts of up to 0.9% (wt/wt) of the total fatty acids, but no α-eleostearic acid was detected. The identities of these unusual fatty acids in S. cerevisiae cultures were established by the mass spectra and gas chromatographic retention times of their methyl esters, both of which were identical to those of methyl α-eleostearic acid or methyl-α-parinaric acid from seed extracts of Impatiens balsamina. No α-eleostearic acid or α-parinaric acid was found in S. cerevisiae cultures lacking exogenous fatty acids or in cells harboring the expression vector without the ImpFadX cDNA insert. Of note, similar experiments conducted with the MomoFadX failed to detect any accumulation of α-eleostearic acid or α-parinaric acid in S. cerevisiae cells. The lack of accumulation of unusual fatty acids with this construct was not pursued further. Overall, the initial result with ImpFadX indicated that diverged forms of Fad2 can catalyze the formation of conjugated double bonds. This was examined further with both MomoFadX and ImpFadX using transgenic soybean embryos, as described below.
Expression of a Momordica Fad2-Related cDNA in Somatic Soybean Embryos.
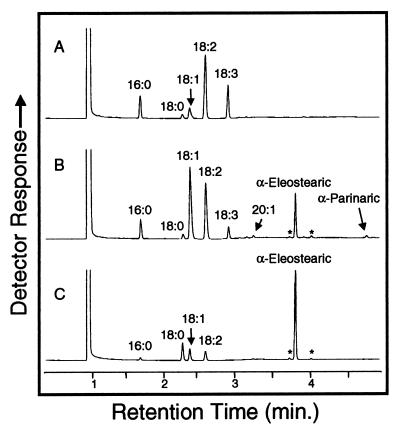
To determine the function of MomoFadX, a cDNA encompassing the complete open-reading frame for this polypeptide was expressed in somatic soybean embryos. Like seeds, somatic soybean embryos are rich in triacylglycerols, and the fatty acid composition of somatic embryos is completely predictive of the fatty acid composition of seeds from plants derived from those embryos (23). Expression of the MomoFadX cDNA in embryos was driven by the strong seed-specific promoter of the gene for the α′-subunit of β-conglycinin (13). Analyses of single transgenic embryos revealed the presence of several fatty acids that were absent from untransformed embryos (Fig. 4). The most abundant of these fatty acids was identified as α-eleostearic acid on the basis of the mass spectrum and gas chromatographic retention time of its methyl ester derivative [supplemental Figs. 5 and 6 (see www.pnas.org)]. This fatty acid accounted for as much as 16% (wt/wt) of the total fatty acids of single soybean embryos. In addition, the transgenic embryos contained small amounts of two fatty acids [≤1% (wt/wt) of the total] that were tentatively identified as cis-trans isomers of α-eleostearic acid. The methyl esters of these fatty acids had similar mass spectra but slightly different retention times than methyl α-eleostearic acid (Fig. 4). These fatty acids were also detected in similar amounts in Momordica seed extracts (Fig. 4). In addition to α-eleostearic acid and its isomers, soybean embryos expressing the MomoFadX cDNA accumulated small amounts of α-parinaric acid (Fig. 4; Table 1). Collectively, fatty acids with conjugated double bonds (eleostearic + parinaric acid isomers) accounted for as much as 18% (wt/wt) of the total fatty acids of single transgenic embryos. Overall, these results provide conclusive evidence for the involvement of MomoFadX in the formation of conjugated double bonds.
Figure 4.
Gas chromatographic analyses of fatty acid methyl esters prepared from an untransformed somatic soybean embryo (A), a transgenic somatic soybean embryo expressing MomoFadX (B), and seeds of M. charantia (C). The asterisks (*) indicate fatty acid methyl esters tentatively identified as cis-trans isomers of methyl α-eleostearic acid. The labeled peaks represent methyl esters of the following fatty acids: 16:0, palmitic acid; 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid; 18:3, α-linolenic acid; and 20:1, eicosenoic acid.
Table 1.
Fatty acid composition of somatic soybean embryos of untransformed lines and transgenic lines expressing cDNAs for MomoFadX and ImpFadX
| Fatty acid | Untransformed (n = 3) | % (wt/wt)
|
|||||
|---|---|---|---|---|---|---|---|
| MomoFadX | ImpFadX | ||||||
| line number
|
line number
|
||||||
| 1* | 2 | 3 | 1 | 2 | 3 | ||
| 16:0 | 15.1 ± 0.3 | 6.9 | 7.9 | 8.4 | 13.3 | 13.2 | 13.5 |
| 18:0 | 3.1 ± 0.2 | 2.4 | 2.1 | 2.3 | 4.2 | 2.5 | 3.1 |
| 18:1 | 9.7 ± 0.5 | 43.1 | 36.8 | 41.9 | 20.8 | 12.2 | 11.1 |
| 18:2 | 48.7 ± 1.6 | 24.1 | 29.4 | 25.1 | 46.3 | 53.8 | 53.0 |
| 18:3 | 22.8 ± 2.0 | 3.1 | 5.1 | 6.1 | 9.2 | 12.5 | 17.3 |
| Eleostearic† | ND | 17.4 | 15.9 | 12.4 | 3.0 | 2.8 | 0.6 |
| α-Parinaric | ND | 1.0 | 1.0 | 1.8 | 2.2 | 2.0 | 1.0 |
| Other§ | <1 | <2 | <2 | <2 | <1 | <1 | <1 |
Analyses were conducted on single embryos from three independent transgenic lines. The fatty acid composition of untransformed samples was obtained from three separate measurements (±SD) of single embryos. Unless indicated, embryos were derived from cultivar A2872. ND, not detected.
Embryos were derived from cultivar Jack.
In the case of embryos expressing MomoFadX, values include tentatively identified cis-trans isomers of α-eleostearic acid, which account for <1% (w/w) of the total fatty acids (see Fig. 4). The remainder is composed of α-eleostearic acid. In the case of embryos expressing ImpFadX, measurements represent amounts of α-eleostearic acid.
Includes primarily 20:0, 20:1, 22:0, and 22:1.
The accumulation of fatty acids with conjugated double bonds was accompanied by large changes in amounts of other fatty acids in transgenic soybean embryos. The most significant of these changes was observed in the content of oleic acid (18:1). In untransformed soybean embryos, oleic acid composed approximately 10% (wt/wt) of the total fatty acids. In contrast, fatty acid extracts from single transgenic embryos contained >40% (wt/wt) oleic acid (Table 1). In addition, amounts of linoleic acid (18:2) decreased from ≈50% (wt/wt) in untransformed embryos to as little as ≈25% (wt/wt) in embryos expressing MomoFadX (Table 1). An approximately 4-fold decrease was also observed in amounts of α-linolenic acid (18:3) in embryos expressing MomoFadX relative to untransformed embryos (Table 1). Furthermore, amounts of palmitic acid (16:0) were also observed to decrease from 15% (wt/wt) in untransformed embryos to less than 7% (wt/wt) in embryos expressing MomoFadX (Table 1). Small increases in amounts of minor fatty acids, particularly 20:1, were also observed in transgenic embryos (Fig. 4).
Expression of an Impatiens Fad2-Related cDNA in Somatic Soybean Embryos.
A full-length cDNA for ImpFadX was expressed in transgenic soybean embryos. As with the experiments described above, expression of the ImpFadX cDNA was driven by the seed-specific promoter of the gene encoding the α′ subunit of β-conglycinin. Accompanying expression of ImpFadX was the accumulation of α-eleostearic acid and α-parinaric acid to amounts of up to 3% (wt/wt) and 2% (wt/wt), respectively, of the total fatty acids of single embryos (Table 1). This finding confirms results obtained with the expression of ImpFadX in S. cerevisiae and further demonstrates that Fad2-related enzymes can catalyze the formation of conjugated double bonds.
The accumulation of α-eleostearic and α-parinaric acids in embryos expressing ImpFadX did not have as large an effect on the overall fatty acid profile as that observed in embryos expressing MomoFadX. In the most extreme cases, oleic acid content increased from 10% (wt/wt) found in untransformed embryos to about 20% (wt/wt) in embryos accumulating α-eleostearic and α-parinaric acid acids (Table 1). In addition, amounts of α-linolenic acid decreased from 20% (wt/wt) found in the total fatty acids of untransformed embryos to as little as 9% (wt/wt) in embryos expressing ImpFadX.
Discussion
The biosynthetic origin of conjugated double bonds in plants has been previously examined by radiolabeling seeds of various species (7, 8, 18). Results from these studies have provided useful information regarding the identity of substrates for conjugated double bond synthesis, but have not revealed the underlying enzymatic nature of this pathway. As an alternative to more traditional biochemical approaches, we used an EST strategy to further characterize the biosynthesis of conjugated double bonds. From ESTs of seeds of two unrelated species (M. charantia and I. balsamina) that accumulate large amounts of fatty acids with conjugated double bonds, we have identified cDNAs for diverged forms of the microsomal Fad2. Expression of these cDNAs in transgenic soybean embryos resulted in the accumulation of α-eleostearic and α-parinaric acids, which contain three and four double bonds in conjugation, respectively. Neither fatty acid was detected in untransformed soybean embryos. These results provide conclusive evidence for the involvement of Fad2-related enzymes in the synthesis of conjugated double bonds in plant fatty acids. This finding also extends a growing list of functions assigned to members of the Fad2 class of enzymes. In addition to conjugated double bond synthesis, these functionalities include fatty acid desaturation, hydroxylation, and epoxidation (19). Given that the activity catalyzed by the MomoFadX and ImpFadX polypeptides has not been previously reported, we suggest the term “conjugase” to describe the function of these enzymes. (This name is not meant to imply any mechanism of action or specific properties of the proteins encoded by these diverged Fad2 genes, simply that they are responsible for the biosynthesis of conjugated double bonds.) Additional research is needed to determine whether Fad2-type conjugase activity also accounts for the conjugated double bonds of other plant fatty acids such as punicic (18:3Δ9cis,11trans,13cis) and catalpic (18:3Δ9trans,11trans,13cis) acids (1).
On the basis of previous reports as well as our results, the conjugated double bonds found in α-eleostearic and α-parinaric acids of Momordica and Impatiens seeds result from the conversion of the cis-Δ12 (or ω6) double bond of linoleic and α-linolenic acids into trans-Δ11 and Δ13 double bonds. For example, Liu et al. (7) have shown that radiolabeled linoleic acid fed exogenously to Momordica seed slices is converted into α-eleostearic acid. In addition, we observed that S. cerevisiae cultures expressing ImpFadX are able to synthesize α-parinaric acid when supplied with α-linolenic acid, but do not accumulate α-eleostearic acid. However, when provided with exogenous linoleic acid, these cells produce α-eleostearic acid but do not accumulate detectable amounts of α-parinaric acid. Taken together, these findings are consistent with linoleic acid serving as the substrate for α-eleostearic acid synthesis and α-linolenic acid serving as the substrate for α-parinaric acid synthesis. Fatty acids are presumably presented as substrates to the Impatiens and Momordica conjugases while bound to phosphatidylcholine, as suggested by the radiolabeling studies described by Liu et al. (7). The results presented here, however, cannot completely exclude the possibility that some portion of the α-parinaric acid accumulated in the transgenic soybean embryos arises from the activity of the Δ15–linoleic acid desaturase (Fad3) on α-eleostearic acid.
One unexpected finding was the presence of small amounts of α-parinaric acid in soybean embryos expressing MomoFadX given that this fatty acid is not detectable in seeds of M. charantia (Fig. 4C). Unlike soybean embryos, though, Momordica seeds do not accumulate α-linolenic acid (Fig. 4C) and therefore lack substrate for the direct conversion of this fatty acid into α-parinaric acid by MomoFadX. Momordica seeds may also lack sufficient Fad3 activity for any potential synthesis of α-parinaric acid from α-eleostearic acid. In contrast to Momordica seeds, Impatiens seeds are enriched in α-parinaric acid (>30% of the total fatty acid) and contain lesser amounts of α-eleostearic acid (≈2% of the total fatty acid) [supplemental Fig. 5; see www.pnas.org)]. These seeds also contain large amounts of α-linolenic acid (30 to 50% of the total fatty acids). Therefore, Impatiens seeds, in contrast to Momordica seeds, likely contain significant substrate pools of α-linolenic acid for conversion to α-parinaric acid. Results from transgenic soybean embryos also suggest that ImpFadX may have a greater ability to synthesize α-parinaric acid from α-linolenic acid than MomoFadX. In this regard, the ratio of eleostearic acid to α-parinaric acid accumulated was as high as 17:1 (Table 1) in soybean embryos expressing MomoFadX. In contrast, the ratio of these two fatty acids ranged from approximately 0.6:1 to 3:2 in soybean embryos expressing ImpFadX (Table 1).
The demonstration that Fad2-type enzymes are involved in the formation of conjugated double bonds adds further complexity to the functional properties of this class of enzymes and their catalytic diiron-oxo centers (19). A notable feature of the Impatiens conjugase relative to other Fad2-related enzymes is its demonstrated ability to use either linoleic acid or α-linolenic acid as substrates. In contrast, Fad2-type desaturases and hydroxylases function on oleic acid, whereas Fad2-type epoxygenases and acetylenases use linoleic acid as their substrate (as reviewed in ref. 19). The ability of the Impatiens conjugase to function on either linoleic acid or α-linolenic acid thus indicates an additional degree of plasticity with regard to the substrate specificity of Fad2-related enzymes.
An interesting observation from this study was the large alteration in the fatty acid composition of soybean embryos that accompanied the expression of MomoFadX. The most significant change was a more than 4-fold increase in the relative amount of oleic acid in these embryos compared with untransformed embryos (Table 1; Fig. 4). A similar but less dramatic effect on fatty acid composition was also reported with the transgenic expression of other Fad2-related enzymes, including the castor and Lesquerella oleic acid hydroxylases (24, 25). The increased oleic acid content of embryos expressing MomoFadX was accompanied by changes in relative amounts of other fatty acids, including a decrease in palmitic acid content. Reduced palmitic acid levels have also been shown to occur along with elevated oleic acid content in soybean seeds with suppressed Fad2 activity (23) and in certain tissues of Arabidopsis fad2 mutants (20).
Finally, the finding that soybean embryos accumulate α-eleostearic and α-parinaric acids on expression of MomoFadX or ImpFadX further demonstrates the ability to produce unusual fatty acids with industrial value in transgenic plants. Vegetable oils that are enriched in α-eleostearic acid are used commercially in coating materials and are obtained primarily from seeds of the tung tree, whose cultivation is limited to subtropical climates. These seed oils have drying properties that are superior to those that contain polyunsaturated fatty acids with methylene-interrupted double bonds (5). The production of α-eleostearic acid in an existing crop species such as soybean via the transgenic expression of MomoFadX or ImpFadX would thus expand the industrial value of the resulting seed oil and open up new markets for its use. However, a more complete understanding of the synthesis and metabolism of α-eleostearic acid will undoubtedly be required to increase the accumulation of this fatty acid in a transgenic crop to levels found in tung oil (>65% of the total fatty acids). Also, generation of transgenic plants from the somatic soybean embryos will be necessary to assess the effect of α-eleostearic acid and α-parinaric acid accumulation on the agronomic properties of the resulting plants.
Supplementary Material
Acknowledgments
We thank Maureen Dolan and the EST group of DuPont Genomics for sequencing of cDNA libraries.
Abbreviations
- EST
expressed sequence tag
- Fad2
Δ12-oleic acid desaturase
Footnotes
References
- 1.Smith C R., Jr . In: Progress in the Chemistry of Fats and Other Lipids. Holman R T, editor. Vol. 11. London: Pergammon; 1970. pp. 137–177. [Google Scholar]
- 2.Conacher H B S, Gunstone F D, Hornby G M, Padley F B. Lipids. 1970;5:434–441. [Google Scholar]
- 3.Gunstone F D, Subbarao R. Chem Phys Lipids. 1967;1:349–359. [Google Scholar]
- 4.Bagby M O, Smith C R, Jr, Wolff I A. Lipids. 1966;1:263–267. doi: 10.1007/BF02531613. [DOI] [PubMed] [Google Scholar]
- 5.Formo M W. In: Bailey’s Industrial Oil and Fat Products. Swern D, editor. New York: Wiley; 1979. pp. 708–709. [Google Scholar]
- 6.Sonntag N O V. In: Bailey’s Industrial Oil and Fat Products. Swern D, editor. New York: Wiley; 1979. p. 440. [Google Scholar]
- 7.Liu L, Hammond E G, Nikolau B J. Plant Physiol. 1997;113:1343–1349. doi: 10.1104/pp.113.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crombie L, Holloway S J. J Chem Soc Perkin Trans I. 1985;1985:2425–2434. [Google Scholar]
- 9.Gunstone F D. Chem Ind (London) 1965;1965:1033–1034. [PubMed] [Google Scholar]
- 10.Jones A, Davies H M, Voelker T A. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1987. [Google Scholar]
- 13.Beachy R N, Chen Z-L, Horsch R B, Rogers S G, Hoffman N J, Fraley R T. EMBO J. 1985;4:3047–3053. doi: 10.1002/j.1460-2075.1985.tb04044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle J J, Schuler M A, Godette W D, Zenger V, Beachy R N, Slightom J L. J Biol Chem. 1986;261:9228–9238. [PubMed] [Google Scholar]
- 15.Gritz L, Davies J. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 16.Finer J J, McMullen M D. In Vitro Cell Dev Biol. 1991;27:175–182. [Google Scholar]
- 17.Sambrook J, Fristch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Crombie L, Holloway S J. J Chem Soc, Chem Commun. 1984;15:953–955. [Google Scholar]
- 19.Shanklin J, Cahoon E B. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 20.Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van de Loo F J, Broun P, Turner S, Somerville C. Proc Natl Acad Sci USA. 1995;92:6743–6747. doi: 10.1073/pnas.92.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, Lenman M, Banas A, Bafor M, Singh S, Schweizer M, Nilsson R, Liljenberg C, Dahlqvist A, Gummeson P O, et al. Science. 1998;280:915–918. doi: 10.1126/science.280.5365.915. [DOI] [PubMed] [Google Scholar]
- 23.Kinney A J. J Food Lipids. 1996;3:273–292. [Google Scholar]
- 24.Broun P, Somerville C. Plant Physiol. 1997;113:933–942. doi: 10.1104/pp.113.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broun P, Boddupalli S, Somerville C. Plant J. 1998;13:201–210. doi: 10.1046/j.1365-313x.1998.00023.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.