Abstract
Vascular endothelium is an important transducer and integrator of both humoral and biomechanical stimuli within the cardiovascular system. Utilizing a differential display approach, we have identified two genes, Smad6 and Smad7, encoding members of the MAD-related family of molecules, selectively induced in cultured human vascular endothelial cells by steady laminar shear stress, a physiologic fluid mechanical stimulus. MAD-related proteins are a recently identified family of intracellular proteins that are thought to be essential components in the signaling pathways of the serine/threonine kinase receptors of the transforming growth factor β superfamily. Smad6 and Smad7 possess unique structural features (compared with previously described MADs), and they can physically interact with each other, and, in the case of Smad6, with other known human MAD species, in endothelial cells. Transient expression of Smad6 or Smad7 in vascular endothelial cells inhibits the activation of a transfected reporter gene in response to both TGF-β and fluid mechanical stimulation. Both Smad6 and Smad7 exhibit a selective pattern of expression in human vascular endothelium in vivo as detected by immunohistochemistry and in situ hybridization. Thus, Smad6 and Smad7 constitute a novel class of MAD-related proteins, termed vascular MADs, that are induced by fluid mechanical forces and can modulate gene expression in response to both humoral and biomechanical stimulation in vascular endothelium.
The entire circulatory system is lined by a continuous single-cell-thick lining consisting of vascular endothelium. This multifunctional tissue is responsive to a vast array of biologically important signals ranging from vasoactive substances derived locally within the vascular wall to hormones, cytokines, and other effectors derived from both local and distant sources. Indeed, the ability of vascular endothelium to act as a mediator and transducer of a broad range of biological effectors is now appreciated as a fundamental property of these cells, and the disruption of these processes is a critical factor in the pathogenesis of vascular disease (1).
As a function of its unique anatomical position, endothelium is constantly exposed to a variety of fluid mechanical forces. One of these, the fluid shear stresses generated at the surface of endothelial cells by the flow of viscous blood, is capable of inducing important phenotypic alterations in endothelial cells, many of which involve changes in gene expression. Indeed, it is now clear that endothelial cells (and possibly other cells such as vascular smooth muscle) can sense and respond to their local hemodynamic environment, and that the resulting phenotypic modulation may be mechanistically important in the pathogenesis of vascular diseases (e.g., the initiation and localization of the early lesions of atherosclerosis) (1, 2).
The transforming growth factor β (TGF-β) family of ligands, receptors, and signal-transducing molecules represents a large complex collection of proteins that mediate many biological effects, ranging from the regulation of cellular proliferation, differentiation, and migration to the elaboration of extracellular matrix and other bioactive substances in many cell types. The diversity of this system is underscored by the multiple TGF-β-like ligands present in animals [e.g., TGF-β isoforms, bone morphogenic protein (BMP) isoforms, and activins] as well as the complex receptor system involving multiple distinct proteins belonging to at least three classes of receptor types. Recently, the events downstream of receptor activation have begun to be elucidated by the identification of a novel class of molecules known as MAD proteins (for mothers against decapentaplegic), which act as second messengers distal to the TGF-β family of receptors. These proteins, originally defined in Drosophila as a component of the decapentaplegic (DPP) signaling pathway, are now known to subserve an analogous role in humans (and other vertebrates) and are likely to be key mediators of signals derived from TGF-β-like molecules in many tissues (3–9).
Utilizing a differential display approach in cultured endothelial cells subjected to multiple soluble and biomechanical stimuli, we have identified two members of the MAD family of proteins, which we have named Smad6 and Smad7. These two species appear to be unique among mammalian MAD-related species in that they are selectively and specifically induced in cultured vascular endothelium by a physiologic fluid mechanical stimulus in vitro, and they appear to manifest an endothelial selective pattern of expression in vivo.
METHODS
Endothelial Cell Culture.
Human umbilical vein endothelial cells (HUVEC) were isolated from multiple segments of normal term umbilical cords, pooled, and cultured in medium 199 supplemented with endothelial cell growth supplement (50 μg/ml; Collaborative Research), heparin (50 μg/ml, porcine intestinal; Sigma), antibiotics (penicillin-G at 100 units/ml, streptomycin at 100 μg/ml; Sigma), and 20% fetal bovine serum. Cells at passage level 2 or 3 were replica-plated on 0.1% gelatin-coated standard Petri dishes or specially designed plates fabricated from the same tissue culture plastic (Costar) and allowed to grow to confluent densities before experimental use.
Shear Stress Apparatus.
The equations and calculations for describing the shear stresses generated in the cone-plate apparatus, and the differential display protocol, have been reported in detail previously (2). Briefly, confluent HUVEC monolayers grown on 17.8-cm-diameter “maxiplates” (approximately 107 cells per plate) were introduced into a cone-plate flow cuvette, consisting of a stainless steel cone rotating over a stationary baseplate. The culture medium present between the cone and the plate was constantly replenished at the rate of 0.5 ml/min during experiments, and the entire apparatus was maintained in a humidified 5% C02/95% air atmosphere. For laminar shear stress (LSS) at 10 dyn/cm2 [1 dyne (dyn) = 10 μN], we utilized a 0.5° cone at a rotational velocity of 100 rpm; for turbulent shear stress we utilized a 3° cone, at 130 rpm, and a minimum radius of 3.5 cm as described previously (2).
Nuclear Runoff Analysis.
Four maxiplates (approximately 107 cells per plate) were simultaneously prepared as above. One served as a control, one was treated with recombinant human interleukin 1β (rhIL-1β) at 20 units/ml for 3 hr, one was subjected to 3 hr of steady LSS at 10 dyn/cm2 and the other to turbulent shear stress (TSS) at the same time-averaged magnitude as above. The runoff assay was performed as previously described (2).
Vectors, Transfections, and Immunoprecipitations.
Epitope-tagged [myc, hemagglutinin (HA)] expression vectors for wild-type Smad6 and Smad7 were prepared in cytomegalovirus (CMV)-based vectors by standard methods. Expression vectors for human Smad1 and Smad2 fused to the FLAG epitope, and Smad4-HA, were kindly provided by J. Wrana (Hospital for Sick Children, Toronto). The mutant forms (Smad6* and Smad7*) were synthesized by site-directed mutagenesis and are described in Fig. 1. Bovine aortic endothelial cells (BAEC) or HUVEC were transfected by Lipofectamine (GIBCO/BRL) or standard calcium phosphate protocols. Antibodies directed against the epitopes used in the immunoprecipitations and Western blots were obtained from commercial suppliers (Boehringer Mannheim, Santa Cruz Biotechnology).
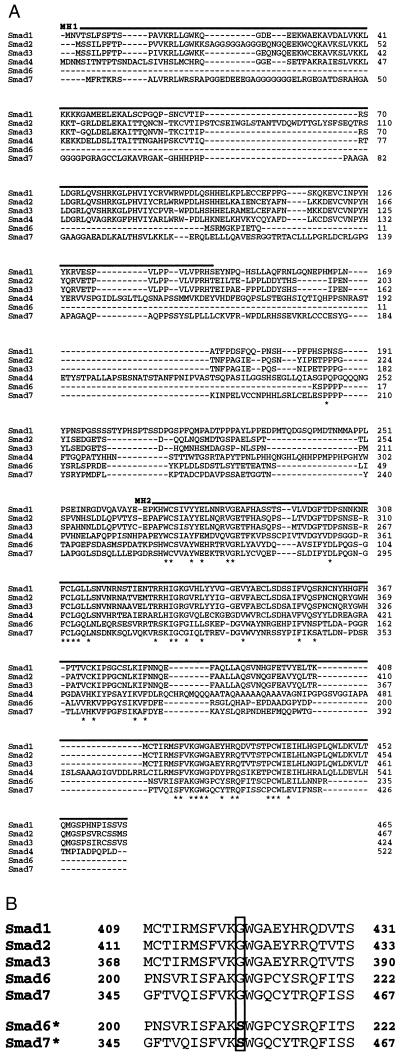
Figure 1.
Sequence alignment of Smad6 and Smad7. (A) The predicted amino acid sequences of Smad1, Smad2, Smad3, Smad4, Smad6, and Smad7 (based on cDNA sequences) are aligned, with residues conserved among all members marked with asterisks, and putative MH1 and MH2 domains with solid overlines. Gaps introduced to enhance alignment are shown as dashes. (B) Mutant forms of Smad6 and Smad7 (Smad6* and Smad7*, respectively) were made by changing a glycine to a serine at position 170 in Smad6 and at position 361 in Smad7 (GGC to AGC codon change in both cases).
Approximately 12 hr after transfection the cells were plated onto duplicate coverslips, allowed to adhere overnight, and then treated with human TGF-β1 (Genzyme, 10 ng/ml) for 18 hr, LSS at 10 dyn/cm2 for 18 hr, or static (no flow) conditions, and luciferase activity was measured.
In Situ Hybridization and Immunohistochemistry.
Human tissue samples were collected at the time of surgery or autopsy, according to established institutional protocols. Sections (4–5 μm) were prepared from formalin-fixed paraffin-embedded tissues for in situ analysis with antisense riboprobe for Smad6 and Smad7, or frozen sections (4–5 μm) were utilized for immunostaining with an affinity-purified rabbit polyclonal antiserum generated against Smad6 expressed as a glutathione S-transferase (GST) fusion protein. As a control for the specificity of this antiserum, excess purified Smad6-GST fusion protein was preincubated with the antiserum overnight prior to its use for tissue staining. The monoclonal antibody to human platelet–endothelial adhesion molecule 1 (PECAM-1) (CD-31) was obtained from Dako. Immunostaining was detected with a secondary antibody conjugated to horseradish peroxidase, using 3-amino-9-ethylcarbazole as substrate, and the slides were counterstained with hematoxylin.
RESULTS AND DISCUSSION
In cultured HUVEC exposed to a physiologic level (10 dyn/cm2) of steady LSS, a nonlaminar fluid mechanical stimulus, TSS of a comparable time-averaged magnitude, or rhIL-1β (10 units/ml), several mRNA transcripts were detected by differential display that manifested a “laminar-selective upregulation” pattern (i.e., up-regulation in response to the steady LSS but not the TSS or cytokine stimuli) (2). Two of these were isolated and used to probe cDNA libraries prepared from shear stress-stimulated endothelial cells. As schematized in Fig. 1, two cDNAs with extensive homology to members of the MAD family (termed Smad6 and Smad7, respectively) were isolated. MAD proteins, which were originally defined in Drosophila, are essential components of the signaling pathway of the TGF-β superfamily of receptors and ligands (3–9). Smad6, whose carboxyl-terminal sequence has been previously reported as an expressed sequence tag (8), encodes a novel MAD-related protein of approximately 35 kDa that exhibits significant homology to other human MAD-related species (Fig. 1). Although the primary stuctures of Smad proteins do not contain significant homology to other proteins in the available database, all Smad molecules identified to date contain highly conserved amino- and carboxyl-terminal domains termed MH1 and MH2, respectively (9). Interestingly, Smad6 is unique in that it appears to lack a putative MH1 domain, a characteristic that may be important in the regulation of its signaling functions. Smad7 is an additional MAD-related protein which most closely resembles Smad4 (DPC4), a MAD species thought to function as a tumor suppressor (10–12). Interestingly, both Smad6 and Smad7 (like Smad4) lack certain carboxyl-terminal serine residues that are conserved among other members of this class of molecules (e.g., Smad2), which function as sites of receptor-mediated phosphorylation and have been demonstrated to be critical for TGF-β/Smad2 signaling in vitro (10, 13). Despite these unique structural features, the overall homology between Smad6 and Smad7 and the other known human MAD-related proteins is strong (Fig. 1A). In addition, somatic cell hybrid analysis has revealed that both the Smad6 (chromosome 15) and Smad7 (chromosome 18) genes are located in regions of the human genome that contain other MAD gene homologues (data not shown) and which have been implicated in the pathogenesis of several human malignancies (8, 14, 15).
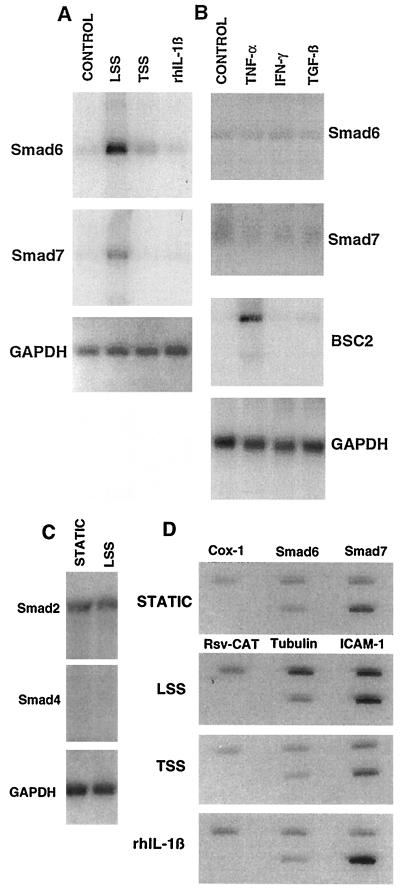
Fig. 2A demonstrates that the mRNAs encoding these two MAD isoforms are selectively up-regulated by the steady LSS stimulus, but not the TSS stimulus or the cytokine stimulus rhIL-1β, in HUVEC. Fig. 2B demonstrates that in addition to rhIL-1β, the cytokines tumor necrosis factor α (TNF-α), interferon-γ (IFN-γ), and active TGF-β1 (each at maximally effective concentrations) had no significant effect on the low basal levels of Smad6 and Smad7 mRNA present in HUVEC cultured under static (no-flow) conditions. Thus, the Smad6 and Smad7 genes in endothelium appear to be selectively responsive to a LSS stimulus, manifesting no response to a nonlaminar fluid mechanical stimulus (TSS) or to any of the humoral stimuli tested. Neither Smad2 nor Smad4, two other human MAD genes, is up-regulated by steady LSS in cultured HUVEC (Fig. 2C).
Figure 2.
Smad6 and Smad7 are regulated genes in vascular endothelium. (A) Northern analysis of RNA from cultured confluent HUVEC monolayers exposed to static (no-flow) conditions, 24 hr of steady LSS (10 dyn/cm2), TSS (10 dyn/cm2), or rhIL-1β (10 units/ml; Biogen) and probed with cDNA for Smad6 or Smad7, or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to control for RNA loading. (B) Northern analysis of RNA from cultured HUVEC treated for 24 hr with tumor necrosis factor α (TNF-α, 200 units/ml; Biogen), interferon-γ (IFN-γ, 500 units/ml; Genzyme), or TGF-β1 (10 ng/ml; Genzyme). All cytokines were of human origin. Bumetanide-sensitive cotransporter 2 (BSC2) is an ion cotransporter that is transcriptionally up-regulated by TNF-α in HUVEC (23). (C) Northern analysis of Smad2 and Smad4 RNA in HUVEC demonstrating that their levels are not significantly altered by 24 hr of steady LSS. (D) Nuclear runoff analysis of HUVEC exposed to static conditions (control), LSS, TSS, or rhIL-1β for 3 hr. The genes encoding Smad6 and Smad7 demon strate a significant increase in their rates of transcription only in response to the LSS stimulus. The cyclooxygenase 1 (Cox-1) and tubulin genes do not demonstrate significant change, and transcription of the intercellular adhesion molecule 1 (ICAM-1) gene is up-regulated by both the LSS and rhIL-1β stimuli as previously reported (2). Rsv-CAT is a negative control plasmid containing no endothelial genes.
Fluid mechanical forces have been shown to regulate the expression of other endothelial genes, a process involving modulation of transcription by means of specific cis-acting promoter elements (2, 16, 17). Fig. 2D, a nuclear runoff analysis of HUVEC stimulated with LSS, TSS, or rhIL-1β, demonstrates that the selective induction of Smad6 and Smad7 in response to LSS in vitro is occurring at least in part at the level of transcriptional activation. Taken together, these data demonstrate that these two MAD-related genes are inducible in vascular endothelial cells, a characteristic of this class of molecules that has not previously been described, and suggest that biomechanical stimuli generated by blood flow may play an important role in the regulation of their expression in vivo. These data also suggest that the regulated expression of relevant intracellular signal transduction proteins in response to physiologic hemodynamic stimuli may be an important mechanism by which endothelial cells integrate diverse humoral and biomechanical stimuli within the cardiovascular system.
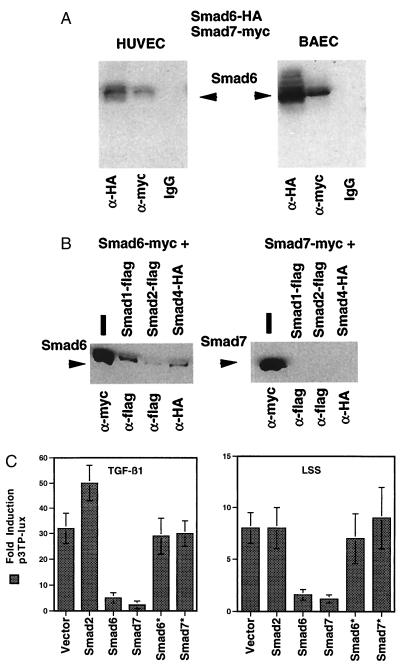
To investigate Smad6 and Smad7 function, expression vectors encoding wild-type Smad6 and Smad7 fused to epitope tags were prepared and transfected into endothelial cells. Fig. 3A demonstrates coimmunoprecipitation of transfected Smad6 and Smad7, indicating that these two endothelial-derived proteins are able to form a stable complex within both human and bovine endothelial cells. Physical interaction of these two MAD-related species was confirmed in a yeast two-hybrid system (Table 1). Smad6 and Smad7 were found to selectively bind each other but not four unrelated control proteins. The Smad6 GAL4 DNA-binding domain (BD) fusion protein bound only to the Smad7 GAL4 transcriptional activation domain (AD) fusion protein, and reciprocally the Smad7 GAL4 BD fusion protein bound only to the Smad6 GAL4 AD fusion protein. Interestingly, when Smad6 and Smad7 were tested by coimmunoprecipitation for their ability to interact with other human MAD-related proteins in endothelial cells, Smad6 interacted with all species tested (Smad1, Smad2, and Smad4), whereas Smad7 interacted only with Smad6 (Fig. 3B).
Figure 3.
Smad6 and Smad7 form complexes in endothelial cells. (A) CMV-driven expression vectors for Smad6 fused to an HA epitope and Smad7 fused to a myc epitope were cotransfected into HUVEC or BAEC, and approximately 16 hr later the monolayers were harvested and immunoprecipitation with anti-HA or anti-myc antibodies or mouse IgG was performed. The precipitated protein was then separated on denaturing SDS gels and probed with anti-HA antibodies to detect precipitated Smad6-HA. As expected, the anti-HA antibody precipitated Smad6-HA; however, immunoprecipitation of Smad7 with anti-myc also precipitated significant Smad6, indicating that Smad7-myc and Smad6-HA were able to form stable complexes in these cells. (B) Cotransfection and immunoprecipation experiments were performed as above to investigate that ability of Smad6 and Smad7 to interact with other human MAD-related proteins. Immunoprecipitation of cotransfected, epitope-tagged Smad1, Smad2, and Smad4 precipitated Smad6, but not Smad7. The expression vectors transfected are listed above the Western blots, and the antibodies used to immunoprecipitate are below. (C) Smad6 and Smad7 can modulate endothelial gene expression. BAEC were cotransfected with p3TP-lux and expression vectors containing no cDNA (some experiments utilized CMV-CAT as controls) or encoding wild-type Smad2, Smad6, Smad7, or mutant forms of these proteins (Smad6*, Smad7*) and stimulated with TGF-β (10 ng/ml, 18 hr), or LSS (10 dyn/cm2, 18 hr). Fold-induction was calculated compared with static controls, and transfection efficiency was normalized by measuring cotransfected β-galactosidase activity (pCMVβ-Gal). The results shown are means for at least three experiments, and error bars represent ± 1 SD.
Table 1.
Smad6 and Smad7 interact specifically in a yeast two-hybrid system
| GAL4 BD construct | GAL4 AD construct | Interaction |
|---|---|---|
| pGBT9-Smad6 | pACTII-Smad7 | + |
| pGBT9-Smad7 | pACTII-Smad6 | + |
| pGBT9-Smad6 | pACTII | − |
| pGBT9-Smad7 | pACTII | − |
| pGBT9 | pACTII-Smad6 | − |
| pGBT9 | pACTII-Smad7 | − |
| pGBT9-Smad6 | pSV40 | − |
| pGBT9-Smad7 | PSV40 | − |
| pIamin C | pACTII-Smad6 | − |
| pIamin C | pACTII-Smad7 | − |
Yeast two-hybrid analysis was performed as previously described (3–7). Briefly, yeast expression vectors producing the GAL4 DNA–binding domain (BD) alone or fusions of the GAL4 BD to Smad6, Smad7, and lamin C and yeast expression vectors producing the GAL4 transcriptional activation domain (AD) only or fusions of the GAL4 AD to Smad6, Smad7, and the simian virus 40 (SV40) large T antigen were cotransformed into two-hybrid reporter yeast strain HF7c in the listed combinations. For each construct combination, three independent cotransformants were subjected to a 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal)-based filter disk β-galactosidase assay, in which yeast strains expressing lacZ develop a blue color, to measure qualitatively the expression of the lacZ reporter gene. The GAL41-147 DNA BD fusion vector was pGBT9 and the GAL4768-881 AD vector was pACTII. pSV40, an SV40 large T antigen84-708 control fish plasmid, and plamin C, a human lamin C67-228 control bait plasmid, were used to confirm specific binding of Smad6 to Smad7.
To determine whether Smad6 and Smad7 can influence endothelial gene expression, we utilized a reporter construct (known as p3TP-lux) derived from the human plasminogen activator 1 promoter that is known to be responsive to TGF-β stimulation (18). As demonstrated in Fig. 3C, when this promoter construct is transfected into cultured BAEC and the cells are then treated with TGF-β1, there is a significant (approximately 30-fold) induction in luciferase activity. Cotransfection of Smad2, a human MAD-related species previously demonstrated to be involved in TGF-β1 signaling, significantly enhances this response, an effect previously documented in other cell types (11, 12). In contrast, cotransfection of either Smad6 or Smad7 resulted in a significant inhibition of the ability of TGF-β1 to activate the p3TP promoter (Fig. 3C). This effect was dose-dependent, with increasing amounts of cotransfected Smad6 or Smad7 expression plasmid resulting in increased inhibition (data not shown). To confirm the specificity of this inhibitory effect, site-specific mutants of both Smad6 and Smad7 (Fig. 1B) were constructed, based on known mutations identified in Drosophila homologues, that would be predicted to disrupt MAD-like signaling functions (3, 6, 19). As shown in Fig. 3C, unlike wild-type Smad6 and Smad7, these mutant proteins were unable to inhibit the activation of the p3TP promoter in response to TGF-β1. The expression levels of mutant and wild-type proteins were comparable (data not shown), indicating that this loss of function was not secondary to protein instabilty. When BAEC transfected with p3TP-lux were subjected to an LSS stimulus at 10 dyn/cm2, the p3TP promoter was significantly induced, although the magnitude of induction was less than that by TGF-β1, and coexpression of Smad2 did not significantly augment this effect. Both Smad6 and Smad7, but not their respective mutant proteins, inhibited the LSS-stimulated induction of p3TP (Fig. 3C). Although LSS has been demonstrated to induce TGF-β in cultured BAEC (20), it is unlikely that an autocrine or paracrine effect is responsible for the shear stress induction of p3TP observed here. The cone-plate flow apparatus utilized in these experiments does not recirculate the culture medium, and conditioned media collected from shear-stressed cells did not up-regulate p3TP activity when applied to static endothelial cells (data not shown).
Together, these results strongly suggest that both Smad6 and Smad7 can function as MAD-related signaling molecules in vascular endothelial cells, and that, in this in vitro overexpression system, they may be acting as dominant-negative effectors. A recent report demonstrated that mutation of the phosphorylation sites present in the carboxyl domain of Smad2 generated a dominant-negative form of this molecule capable of blocking TGF-β-dependent transcriptional responses (13). Smad6 may be functioning as a negative regulator by forming nonproductive heterodimers with other endogenous MAD species, given its ability to interact widely with these proteins (Fig. 3B). Since Smad7 does not appear to interact specifically with MAD species other than Smad6, its inhibitory effect may be mediated by its ability to interact with other factors (i.e., receptors, transcriptional activators) necessary for the induction of the p3TP promoter (21). Indeed, recent data indicate that Smad7 can stably interact with an activated form of the TGF-β type 1 receptor and inhibit its association with and phosphorylation of Smad2 in vitro (22). Thus Smad 7 may function as an endogenous inhibitor of TGF-β signaling, and its flow-mediated expression may be an important mechanism regulating the responsiveness of vascular endothelium to the TGF-β superfamily of growth factors. Furthermore, the ability of Smad6 and Smad7 to inhibit both TGF-β- and LSS-induced p3TP-lux expression in vitro suggest that the signal transduction pathways linking humoral and biomechanical stimulation to gene-regulatory events in endothelial cells share common components (17).
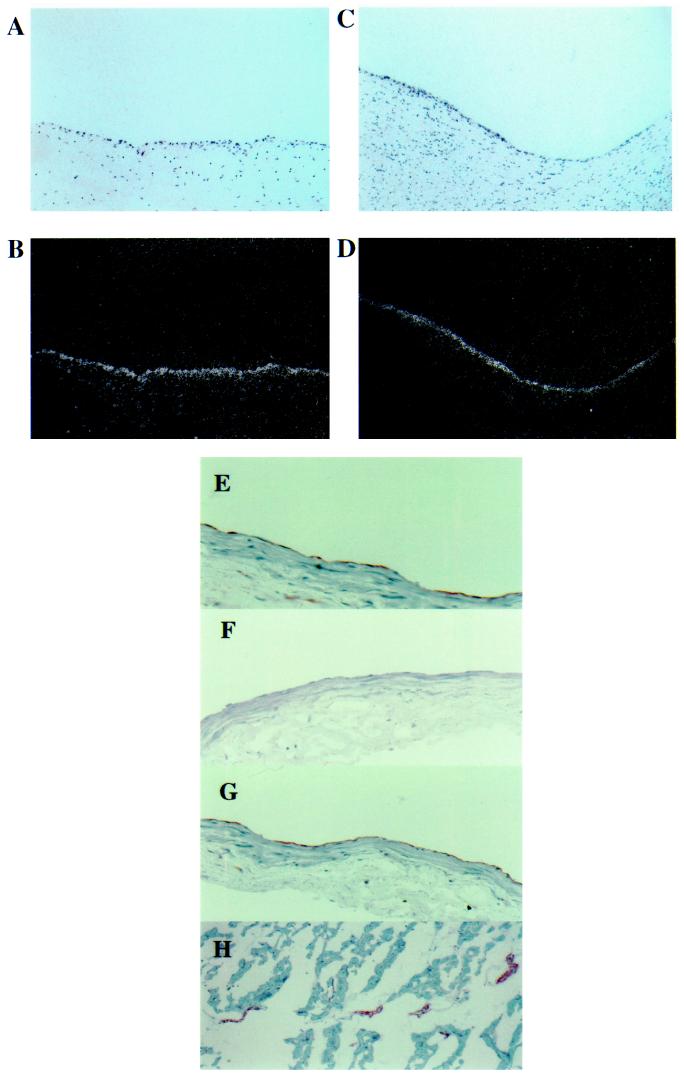
To determine whether Smad6 and Smad7 are expressed in vascular endothelium in vivo, immunohistochemical staining and in situ hybridization studies were performed on human tissues. As demonstrated in Fig. 4, both Smad6 and Smad7 are readily detectable in the endothelial lining of human atherosclerotic carotid artery specimens by in situ hybridization (Fig. 4 A–D). A rabbit polyclonal antiserum generated against Smad6 also prominently and selectively stains the endothelium of large vessels such as the epicardial portions of undiseased human coronary arteries (E), as well as smaller vessels present within human myocardium (H). An antibody to PECAM-1 (CD31), an endothelial-specific molecule, was used as a control marker for endothelium (G), and the ability of recombinant Smad6 fusion protein to block endothelial staining confirmed the specificity of the rabbit polyclonal antiserum (F). Thus Smad6 and Smad7 demonstrate an endothelial-selective pattern of expression and can be detected in both atherosclerotic and normal vessels (Fig. 4 and data not shown). Although the signaling pathways, including particular receptors/ligands operative upstream of Smad6 and Smad7, and the genes regulated by these molecules in vivo remain to be elucidated, this demonstration of cell type-restricted expression of MAD-related isoforms suggests that an additional, previously unappreciated, level of complexity may exist for the TGF-β superfamily of effectors. Regulated expression of individual MAD isoforms in a tissue- or cell type-specific manner may represent an important mechanism contributing to both the specificity and diversity of effects characteristic of this family of molecules.
Figure 4.
Smad6 and Smad7 are expressed in human vascular endothelium in vivo. (A–D) In situ hybridization for Smad6 (B) and Smad7 (D) mRNA in sections of endarterectomy specimens of atherosclerotic human carotid artery with corresponding hematoxylin- and eosin-stained sections (A and C) (×80.) (E–G) Immunohistochemical analysis of normal human coronary artery (left anterior descending, midepicardial segment) stained with anti-Smad6 antiserum (E); anti-Smad6 antiserum preincubated with recombinant Smad6 fusion protein (F); and a monoclonal antibody to PECAM-1 (CD31), an endothelial-specific marker (G). (H) Section of human myocardium stained with anti-Smad6. Note the selective staining of the small arteries and veins present within the myocardial tissue. The extensive capillary network present within this tissue does not demonstrate significant immunostaining for Smad6. (×40.)
In summary, Smad6 and Smad7 appear to define a novel class of MAD proteins. These vascular MADs have the following unique characteristics: (i) In vitro they are induced by a physiologically relevant fluid mechanical stimulus, steady LSS. (ii) They demonstrate a predominantly vascular endothelial-selective pattern of expression in vivo. (iii) They possess unique structural features, the most striking of which is a truncation of their carboxyl domains resulting in a lack of putative phosphorylation sites, suggesting that the regulation of their function(s) may be distinct from other MAD species described to date. (iv) They are capable of modulating endothelial gene expression in response to both humoral (TGF-β) and biomechanical (LSS) stimuli in vitro.
The TGF-β family of growth factors has been implicated in a variety of physiological and developmental processes within the cardiovascular system. Similarly, a growing body of evidence now points to the importance of hemodynamic stimuli generated by blood flow in the regulation of vascular structure and function in vivo. Vascular MADs represent a class of signaling molecules selectively expressed in vascular endothelium that may function in mediating responses to both the TGF-β superfamily of humoral factors and biomechanical forces within the vasculature. Vascular MADs may play a critical role in the ability of endothelium to integrate these diverse stimuli and thus be relevant to a variety of physiologic and pathophysiologic processes in the cardiovascular system (23).
Acknowledgments
We thank K. Case, W. Atkinson, M. DiChiara, S. Wasserman, J. Kiely, T. Nagel, G. Stavrakis, S. Squazzo, K. Harvey, J. Morgenstern, and P. Libby for advice and assistance, and J. Wrana for providing reagents. J.N.T. is a recipient of the Howard Hughes Medical Institute Research Fellowship for Physicians. This work was supported in part by grants from the National Heart, Lung, and Blood Institute (R37-HL51150, and P50-HL56985, to M.A.G.), and a sponsored research agreement to the Brigham and Women’s Hospital from Millennium Pharmaceuticals, in collaboration with Eli Lilly.
ABBREVIATIONS
- BAEC
bovine aortic endothelial cells
- CMV
cytomegalovirus
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HA
hemagglutinin
- HUVEC
human umbilical vein endothelial cells
- LSS
laminar shear stress
- MAD
mothers against decapentaplegic
- PECAM-1
platelet–endothelial adhesion molecule 1
- rhIL-1β
recombinant human interleukin 1β
- TGF-β
transforming growth factor β
- TNF-α
tumor necrosis factor α
- TSS
turbulent shear stress
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. for Smad6, HSV59914; for Smad7, AF010193).
References
- 1.Gimbrone M A., Jr . In: Molecular Cardiovascular Medicine. Haber E, editor. New York: Scientific American Medicine; 1995. pp. 49–61. [Google Scholar]
- 2.Topper J N, Cai J, Falb D, Gimbrone M A., Jr Proc Natl Acad Sci USA. 1996;93:10417–10422. doi: 10.1073/pnas.93.19.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekelsky J J, Newfeld S J, Raftery L A, Chartoff E H, Gelbart W M. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raftery L A, Twombly V, Wharton K, Gelbart W M. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newfeld S J, Chartoff E H, Graff J M, Melton D A, Gelbart W M. Development. 1996;122:2099–2108. doi: 10.1242/dev.122.7.2099. [DOI] [PubMed] [Google Scholar]
- 6.Wiersdorff V, Lecuit T, Cohen S M, Mlodzik M. Development. 1996;122:2153–2162. doi: 10.1242/dev.122.7.2153. [DOI] [PubMed] [Google Scholar]
- 7.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 8.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 9.Massague J. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- 10.Hahn S A, Schutte M, Hoque A T M S, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern SE. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 11.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Feng X, We R, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 13.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 14.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L, Bapat B, Gallinger S, Andrulis I L, Thomsen G H, Wrana J L, Attisano L. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 15.Serra R, Moses H L. Nat Med. 1996;2:390–391. doi: 10.1038/nm0496-390. [DOI] [PubMed] [Google Scholar]
- 16.Resnick N, Gimbrone M A., Jr FASEB J. 1995;9:874–882. doi: 10.1096/fasebj.9.10.7615157. [DOI] [PubMed] [Google Scholar]
- 17.Gimbrone M A, Jr, Nagel T, Topper J N. J Clin Invest. 1997;99:1809–1813. doi: 10.1172/JCI119346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrana J L, Attisano L, Wieser R, Ventura F, Massague J. Nature (London) 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 19.Hoodless P A, Haerry T, Abdollah S, Stapleton M, O’Conner M B, Attisano L, Wrana J L. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 20.Ohno M, Cooke J P, Dzau V D, Gibbons G H. J Clin Invest. 1995;95:1363–1369. doi: 10.1172/JCI117787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Rubock M J, Whitman M. Nature (London) 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi H, Abdollah S, Cai J, Qiu Y, Yong-Yao X, Topper J N, Gimbrone M A, Jr, Wrana J L, Falb D. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 23.Topper J N, Wasserman S M, Anderson K R, Cai J, Falb D, Gimbrone M A., Jr J Clin Invest. 1997;99:2941–2949. doi: 10.1172/JCI119489. [DOI] [PMC free article] [PubMed] [Google Scholar]