Summary
Identification of the RNA polymerase (RNAP) binding site for ppGpp, a central regulator of bacterial transcription, is crucial for understanding its mechanism of action. A recent high resolution x-ray structure defined a ppGpp binding site on T. thermophilus RNAP. We report here effects of ppGpp on ten mutant E. coli RNAPs with substitutions for the analogous residues within 3–4Å of the ppGpp binding site in the T. thermophilus cocrystal. None of the substitutions in E. coli RNAP significantly weakened its responses to ppGpp. This result differs from the originally-reported finding of a substitution in E. coli RNAP eliminating ppGpp function. The E. coli RNAPs used in that study likely lacked stoichiometric amounts of ω, an RNAP subunit required for responses of RNAP to ppGpp, in part explaining the discrepancy. Furthermore, we found that ppGpp did not inhibit transcription initiation by T. thermophilus RNAP in vitro or shorten the lifetimes of promoter complexes containing T. thermophilus RNAP, in contrast to the conclusion in the original report. Our results suggest that the ppGpp binding pocket identified in the cocrystal is not the one responsible for regulation of E. coli rRNA transcription initiation and highlight the importance of inclusion of ω in bacterial RNAP preparations.
Keywords: ppGpp, ppGpp binding site, RNA polymerase, Omega, DksA
Introduction
The unusual nucleotides guanosine pentaphosphate and tetraphosphate (guanosine-3′,5′-bispyrophosphate), collectively referred to here as ppGpp (“magic spot”), are signaling alarmones produced in bacterial cells by the ribosome-associated synthase RelA and/or the hydrolase/synthase SpoT.1 The concentration of ppGpp is inversely proportional to the cellular growth rate and increases rapidly in response to nutritional downshifts and starvations.1, 2 Together with the cofactor DksA, ppGpp can regulate cellular promoters either negatively or positively. In E. coli, it directly inhibits the synthesis of rRNAs, tRNAs, and some mRNAs3, 4 and it directly and indirectly stimulates the synthesis of several amino acids and a number of other important gene products required for growth, stress responses, and pathogenesis in E. coli and other bacteria.5, 6, 7, 8, 9 Following amino acid starvation (when the substrates for translation are unavailable), direct and indirect effects of ppGpp on transcription initiation help the bacterial cell adjust to its nutritional status by reducing production of ribosomes and increasing amino acid biosynthesis and transport. 8, 10, 11
In E. coli, ppGpp acts directly on RNA polymerase (RNAP) by decreasing the lifetime of competitor-resistant complexes formed between RNAP and all promoters studied to date.12 At most promoters, this does not lead to inhibition of transcription, because RNAP escapes into its elongation mode before ppGpp significantly reduces the occupancy of the promoter complex. However, because rRNA promoters form intrinsically very short-lived complexes with RNAP, there is a kinetic competition between DNA strand collapse and addition of the initial NTP (iNTP) during transcription initiation at these promoters.12 In support of this model for control by ppGpp, there is a strong correlation between promoters that are inhibited by ppGpp and those that form short-lived complexes with RNAP.12, 13, 14
The 17 kDa DksA protein is required for full regulation of transcription initiation by ppGpp at rRNA promoters. DksA binds in the secondary channel of RNAP and greatly amplifies effects of ppGpp on transcription initiation in vitro and in vivo.4, 15 Like ppGpp, DksA decreases the lifetime of complexes formed by promoters with E. coli RNAP and inhibits transcription from rRNA promoters in vitro.4 DksA and ppGpp together strongly and synergistically inhibit rRNA promoters directly and stimulate a class of amino acid biosynthetic promoters both directly and indirectly.8,10
Despite our understanding of the effects of ppGpp on the kinetics of transcription initiation by E. coli RNAP, the structural basis by which ppGpp affects the kinetics of promoter-RNAP complexes is not well-understood. Identification of the ppGpp binding site on RNAP would contribute greatly to our understanding of the mechanism of ppGpp action. Several attempts have been made previously to determine the ppGpp binding site on E. coli RNAP. Mutations that conferred resistance to high levels of ppGpp in vivo were mapped to rpoB, the β subunit of RNAP, but were not localized further.16 Analysis of fluorescence quenching upon addition of ppGpp to RNAP suggested that ppGpp binds to a single site on RNAP.17 A crosslink to 8-azido-ppGpp was identified within the C-terminal half of β.18 A crosslink between 6-thio-ppGpp and RNAP was localized to the N-terminal ~102 residues of β′.19 One interpretation of these seemingly conflicting results is that ppGpp resides at an interface of β and β′.
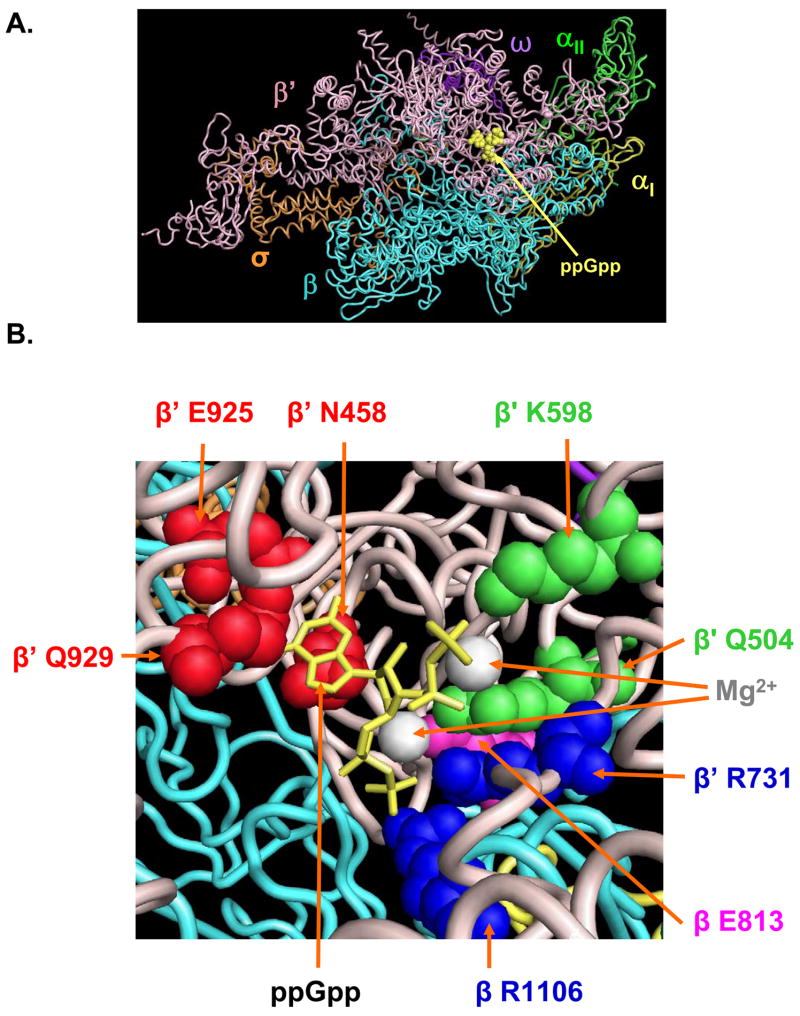
More recently, an x-ray structure of Thermus thermophilus RNAP in complex with ppGpp placed the ppGpp binding site adjacent to, but not overlapping, the RNAP active site (Fig. 1A).20 Two different orientations of ppGpp were present in the two complexes in the cocrystal’s asymmetric unit, with ppGpp “flipped” in one complex relative to the other. The 5′-diphosphate was located closer (proximal) to the active site Mg2+ in one orientation, and the 3′-diphosphate was closer to the active site Mg2+ in the other orientation. Nevertheless, the same RNAP residues contacted the same three specificity determinants in ppGpp (the two diphosphates and the guanine base) in both ppGpp orientations. No contacts were observed to the ribose.
Figure 1.
ppGpp binding site in T. thermophilus RNAP. (A) The T. thermophilus ppGpp-RNAP x-ray structure (ref.20; PDB coordinates 1SMY) is displayed using PyMol (DeLano Scientific). Subunits are colored as follows: ω, dark purple; αI, yellow; αII, green; β, cyan; β′, pink; σ, orange. ppGpp is in yellow spacefill. (B) T. thermophilus RNAP amino acids in close proximity to ppGpp (E. coli RNAP residue numbering). ppGpp is in yellow stick form. Mg2+ ions predicted to be coordinated by the proximal and distal (with respect to the active site) ppGpp diphosphates are shown as white spheres. Residues predicted to contact the guanine base of ppGpp (β′ N458, β′ E925, and β′Q929) are in red spacefill, to contact the distal phosphates (β′ K598 and β′ Q504) are in green, to contact the proximal phosphates (β′ R731 and β R1106) are in dark blue, and to contact a ppGpp-coordinated Mg2+ (β E813) is in magenta. Substitutions were made for each of these residues and for β E814 and β′ K599 as well (see text), but the T. thermophilus residues corresponding to these amino acids are not pictured because they do not contact ppGpp in the cocrystal. T. thermophilus residues corresponding to the E. coli amino acids in the figure are in parentheses:β′ N458 (N737), β′ Q504 (R783), β′ K598 (K908), β′ R731 (R1029), β′ E925 (E1231), β′ Q929 (Q1235), β R1106 (R879), and β E813 (E685).
The residues contacting ppGpp in the T. thermophilus cocrystal are highly conserved in E. coli RNAP, far from the N-terminal region of β′ in E. coli RNAP proposed to crosslink to ppGpp.19, 20, 21, 22, 23 It was claimed that T. thermophilus RNAP responded directly to ppGpp in vitro, and that an N458S substitution in the β′ subunit of E. coli RNAP, corresponding to a residue contacting ppGpp in the T. thermophilus RNAP cocrystal, decreased the ability of the promoter complex to respond to ppGpp.20 Finally, it was proposed that the guanine base in ppGpp pairs directly to cytosines on the nontemplate DNA strand in the open complex, just upstream of the transcription start site.
Several observations led us to reevaluate the significance of the ppGpp binding site defined in the T. thermophilus RNAP cocrystal for regulation of E. coli RNAP. First, the E. coli RNAPs used in the previous mutational analysis of the ppGpp binding site were prepared by overexpression of the α, β, and β′ subunits in vivo.20, 24 RNAPs prepared by this method are grossly undersaturated with the ω subunit of RNAP. We have shown previously that RNAPs lacking ω, either because they were prepared by this method or because the strain used to prepare RNAP lacked the gene encoding ω, were not inhibited by ppGpp in vitro.25 Since the wild-type and mutant E. coli RNAPs used for testing the biological significance of the ppGpp binding site identified in the T. thermophilus cocrystal lacked ω,20 conclusions based on comparisons of their responses to ppGpp were subject to question.
Second, recent studies indicate that the site of ppGpp binding in the T. thermophilus RNAP cocrystal can accommodate a number of negatively-charged molecules. Not only can ppGpp bind in two different orientations, but this site overlaps a binding site for an NTP in yeast Pol II and T. thermophilus transcription elongation complexes 26, 27, 28 as well as a binding site for the antibiotic tagetitoxin.29 It is unclear that this NTP binding site plays a role in initiation complex formation,30 but these results nevertheless are consistent with the possibility that ppGpp might be occupying a positively-charged pocket in the cocrystal, but not the pocket physiologically significant for regulation of transcription initiation.
Third, although relA homologs are present in most or all bacterial genomes, not all bacterial RNAPs respond directly to ppGpp. ppGpp synthesis is induced in Bacillus subtilis, as in E. coli, in response to amino acid deprivation, concurrent with a large decrease in rRNA transcription.31 However, in contrast to its direct inhibition of E. coli RNAP, ppGpp only indirectly inhibits transcription by B. subtilis RNAP, probably by reducing the concentration of GTP, the initial NTP for initiation at all 20 B. subtilis rRNA promoters.31 A recent study concluded that ppGpp also decreases T. thermophilus transcription indirectly.32 Therefore, not only is it unclear that the binding site identified in the T. thermophilus RNAP-ppGpp complex represents the one responsible for regulating transcription in E. coli, but it is also unclear whether the ppGpp binding site identified in the T. thermophilus RNAP-ppGpp cocrystal is biologically significant even in that organism.
These uncertainties prompted us to test whether the site in E. coli RNAP analogous to the ppGpp binding site in the T. thermophilus RNAP cocrystal is the one responsible for effects of ppGpp on E. coli rRNA transcription initiation. Single and multiple substitutions were constructed for residues predicted to make either direct contacts to ppGpp or indirect contacts to ppGpp through a bound Mg2+. None of the substitutions reduced the responses of RNAPs (containing ω) to ppGpp, either in the presence or absence of DksA. Furthermore, ppGpp did not compete with the initiating NTP for binding to E. coli RNAP in vitro, nor did ppGpp inhibit transcription initiation by T. thermophilus RNAP in vitro. Taken together, our results indicate that models for the mechanism of transcription regulation by ppGpp, DksA, and ω based on the position of ppGpp in the T. thermophilus RNAP cocrystal should be reevaluated.
Results
Choice of substitutions in the ppGpp binding pocket
A sequence alignment of E. coli and T. thermophilus RNAP was used to identify E. coli amino acid residues analogous to those contacting ppGpp in the cocrystal. Ten mutant RNAPs with substitutions for residues within 3–4 Å of ppGpp were constructed and purified (see Materials and Methods and Fig. 1). The mutant RNAPs are designated by the wild-type amino acid before the number of the residue in the appropriate E. coli subunit, followed by the identity of the altered amino acid. Eight RNAPs contained single or multiple substitutions in the β′ subunit (N458S, Q504E, Q504Y, K598A/K599A, R731A, E925A, Q929A, K598A/K599A/E925A), and two RNAPs contained changes in the β subunit (E813A/E814A, R1106A) (Fig. 1B; the numbers of the analogous residues in T. thermophilus RNAP, as well as the ppGpp determinant contacted by these amino acids in the structure, are provided in the figure legend). Three of these residues (β′ Q504, β′ R731, and β R1106) are in the “basic rim” that surrounds the NTP phosphates for substrate loading in the T. thermophilus transcription elongation complex.26
The alanine substitutions removed all side chain atoms beyond Cβ and thus all potential side chain contacts of that residue to ppGpp. β′ N458S was created rather than β′ N458A for comparison with the effects of the substitution reported previously.20 β′ Q504E was used because this negatively-charged glutamate substitution would be predicted to interfere with contact(s) to the negatively-charged ppGpp phosphates. β′ Q504Y was created because the tyrosine side chain would be predicted to cause a steric clash with ppGpp phosphates. Since β′ K598 in E. coli RNAP is adjacent to another lysine, K599, we constructed a double substitution, β′ K598A/K599A, to prevent potential compensation by one lysine side chain for the other. The same double substitution was also made in conjunction with β′ E925A, eliminating three potential H-bonds to ppGpp. Finally, we also tested the double substitution mutant β E813A/E814A, since the cocrystal predicted that β E813 contacted a magnesium ion coordinated by the ppGpp proximal phosphates.
Substitutions in the binding pocket defined in the T. thermophilus cocrystal do not weaken the response of E. coli RNAP to ppGpp
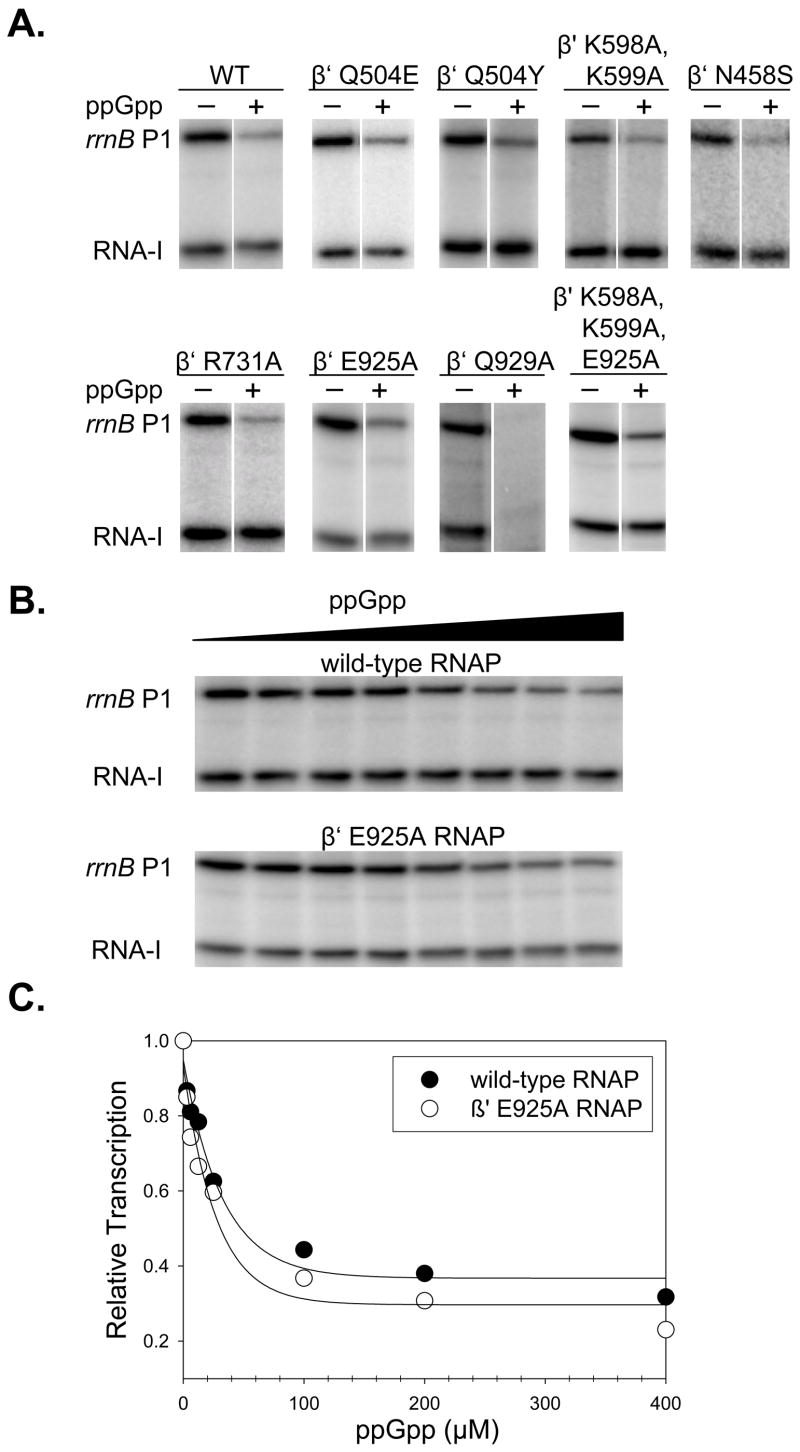
We tested the effects of ppGpp on transcription initiation by each of the mutant RNAPs, using a supercoiled plasmid template containing the rRNA promoter rrnB P1 and a control promoter, RNA-I. All but two of the mutant RNAPs, β E813A/E814A and β R1106A, were active in transcription. 400 μM ppGpp inhibited transcription at least 3-fold with each of the catalytically active RNAPs (Fig. 2A), and inhibition was specific to rrnB P1 for all but β′ Q929A RNAP (see Discussion).
Figure 2.
Effects of RNAP substitutions on transcription inhibition by ppGpp. (A) Transcription inhibition at saturating ppGpp concentration (400 μM). Multi-round transcription from the rrnB P1 and RNA-I promoters on plasmid pRLG6798 was performed as described (see Materials and Methods). Lanes ± ppGpp are from the same gel in the same experiment. WT = wild-type E. coli RNAP. (B) Transcription was as described in (A) but with 0 to 400 μM ppGpp with wild-type and β′ E925A RNAP. (C) Determination of IC50 for ppGpp and mutant RNAPs. Transcription from rrnB P1 in the experiment shown in (B) was normalized to transcription from RNA-I at each ppGpp concentration (to correct for errors in gel loading) and expressed as a fraction of transcription without ppGpp. The plot allows calculation of the maximal extent of inhibition by ppGpp and the IC50 (ppGpp concentration at which inhibition is half-maximal). Plots for other transcriptionally-active mutants are in Supplementary Fig. 1, and the data are compiled in Table 1.
Since potential effects of the loss of a side chain-ppGpp contact might have been masked at the high ppGpp concentration used in Fig. 2A, inhibition of rrnB P1 at a range of ppGpp concentrations was measured for each RNAP (representative gels and plots for wild-type and a mutant RNAP are shown in Figs. 2B and 2C; plots for other transcriptionally-active RNAPs are in Supplementary Fig. 1; and the averages and standard deviations for the effects of ppGpp on transcription in multiple experiments are provided in Table 1). This allowed not only a more precise estimate of the fraction of transcription at saturating ppGpp concentrations (column 2, Table 1), but also calculation of a half-maximal inhibitory ppGpp concentration, the IC50 (Fig. 2C; column 3, Table 1). For wild-type RNAP at 30°C, the IC50 was ~25 μM ppGpp, in agreement with the IC50 obtained previously (Barker et al. 2001b). In no case (including the β′ N458S RNAP concluded previously to be deficient in responding to ppGpp20) was the IC50 for a mutant RNAP greater than that for wild-type RNAP. In fact, several mutants had an IC50 ratio less than 1.0 relative to wild-type RNAP, indicating that the mutant RNAP was more sensitive to ppGpp than the wild-type. Some of the substitutions also increased the extent of inhibition by saturating ppGpp concentrations (Tables 1 and 2). It is possible that these substitutions might reduce the energy barrier needed for making the ppGpp-induced conformational changes in the enzyme, thus increasing the apparent effects of ppGpp. In any case, in contrast to the conclusions of the previous study,20 we conclude that ppGpp inhibits transcription initiation by E. coli RNAPs containing single or multiple substitutions in the ppGpp binding site identified in the cocrystal.
Table 1.
Effects of Substitutions in RNAP on Transcription Inhibition by ppGpp
| RNAP substitution (E. coli numbering) | Relative transcription at saturating ppGppa | Relative IC50 (mutant IC50 / WT IC50)b |
|---|---|---|
| Wild-type RNAP | 0.32 ± 0.05 | 1 |
| B′ Q504E | 0.35 ± 0.02 | 0.59 ± 0.13 |
| B′ Q504Y | 0.36 ± 0.05 | 0.60 ± 0.14 |
| β′ K598A/K599A | 0.22 ± 0.02 | 0.71 ± 0.20 |
| β′ N458S | 0.20 ± 0.01 | 0.45 ± 0.04 |
| β′ R731A | 0.24 ± 0.04 | 0.36 ± 0.03 |
| β′ E925A | 0.20 ± 0.06 | 0.91 ± 0.43 |
| β′ K598A/K599A/E925A | 0.32 ± 0.02 | 0.99 ± 0.19 |
Transcription at saturating ppGpp is relative to that without ppGpp as illustrated in Fig. 2 and as described in Results and Materials and Methods. Saturating ppGpp concentration is that at the plateau in the plots shown in Fig. 2 and Supplemental Fig. 1. Reported errors are from at least three experiments. Results for the β′ Q929A holoenzyme are not included in the table, because the inhibition by ppGpp was too strong to be quantified and was not specific to rrnB P1 (see Fig. 2A).
Calculation of the IC50 (the concentration of ppGpp at which inhibition is half-maximal) is described in Results and in Fig. 2 legend. The relative IC50 is the IC50 for the mutant RNAP / IC50 for wild-type RNAP.
Table 2.
Effects of substitutions in RNAP on reduction of promoter complex half-life by ppGpp.
| Substitution (E. coli RNAP numbering) | Relative lifetime without ppGppa | Relative lifetime at saturating ppGppb | Relative IC50 (mutant IC50 / WT IC50)c |
|---|---|---|---|
| wild-type RNAP | 1 | 0.47 ± 0.08 | 1 |
| β′ Q504E | 0.99 ± 0.11 | 0.48 ± 0.04 | 1.56 ± 0.20 |
| B′ K598A/K599A | 1.14 ± 0.08 | 0.40 ± 0.08 | 0.57 ± 0.00 |
| β′ N458S | 0.90 ± 0.08 | 0.40 ± 0.02 | 0.43 ± 0.13 |
| β′ R731A | 1.86 ± 0.12 | 0.40 ± 0.03 | 0.25 ± 0.02 |
| β′ E925A | 1.68 ± 0.10 | 0.24 ± 0.05 | 0.92 ± 0.20 |
| β′ Q929A | 1.23 ± 0.10 | 0.26 ± 0.06 | 0.46 ± 0.10 |
| β E813A/E814A | 0.76 ± 0.02 | 0.36 ± 0.00 | 0.60 ± 0.14 |
| β R1106A | 0.98 ± 0.10 | 0.32 ± 0.02 | 0.52 ± 0.15 |
Half-lives were measured on the lacUV5 promoter using a filter-binding assay as described in Materials and Methods and Fig. 3 legend. Reported half-lives are from at least two titrations, each including ≥ 5 ppGpp concentrations.
The values reported are the lifetimes of the promoter complexes at saturating ppGpp concentration relative to the same complexes without ppGpp, taken from the plateau values on plots like those illustrated in Fig. 3 and Supplemental Fig. 2. The reported values are from the same experiments as the intrinsic half-lives (i.e. without ppGpp).
The relative IC50 is the concentration of ppGpp resulting in a half-maximal decrease in complex lifetime with the mutant RNAP, relative to that with the wild-type RNAP. The half-lives were taken from plots like those shown in Fig. 3 and Supplemental Fig. 2, representing ≥ 2 determinations at each ppGpp concentration.
The half-lives of competitor-resistant complexes formed by mutant RNAPs are decreased by ppGpp
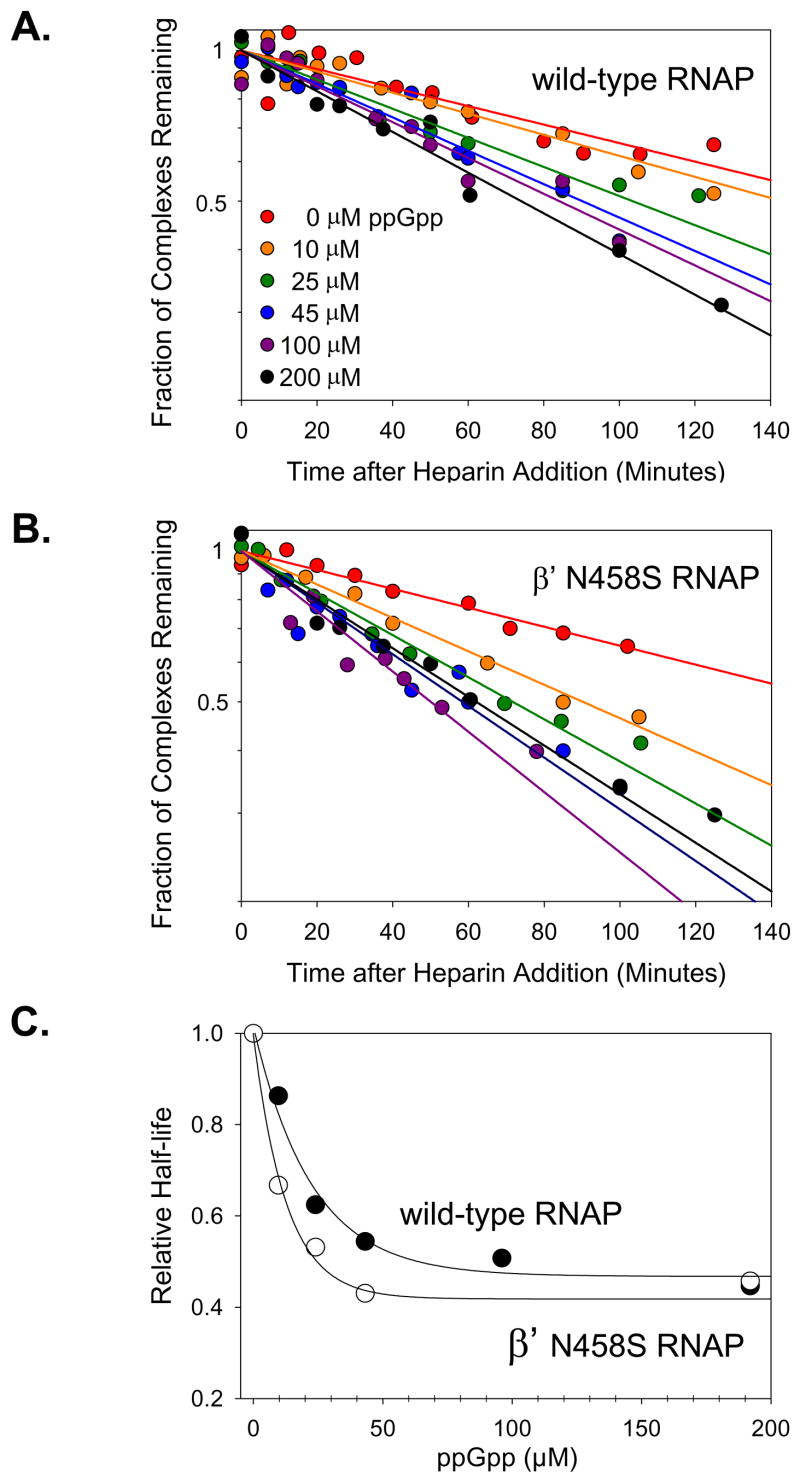
As indicated above, ppGpp decreases the lifetimes of competitor-resistant complexes formed between E. coli Eσ70 and all promoters that have been assayed.12 Since promoter-specific effects of ppGpp on three of the mutant RNAPs could not be quantified by transcription inhibition (β R1106A, β E813A/E814A, and β′ Q929A; see Table 1 legend), we examined the responses to ppGpp of these mutant RNAPs (as well as five others) using a promoter complex lifetime assay that did not require that the enzyme be catalytically active. Complexes formed by the lacUV5 promoter and each of the eight mutant RNAPs were challenged with heparin, a competitor for RNAP, and the fraction of complexes bound to filters was plotted versus time. The time required for half of the complexes to dissociate was determined at a range of ppGpp concentrations for the wild-type and mutant RNAPs (Fig. 3 and Supplementary Fig. 2).
Figure 3.
Effects of ppGpp on promoter complex lifetime of wild-type and β′ N458S mutant RNAP. Fraction of lacUV5 complexes remaining as a function of time after heparin addition at different ppGpp concentrations (see Materials and Methods). Semilog plots for representative experiments: (A) wild-type RNAP, (B) β′ N458S RNAP. (C) Half-lives at each ppGpp concentration. Comparisons of the relative complex half-lives for wild-type RNAP and the other mutant RNAPs are shown in Supplementary Fig. 2, and the data are compiled in Table 2.
The averages and standard deviations for the effects of ppGpp on the half-lives of promoter complexes containing each of eight mutant RNAPs are provided in Table 2. The intrinsic lifetimes of the complexes formed by the mutant RNAPs (i.e. without ppGpp) were all within ~2-fold of that with wild-type RNAP; column 2, Table 2). In agreement with previous results,12 ppGpp decreased the half-lives of the complex made with wild-type RNAP ~2-fold at a saturating concentration of ppGpp (~200 μM) (Fig. 3). Similar results were obtained with β′ N458S RNAP (Figs. 3B and 3C), the one mutant RNAP examined previously (in ref. 20; see Discussion). Saturating concentrations of ppGpp also decreased the half-lives of the complexes formed by the seven other mutant RNAPs that were tested, and the effects were at least as great as with the wild-type and β′ N458S RNAPs (column 3, Table 2; for additional plots, see Supplementary Fig. 2).
In agreement with previous results 12, the concentration of ppGpp at which its effect on complex lifetime was half-maximal (the IC50) was ~17 μM for wild-type RNAP (Fig. 3A and 3C). The IC50 was also determined for each mutant RNAP (Fig. 3B and 3C; Supplementary Fig. 2) and expressed relative to the IC50 for wild-type RNAP (column 4, Table 2). For all but one mutant (β′ Q504E RNAP), the relative IC50 was as low as, or lower than, that for the wild-type complex; i.e the same or a lower concentration of ppGpp was needed to reduce the lifetime of the mutant complex. For β′ Q504E RNAP, there was a slight (~50%) increase in the ppGpp concentration required for the half-maximal effect on complex lifetime, but this small increase, even if statistically significant, apparently has no functional consequence for transcription inhibition; see Table 1). In conjunction with the results of the transcription inhibition experiments (Fig. 2, Table 1, Supplementary Fig. 1), we conclude that none of the side chain contacts to ppGpp predicted by the T. thermophilus RNAP-ppGpp cocrystal are required for effects of ppGpp on E. coli RNAP-promoter complexes.
RNAP mutants respond to ppGpp in the presence of DksA
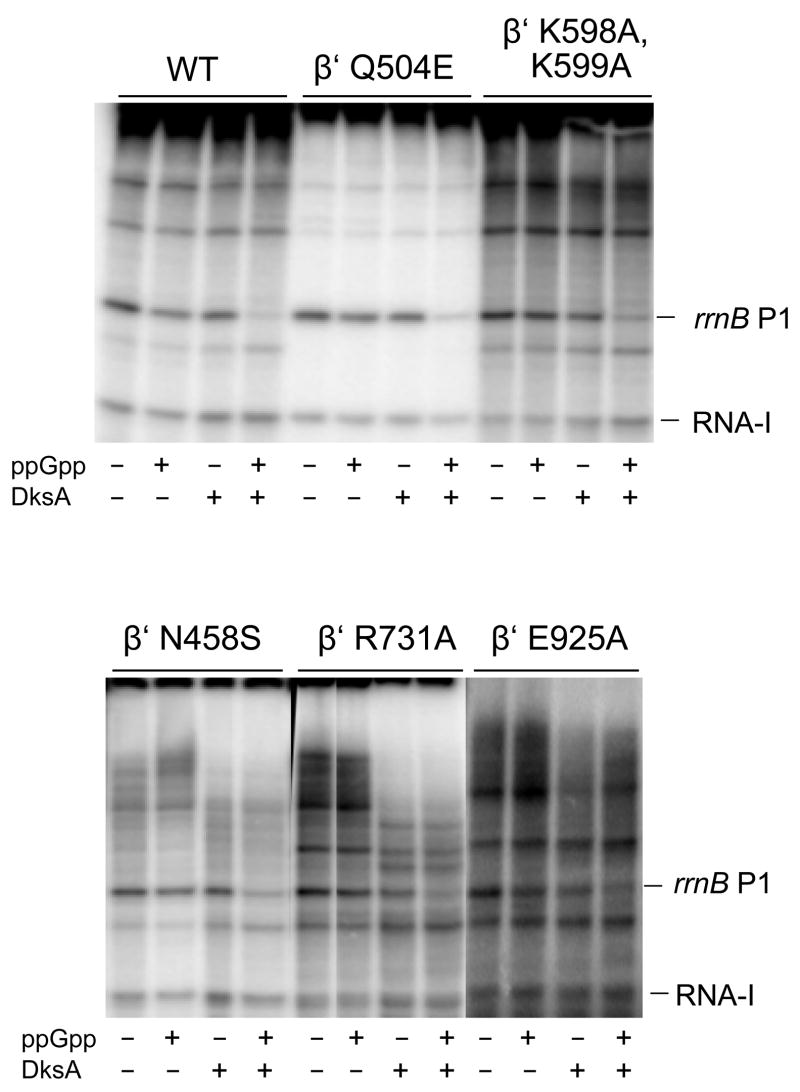
We demonstrated previously that the effect of ppGpp on transcription initiation is strongly amplified by DksA in vitro and in vivo.4 Therefore, we addressed the formal possibility that side chain contacts to ppGpp identified in the T. thermophilus cocrystal might be utilized by E. coli RNAP when DksA is present, even though they are dispensable for ppGpp function in the absence of DksA. Representative results from transcription of rrnB P1 by five of the mutant RNAPs are shown in Fig. 4 under conditions where the synergistic effects of ppGpp and DksA could be observed since the independent effects of each alone were small (i.e. at relatively low salt concentrations that resulted in relatively stable promoter complexes; see also Materials and Methods).
Figure 4.
Inhibition of mutant RNAPs by ppGpp in the presence of DksA. Single-round transcription was performed as described (see Materials and Methods) with neither ppGpp nor DksA (first lane in each panel), with ppGpp alone (100 μM; second lane in each panel), with DksA alone (third lane in each panel; for concentrations see Materials and Methods), or with both together (fourth lane in each panel). WT = wild-type RNAP. β′ N458S, β′ E925A, and β′ Q929A RNAPs (not shown) exhibited transcription elongation defects, resulting in some incomplete extension products under these conditions (see Materials and Methods).
At concentrations where little or no inhibition was observed with ppGpp by itself (second lane in each panel) and where DksA by itself inhibited rrnB P1 transcription only ~twofold (third lane in each panel), DksA and ppGpp together much more strongly inhibited transcription (fourth lane in each panel). Thus, for each of the five RNAPs tested, side chain contacts to ppGpp identified in the T. thermophilus cocrystal are dispensible for ppGpp function in the presence of DksA.
The iNTP and ppGpp do not compete for binding to RNAP during transcription initiation
The ppGpp binding site on T. thermophilus RNAP overlaps site(s) that have been proposed for substrate binding on the pathway to the active site during transcription elongation.26, 27, 28 If ppGpp binding also overlapped an entry site for the first NTP (iNTP) into the open complex, this might lead to competition and an increase in the iNTP concentration needed for transcription initiation. Since rrnB P1 (but not RNA-I) activity is very sensitive to the concentration of the iNTP, ATP,33 we measured transcription from a plasmid containing these promoters at increasing ATP concentrations in the presence or absence of a high concentration of ppGpp (1 mM). As expected, transcription from rrnB P1 (but not from RNA-I) increased as the ATP concentration increased, and ppGpp inhibited rrnB P1 (but not RNA-I) activity at each ATP concentration. However, ppGpp did not affect the relative increase in transcription from rrnB P1 at each ATP concentration (Fig. 5A). These results suggest that ppGpp does not affect NTP entry into E. coli RNAP during transcription initiation, whatever the pathway by which the iNTP approaches the active site.
Figure 5.
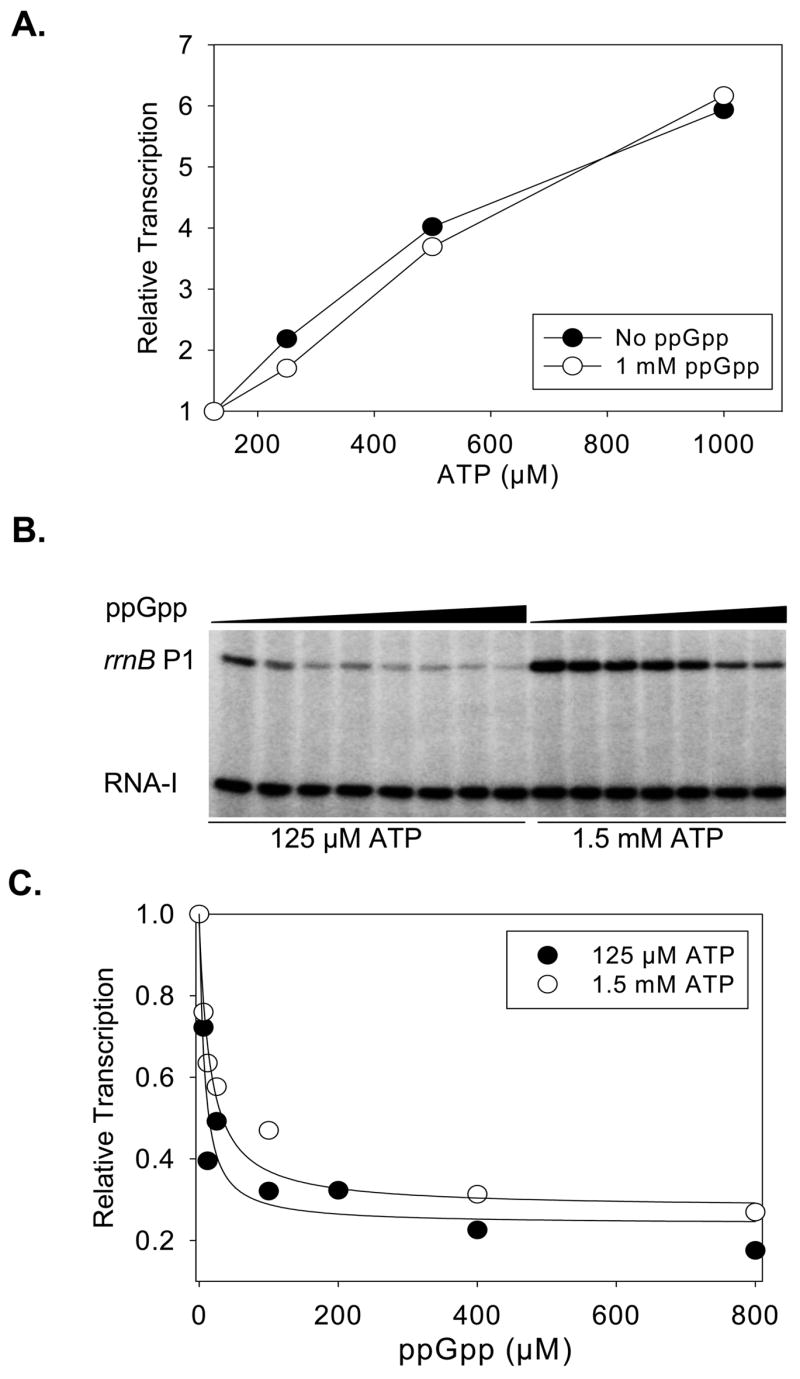
Tests of competition between ppGpp and the iNTP. (A) ppGpp does not change the effect of the concentration of the first NTP on transcription from rrnB P1. Multi-round transcription from plasmid pRLG6798 containing rrnB P1 was performed ± 1 mM ppGpp at increasing concentrations of ATP (the iNTP) and plotted relative to transcription at 125 μM ATP. (B) iNTP concentration does not affect relative inhibition by ppGpp. Representative gel showing multi-round transcription from rrnB P1 at increasing concentrations of ppGpp with 125 μM or 1500 μM ATP. (C) Transcription from (B) at each ppGpp concentration is plotted as a fraction of transcription without ppGpp.
We also examined whether a high concentration of the iNTP would compete with binding of ppGpp and decrease its effect on RNAP. At increasing concentrations of ppGpp, transcription inhibition was measured at a relatively low (125 μM) and a relatively high (1500 μM) concentration of ATP (Fig. 5B). The extent of inhibition by ppGpp at saturating ppGpp concentration and the ppGpp concentration required for half-maximal inhibition were virtually identical at both ATP concentrations (Fig. 5C). Although these results do not address the identity of the binding site of ppGpp on E. coli RNAP directly, they do not support a competitive inhibition mechanism for ppGpp function during transcription initiation.
ppGpp affects the lifetime of the promoter complex independent of the identitiy of the bases at promoter positions -1 and -2
It was suggested previously 20 that the position of ppGpp in the cocrystal with T. thermophilus RNAP provided a potential mechanism to explain the effects of ppGpp on the promoter complex: base pairing of ppGpp directly with nontemplate strand C residues at positions -1 and/or -2 in ppGpp-sensitive promoters such as rrnB P1. Therefore, we tested whether mutations at C-1, C-2, or both prevented effects of ppGpp on rrnB P1 promoter complexes, using the half-life assay to evaluate ppGpp function. As summarized in Table 3, ppGpp decreased the lifetimes of all the mutant promoter complexes at least as much as it decreased the lifetime of the wild-type promoter complex. These data, in conjunction with several other lines of evidence (see Discussion), do not support the proposal that ppGpp pairs with C-1 or C-2 in rrn P1 promoter complexes in order to inhibit transcription iniitiation. Whether or not ppGpp actually decreases transcription initiation depends on a promoter’s intrinsic kinetic properties: only those promoters that make short-lived competitor-resistant complexes with RNAP are subject to inhibition (see Introduction and ref. 14). Since several of the substitutions for C-1 and C-2 increased the absolute lifetime of the rrnB P1 promoter complex (Table 3),14 the effect of ppGpp on transcription initiation from the mutant promoters was not tested directly.
Table 3.
Effect of ppGpp on lifetimes of promoter complexes containing substitutions at positions -1 and/or -2
| Plasmid | Promoter a | -2 b | -1 c | Half-Lifed | ||
|---|---|---|---|---|---|---|
| - ppGpp | + ppGpp | + / - ppGpp e | ||||
| pRLG3733 | rrnB P1 WT | C | C | 25 sec | 15 sec | 0.61 ± 0.06 |
| pRLG6755 | rrnB P1 C-1G | C | G | 84 sec | 51 sec | 0.60 ± 0.10 |
| pRLG6754 | rrnB P1 C-2G | G | C | 64 sec | 42 sec | 0.66 ± 0.02 |
| pRLG7752 | rrnB P1 AA | A | A | 326 sec | 123 sec | 0.38 ± 0.05 |
| pRLG7753 | rrnB P1 GG | G | G | 97 sec | 47 sec | 0.48 ± 0.02 |
| pRLG7754 | rrnB P1 TT | T | T | 551 sec | 223 sec | 0.40 ± 0.19 |
Promoter is named by the identity of the base on the nontemplate strand at positions -1 and -2.
Identity of nontemplate base at -2.
Identity of nontemplate base at -1.
Half-lives were determined as described in Materials and Methods.
The values and standard deviations are the averages from three separate ratios of the half-lives in the presence of ppGpp to those in the absence of ppGpp.
T. thermophilus RNAP is not inhibited by ppGpp like E. coli RNAP in vitro
Our previous studies showed that not all bacterial RNAPs are inhibited directly by ppGpp.31 Since the RNAP-ppGpp cocrystal contained T. thermophilus RNAP, we investigated whether ppGpp affects T. thermophilus RNAP function in the same manner as E. coli RNAP.
Transcription from a T. thermophilus rRNA promoter by T. thermophilus RNAP was inhibited only slightly, if at all (1.3-fold ± 0.2), by 1 mM ppGpp (Fig. 6A), and this slight inhibition was observed with the RNA-I promoter as well, indicating it was not promoter-specific. Similar results were obtained at a variety of solution conditions (i.e. different temperatures and salt concentrations; data not shown).
Figure 6.
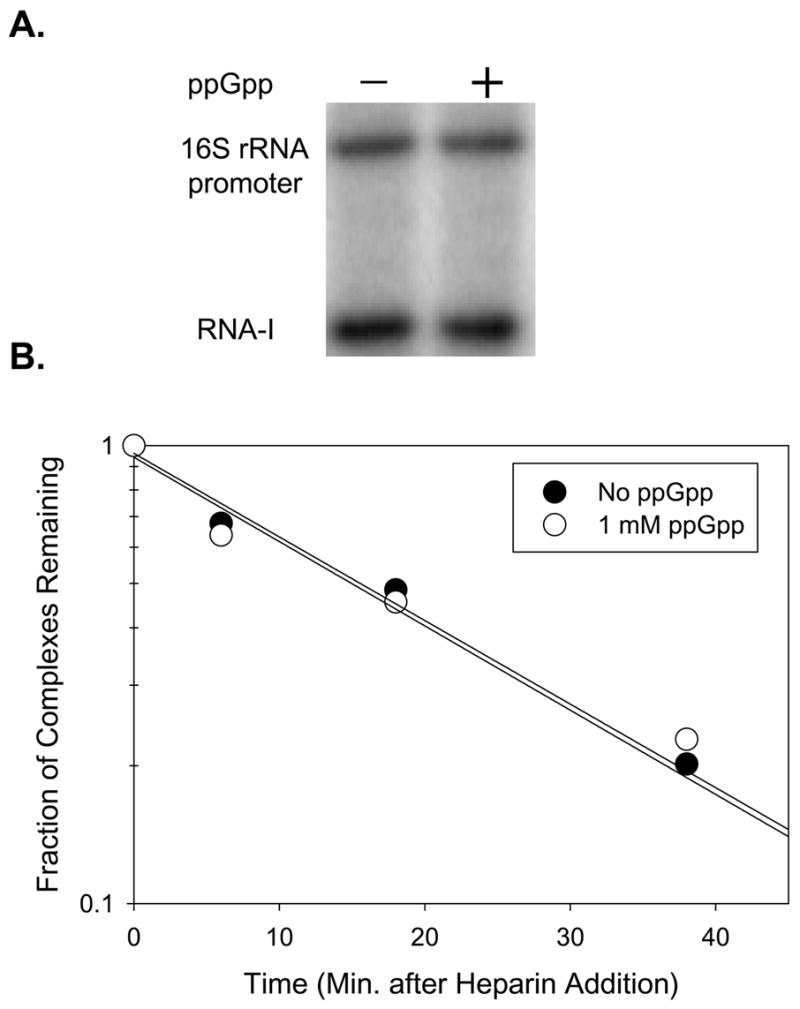
Effect of ppGpp on transcription initiation by T. thermophilus RNAP. (A) ppGpp (1 mM) does not inhibit transcription by T. thermophilus RNAP. Multi-round transcription from pRLG6770 was measured in 170 mM NaCl transcription buffer at 65°C on a supercoiled template carrying the T. thermophilus 16S rRNA promoter and vector-derived RNA-I promoter (see Materials and Methods). (B) ppGpp (1 mM) does not reduce the lifetime of a promoter complex containing T. thermophilus RNAP. Representative plots show the fraction of heparin-resistant complexes containing T. thermophilus RNAP and the RNA-I promoter on a supercoiled plasmid as a function of time after heparin addition (55°C in 100 mM KCl transcription buffer; see Materials and Methods). The mean ratio of the observed half-lives with/without ppGpp was 0.84 ± 0.17 (3 experiments).
We also measured the effect of ppGpp on the half-life of promoter complexes formed by T. thermophilus RNAP. The half-lives of complexes containing T. thermophilus RNAP with the RNA-I promoter, λPR (a phage promoter), and a synthetic promoter, -35 con34 were unaffected by ppGpp (Fig. 6B and data not shown), in contrast to the conclusion reached previously (in ref. 20; see Discussion). Promoter complexes formed by RNAP from another Thermus species, T. aquaticus, were also unaffected by ppGpp (data not shown). In addition, we note that dksA homologs are not apparent in the T. thermophilus and T. aquaticus genome sequences. Taken together, our results indicate that E. coli RNAP and T. thermophilus RNAP respond quite differently to ppGpp.
Discussion
Substitutions in E. coli RNAP that should have eliminated single or multiple contacts to ppGpp bound at the site identified in the cocrystal failed to reduce transcription inhibition by ppGpp in the presence or absence of DksA or to alter the effect of ppGpp on promoter complex lifetime. In general, we expected ≥ fivefold defects in ppGpp function from substitutions contributing to the functionally-significant ppGpp binding site. For example, the responses of RNAP to the small ligands microcin and rifampicin have been studied in detail; single amino acid substitutions in these binding pockets increased the IC50 five to 100-fold. 35, 36 Binding sites for some small ligands display a degree of plasticity, causing smaller than expected effects of certain mutations.26 However, in most cases, substitutions like those reported here that alter the charge of a side chain, remove a side chain interaction, or create a steric clash, strongly decrease ligand binding.37
We observed no increases at all in the concentrations of ppGpp required for inhibiting each of the ten mutant E. coli RNAPs tested, we observed no competition of ppGpp with the iNTP during transcription initiation from an E. coli rRNA promoter complex, and we found that ppGpp did not affect transcription initiation by T. thermophilus RNAP. Taken together, our results strongly suggest that the ppGpp binding site in the T. thermophilus RNAP-ppGpp cocrystal is unlikely to be the one responsible for direct inhibition of E. coli transcription initiation. Rather, the presence of ppGpp at this position in the cocrystal may reflect simply that ppGpp shares binding determinants with NTPs, and that the positively-charged NTP binding pocket in T. thermophilus RNAP can bind ppGpp under the conditions used for cocrystal formation.
Even though relA is widely distributed among bacteria1 and ppGpp has been identified even in chloroplasts,38 ppGpp does not appear to function in the same manner in all organisms. Consistent with the hypothesis that the T. thermophilus ppGpp binding pocket identified in the cocrystal is not the one responsible for promoter-specific regulation, it was reported recently that ppGpp concentrations in T. thermophilus are insufficient to affect transcription initiation directly.32 Rather, these investigators proposed that control of T. thermophilus rRNA transcription by ppGpp in vivo might work indirectly by reduction of GTP levels, similar to the mechanism suggested for control of B. subtilis rRNA synthesis.31 Therefore, T. thermophilus RNAP may not be the appropriate model system for determining the location of the ppGpp binding site to E. coli RNAP.
ppGpp reduced the synthesis of some abortive products by T. thermophilus RNAP at high ppGpp and very low NTP concentrations.20 We (ref. 12 and data not shown) and others39 have observed competition between ppGpp and substrate NTPs with E. coli RNAP at high ppGpp : NTP ratios, and small effects of ppGpp on transcription elongation have been reported in the literature.40 This competitive effect on elongation is not the promoter-specific mechanism for rRNA transcription inhibition by ppGpp that occurs during the stringent response in E. coli, but could, in principle, result from binding of ppGpp to the site in RNAP defined by the T. thermophilus cocrystal.
Although none of the RNAP substitutions reduced effects of ppGpp on transcription initiation, β′ Q929A RNAP, and to a lesser degree β′ N458S RNAP, resulted in promoter-nonspecific inhibition by ppGpp, suggesting that these substitutions might allow ppGpp to affect transcription elongation. β′ Q929 is in the trigger loop, which is central to the mechanism of nucleotide incorporation during transcription elongation27, 41 and β′ N458 affects deoxyribo- vs. ribo-nucleotide discrimination.42 We suggest that by binding to RNAP at the site identified in the cocrystal, ppGpp may compete with NTP incorporation when the latter is severely compromised by mutation.
Our conclusion that the β′ N458S mutant RNAP responded at least as well as wild-type RNAP to ppGpp in both promoter complex lifetime and transcription inhibition assays for ppGpp function in vitro directly contradicts the conclusion reached previously.20 We suggest that neither the wild-type nor the mutant E. coli promoter complexes were actually affected by ppGpp in the previous work (Fig. 4B in ref. 20), since the overall slopes were virtually superimposable in the presence and absence of ppGpp (small differences in times of sample processing can lead to large differences in apparent slopes estimated from only the initial time points). We further suggest that there is a relatively straightforward explanation for the failure to observe a response to ppGpp by the RNAPs used in the previous report: those RNAPs were made by overexpression of core RNAP subunits without concurrent overexpression of the ω subunit. We have shown previously that RNAP made in this manner lacks ω and is therefore unable to respond to ppGpp.25 Therefore, in addition to its implications for the mechanism of ppGpp action, our findings highlight the importance of inclusion of ω in preparations of RNAP (unless an ω requirement has been ruled out explicitly).
We found that nontemplate C residues at positions -1 and/or -2 are not required for ppGpp to reduce the lifetimes of rrn P1 or λPR promoter complexes (Table 3 and data not shown). These results also contradict results reported by Artsimovitch and coworkers.20 We suggest that the absence of an effect of ppGpp on promoter complexes lacking C residues at -1 and/or -2 reported in the previous study was not indicative of a requirement for these bases for a response to ppGpp. Rather, we suggest that neither the wild-type nor the mutant promoters responded to ppGpp in the previous report because the RNAP lacked ω.20 We also showed previously that an rrnB P1 promoter with a C-1T substitution was still strongly inhibited in vivo following amino acid starvation,43 and that ppGpp reduced the half-lives of other promoter complexes lacking C residues at C-1 and/or C-2.12 Furthermore, the proposed path of the nontemplate strand in the ppGpp base pairing model appears inconsistent with models based on the T. aquaticus RNAP cocrystal with fork-junction promoter DNA21, 44 and on recent cross-linking studies.14 These models place nontemplate strand bases -1 and -2 quite distant from the position of ppGpp defined in the T. thermophilus RNAP cocrystal.
The previous report20 also concluded that ppGpp reduced the half-life of promoter complexes formed by T. thermophilus RNAP, in contrast to the results reported here (Fig. 6). Re-examination of Fig. S3B in ref. 20, however, suggests that the presence of ppGpp did not actually change the responses of promoter complexes formed by T. thermophilus RNAP. The reported half-life curves were biphasic, and a difference in slope was observed only after 90–99% of the complexes in the population had decayed. We suggest that the difference in slope attributed to an effect of ppGpp may have resulted from a low signal : noise ratio at that point in the decay curve, or that ppGpp affected only some small subpopulation of complexes in the reaction.
Based on the position of ppGpp defined in the cocrystal, it was suggested that acidic residues at the tip of the coiled-coil of DksA (the cofactor that occupies the secondary channel of RNAP; ref. 15; I. Toulokhonov and R.L.G. unpublished data) reposition a Mg2+ ion coordinated to ppGpp. This model thereby suggested a mechanism for DksA action.15 However, we note that DksA affects RNAP function even in the absence of ppGpp.4 Therefore, we suggest that if the mechanism of DksA action includes repositioning a Mg2+ ion by the DksA coiled-coil tip, this does not depend on the presence of ppGpp. In any case, since the functionally-significant ppGpp binding site in E. coli RNAP does not appear to be located at the position defined by the T. thermophilus cocrystal, the proposed model for DksA function15 requires reevaluation.
A subset of the substitutions in β and β′ reported here reduced the effect of DksA on E. coli RNAP (data not shown), necessitating inclusion of high DksA concentrations in some of the transcription reactions (Fig. 4 legend). These results will be published separately as part of an investigation of the RNAP determinants of DksA function. These amino acids in RNAP may be important for DksA binding, either directly or indirectly, even though they do not define the functionally-significant ppGpp binding site in E. coli RNAP.
The central role of ppGpp in the regulation of bacterial gene expression, its importance in bacterial pathogenesis, and its usefulness as a model system for understanding allosteric control of RNAP by small molecules justify continued interest in identifying the ppGpp binding site in E. coli RNAP. The discovery that the ppGpp binding site identified in the cocrystal is unlikely to be the one responsible for regulating rRNA transcription in E. coli was a necessary first step, but it is only the first step, in the solution to this problem.
Materials and Methods
RNA polymerases
Strains and plasmids are listed in Supplementary Table 1. Wild-type RNAPs were purified by standard procedures or by overexpression of core RNAP subunits as described below for the mutant RNAPs (see also ref. 25). No differences were observed among the wild-type preparations in any of our assays. We emphasize that enzymes purified without overexpression of RNAP subunits are sensitive to inhibition by ppGpp, and RNAP purified by overexpression of subunits is sensitive to inhibition by ppGpp so long as ω is co-overexpressed with the other core subunits in vivo or purified ω is reconstituted with the overexpressed core RNAP in vitro.25
Mutant RNAPs, with the exception of Q504E (see below), were purified by overproduction of α, β, and β′ from plasmids pIA299 or pIA333, carrying rpoA, rpoB, and rpoC under T7 promoter control.21 pIA299 encodes C-terminal hexahistidine-tagged β′, and pIA333 encodes N-terminal hexahistidine tagged β. Substitutions were made by oligonucleotide-directed mutagenesis, with silent restriction sites introduced adjacent to the mutations to facilitate screening. The mutagenized regions were always subcloned into unmutagenized pIA299 or pIA333, and the subcloned region was sequenced to confirm that only the desired change(s) were present. Plasmid DNAs were analyzed for the desired mutation at the time of induction.
RNAP purification after overexpression of RNAP subunits has been described.25 Briefly, the ω subunit was added in 10-fold molar excess to overexpressed core RNAP by incubation at 30°C for 30′. σ70 was added to overexpressed core RNAP in 2-fold molar excess by incubation at 30°C for 30′. For β′ K598A/K599A core RNAP, the mutant β′ was expressed with wild-type α and β from a pIA299 derivative in BL21λDE3, and ω was co-expressed from plasmid pCDFω.25 RNAP containing his-tagged β′ Q504E was purified as holoenzyme from a plasmid encoding only the β′ subunit.36 Since the other subunits were not overproduced in this case, supplementation with ω was not necessary.
Wild-type Thermus thermophilus RNAP was provided by D. Vassylyev (Univ. of Alabama-Birmingham), purified without overexpression as described previously.23 The presence of a protein migrating at the size expected for ω was confirmed by SDS-PAGE.
In vitro transcription assays
Since effects of substitutions in β and β′ were evaluated from assays for ppGpp function and not from assays measuring direct binding of ppGpp to RNAP, the transcription or filter-binding activity of a mutant RNAP is always expressed as a ratio to the activity of the same enzyme in the absence of ppGpp, and the effect of ppGpp on wild-type RNAP was always measured in parallel. Some mutant RNAPs (β E813A/E814A and β R1106A) were catalytically inactive, and others (β′ N458S and β′ Q929A) had reduced transcription activities even in the absence of ppGpp and/or produced shorter transcripts in addition to the full-length product under some solution conditions. Given the locations of these substitutions in RNAP and their roles in catalysis and/or elongation, these effects were not unexpected (see Discussion).
Multi-round transcription with E. coli RNAPs was carried out in 40 mM Tris-HCl pH 7.9, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, and 170 mM NaCl at 30°C.22 Concentrations of mutant RNAPs were adjusted to provide about the same amount of transcription from rrnB P1 as 10 nM wild-type RNAP, even though the concentration of wild-type RNAP did not affect the magnitude of the effect of ppGpp (data not shown). Reactions containing NTPs (200 μM ATP, GTP, and CTP; 10 μM UTP, and 1 μCi of [α-32P UTP]), 1 nM supercoiled plasmid template carrying the rrnB P1 and RNA-I promoters (pRLG6798, rrnB P1 endpoints -66 to +50; ref. 22), and 0–400 μM ppGpp (TriLink Biotechnologies) were initiated by addition of RNAP and stopped after 30 min by addition of an equal volume of urea stop buffer.25 Reactions were separated by gel electrophoresis and analyzed by phosphorimaging.
For T. thermophilus RNAP, multi-round transcription was performed on a supercoiled plasmid pRLG6770. This plasmid carries the RNA-I promoter and a T. thermophilus rRNA promoter and was constructed by annealing oligonucleotides with endpoints corresponding to T. thermophilus rRNA promoter sequence -70 to +15 (NCBI accession number AE017221; template strand sequence 1766683–1766768) and insertion into pRLG770.45 Reactions were carried out for 15 min at 65°C in transcription buffer containing 170 mM NaCl or for 15 min at 60°C in transcription buffer containing 100 mM NaCl (with the same results).
Promoter complex half-life assays
E. coli RNAP-promoter complex half-lives were measured by filter binding.12 lacUV5 promoter sequences corresponding to -59 to +40 with respect to the transcription start site were embedded within a DNA fragment with endpoints from -140 to +68. The fragment was excised from pRLG4264,45 end-labeled with α-32P TTP, and 8–30 nM RNAP was incubated with DNA for 17 min at 30°C in transcription buffer containing 100 mM KCl and 0–200 μM ppGpp. After heparin (Sigma) addition to 10 μg/ml, aliquots were removed at intervals and filtered through nitrocellulose discs. The filters were washed with 100 mM NaCl, 10 mM Tris-HCl pH 8.0, 1 mM EDTA, dried, and quantified by phosphorimaging. Half-lives at each ppGpp concentration were determined from semi-log plots of the fraction of filter-retained complex at each time point. Time zero was defined as 30 sec after heparin addition. In the absence of ppGpp, the lifetimes of the mutant complexes were always within twofold of the lifetime of the wild-type complex (Table 2). The relative effects of ppGpp were very reproducible (Table 2), but the absolute lifetimes of RNAP complexes varied slightly from day to day (presumably from slight changes in solution conditions). Therefore the effect of ppGpp on each mutant RNAP was always compared to the effect on WT RNAP in the same experiment.
Half-lives of the E. coli RNAP-promoter complexes reported in Table 3 were determined using transcription as a readout. The fraction of competitor-resistant complexes remaining at various times was determined essentially as described in ref. 12. 20 nM RNAP was added to 1 nM supercoiled plasmid template in 40 mM Tris-HCl pH 7.9, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 30 mM KCl, and 400 μM ppGpp (where indicated) at 22°C for 15 min. Heparin (40 μg/ml) was added at time zero, and 10 μl aliquots were removed at intervals from 10 sec to 20 min to a tube containing 200 μM ATP, 200 μM GTP, 200 μM CTP, 10 μM UTP, and 1 μCi [α-32P] UTP. After 15 min, reactions were stopped by addition of an equal volume of urea stop buffer, and the transcripts were separated by gel electrophoresis and quantified by phosphorimaging. Differences in the absolute lifetimes of the wild-type and mutant promoter complexes reported here and those reported previously14 are likely attributable to differences in solution conditions, as described previously.12
Lifetimes of complexes formed with T. thermophilus RNAP were also measured using transcription to monitor the fraction of complexes remaining at times after heparin (100 μg/ml) addition. Supercoiled plasmids carrying RNA-I (and/or other promoters) were incubated with RNAP for either 20 min in 40 mM Tris-HCl, pH 7.9, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, and 100 mM KCl at 55°C or 30 mM KCl at 65°C (with the same results) in the presence or absence of 1 mM ppGpp. After heparin addition, aliquots were removed at intervals to an NTP mixture (final 200 μM ATP, GTP, and CTP; 10 μM UTP, and 1 μCi [α-32P UTP]) for 15 min. Reactions were stopped by addition of an equal volume of urea buffer and analyzed on gels (see above).
Effects of DksA on responses of RNAPs to ppGpp
To evaluate effects of ppGpp in the presence of DksA (Fig. 4), low salt conditions were used to stabilize the promoter complex, resulting in only small effects of either DksA or ppGpp alone on transcription.4,8 DksA and ppGpp together strongly inhibited transcription under these conditions. Single-round transcription reactions contained 1 nM supercoiled plasmid template (pRLG6798, rrnB P1 endpoints -66 to +50) carrying the rrnB P1 and RNA-I promoters, 40 mM Tris-HCl pH 7.9, 10 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, and 30 mM NaCl at 30°C. RNAP was added for 20 min, followed by addition of heparin to 10 μg/ml, of ATP, GTP, and CTP (200 μM each), UTP (10 μM), and 1 μCi of [α-32P UTP] for 30 min.4 ppGpp was present at 100 μM when indicated. Different concentrations of DksA were used with different mutant RNAPs, since some of the substitutions affected DksA function (see text; [β′ Q504E, 2 μM DksA; β′ K598A/K599A and β′ R731A, 4 μM; β′ N458S, β′ E925A, β′ Q929A, and wild-type RNAP, 0.5 μM)]. Reactions were processed and analyzed as described above. (The catalytically-defective β E813A/E814A and β R1106A RNAPs could not be evaluated using this assay, β′ Q929A RNAP was inhibited too strongly by ppGpp alone under these conditions to allow synergistic effects of DksA to be evaluated, and β′ Q504Y RNAP and the triple mutant were not tested).
iNTP-ppGpp competition assays
Effects of 125–1000 μM ATP (the first NTP in the transcript) on rrnB P1 transcription were measured by multi-round transcription as described above ± 1 mM ppGpp. Reactions contained 50 μM GTP, 25 μM CTP, 10 μM UTP, 1 μCi of [α-32UTP], and ~10 nM wild-type RNAP. Effects of 0–800 μM ppGpp were determined by multi-round transcription from rrnB P1 at high (1500 μM) and low (125 μM) ATP concentration.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R37-GM37048 (to R.L.G.), a predoctoral fellowship from the Howard Hughes Medical Institute (to C.E.V.), NIH Molecular Biosciences Training Grant predoctoral fellowships (to S.T.R. and M.B.B.), and a Department of Bacteriology predoctoral fellowship (to S.P.H.). We thank D. Vassylyev for discussions during the early stages of this study and for the T. thermophilus RNAP used in Fig. 6. We also thank R. Landick, R. Ebright, R. Brennan, S. Darst, P. Rice, T. Steitz, and I. Artsimovitch for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cashel M, Gentry DR, Hernandez VH, Vinella D. The Stringent Response. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D.C: 1996. pp. 1458–1496. [Google Scholar]
- 2.Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 3.Mallik P, Paul BJ, Rutherford ST, Gourse RL, Osuna R. DksA is required for growth phase-dependent regulation, growth rate-dependent control, and stringent control of fis expression in Escherichia coli. J Bacteriol. 2006;188:5775–5782. doi: 10.1128/JB.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Bernardo LM, Johansson LU, Solera D, Skarfstad E, Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription. Mol Microbiol. 2006;60:749–764. doi: 10.1111/j.1365-2958.2006.05129.x. [DOI] [PubMed] [Google Scholar]
- 6.Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigma E, by guanosine 3′ 5′ bisphosphate, ppGpp. J Bacteriol. 2006;188:4589–4591. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorragic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- 8.Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma AK, Payne SM. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol Microbiol. 2006;62:469–479. doi: 10.1111/j.1365-2958.2006.05376.x. [DOI] [PubMed] [Google Scholar]
- 10.Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 11.Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 13.Barker MM, Gourse RL. Regulation of rRNA transcription correlates with NTP-sensing. J Bacteriol. 2001;183:6315–6323. doi: 10.1128/JB.183.21.6315-6323.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 15.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 16.Tedin K, Bremer H. Toxic effects of high levels of ppGpp are relieved by rpoB mutations. J Biol Chem. 1992;267:2337–2344. [PubMed] [Google Scholar]
- 17.Reddy PS, Raghavan A, Chatterji D. Evidence for a ppGpp binding site on Escherichia coli RNA polymerase: proximity relationship with the rifampicin-binding domain. Mol Microbiol. 1995;15:255–265. doi: 10.1111/j.1365-2958.1995.tb02240.x. [DOI] [PubMed] [Google Scholar]
- 18.Chatterji D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 19.Toulokhonov I, Shulgina I, Hernandez VJ. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and oocurs near the N-terminus of the β′-subunit. J Biol Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- 20.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 21.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 22.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 23.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 24.Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 25.Vrentas CE, Gaal T, Ross WE, Ebright RE, Gourse RL. Response of RNA polymerase to ppGpp: requirement for omega and relief of this requirement by DksA. Genes Dev. 2005;19:2378–2387. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Vassylyev DG, Svetlov V, Vassylyeva MN, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Artsimovitch I. Structural basis for transcription inhibition by tagetitoxin. Nat Struct Mol Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landick RL. NTP-entry routes in multi-subunit RNA polymerases. Trends Biochem Sci. 2005;30:651–654. doi: 10.1016/j.tibs.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Krasny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasai K, Nishizawa T, Takahashi K, Hosaka T, Aoki H, Ochi K. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J Bacteriol. 2006;188:7111–7122. doi: 10.1128/JB.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 34.Gaal T, Ross W, Estrem ST, Nguyen LH, Burgess RR, Gourse RL. Promoter recognition and discrimination by EsigmaS RNA polymerase. 2001;42:939–954. doi: 10.1046/j.1365-2958.2001.02703.x. [DOI] [PubMed] [Google Scholar]
- 35.Jin D, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 36.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 38.Givens RM, Lin MH, Taylor DJ, Mechold U, Berry JO, Hernandez VJ. Inducible expression, enzymatic activity, and origin of higher plant homologues of bacterial RelA/SpoT stress proteins in Nicotiana tabacum. J Biol Chem. 2004;279:7495–7504. doi: 10.1074/jbc.M311573200. [DOI] [PubMed] [Google Scholar]
- 39.Jores L, Wagner R. Essential steps in the ppGpp-dependent regulation of bacterial ribosomal RNA promoters can be explained by substrate competition. J Biol Chem. 2003;278:16834–16843. doi: 10.1074/jbc.M300196200. [DOI] [PubMed] [Google Scholar]
- 40.Sorensen MA, Jensen KF, Pedersen S. High concentrations of ppGpp decrease the RNA chain growth rate. Implications for protein synthesis and translational fidelity during amino acid starvation in Escherichia coli. J Mol Biol. 1994;236:441–454. doi: 10.1006/jmbi.1994.1156. [DOI] [PubMed] [Google Scholar]
- 41.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Svetlov V, Vassylyev DG, Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J Biol Chem. 2004;279:38087–38090. doi: 10.1074/jbc.C400316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Josaitis CA, Gaal T, Gourse RL. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci USA. 1995;92:1117–21. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross W, Gourse RL. Sequence-independent upstream DNA-alphaCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc. Natl. Acad. Sci. USA. 2005;102:1–296. doi: 10.1073/pnas.0405814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.