Abstract
The β-chemokine receptor CCR-5 is essential for the efficient entry of primary macrophage-tropic HIV-1 isolates into CD4+ target cells. To study CCR-5-dependent cell-to-cell fusion, we have developed an assay system based on the infection of CD4+ CCR-5+ HeLa cells with a Semliki Forest virus recombinant expressing the gp120/gp41 envelope (Env) from a primary clade B HIV-1 isolate (BX08), or from a laboratory T cell line-adapted strain (LAI). In this system, gp120/gp41 of the “nonsyncytium-inducing,” primary, macrophage-tropic HIV-1BX08 isolate, was at least as fusogenic as that of the “syncytium-inducing” HIV-1LAI strain. BX08 Env-mediated fusion was inhibited by the β-chemokines RANTES (regulated upon activation, normal T cell expressed and secreted) and macrophage inflammatory proteins 1β (MIP-1β) and by antibodies to CD4, whereas LAI Env-mediated fusion was insensitive to these β-chemokines. In contrast soluble CD4 significantly reduced LAI, but not BX08 Env-mediated fusion, suggesting that the primary isolate Env glycoprotein has a reduced affinity for CD4. The domains in gp120/gp41 involved in the interaction with the CD4 and CCR-5 molecules were probed using monoclonal antibodies. For the antibodies tested here, the greatest inhibition of fusion was observed with those directed to conformation-dependent, rather than linear epitopes. Efficient inhibition of fusion was not restricted to epitopes in any one domain of gp120/gp41. The assay was sufficiently sensitive to distinguish between antibody- and β-chemokine-mediated fusion inhibition using serum samples from patient BX08, suggesting that the system may be useful for screening human sera for the presence of biologically significant antibodies.
Penetration of the HIV-1 into target cells involves the fusion of the virus envelope (Env) with the cell membrane. Similary, cell-to-cell fusion of infected cell membranes with those of other cells, which results in the formation of syncytia, requires the expression of the viral Env on the infected cell surface. The HIV-1 Env glycoprotein (gp160) comprises an external surface domain (gp120) and a transmembrane anchor domain (gp41) which remain associated in a noncovalent fashion. The primary receptor for HIV is CD4, a differentiation marker expressed on the surface of T lymphocytes, dendritic cells and macrophages (1). It appears that binding of HIV to CD4 induces conformational rearrangements in the gp120/gp41 molecule (2, 3) that lead to fusion of the virus and cell membranes (2) by a process that is mediated by the N-terminal fusion peptide of gp41. Recent evidence has indicated that different chemokine receptors collaborate with CD4 to facilitate HIV-1 entry into CD4+ target cells. T cell line-adapted (TCLA) strains and T-cell-tropic, “syncytium-inducing,” primary isolates use CXCR-4 (LESTR, fusin) as their coreceptor (4, 5), whereas macrophage-tropic, so-called “nonsyncytium-inducing” primary isolates principally use CCR-5 (5–10). These receptors belong to the family of G protein-coupled, seven-transmembrane-domain proteins. It has been shown that expression of CCR-5 renders CD4+ cells capable of being infected by the macrophage-tropic HIV-1 strains ADA, Bal (9), JR-FL, or SF162 (10). Furthermore, SF162 can cause the formation of small syncytia in CCR-5+ CD4+ HeLa cells (10). Similarly, cell-to-cell fusion can be demonstrated by coculture of permissive cells with HeLa cells expressing the Env glycoproteins of JR-FL (9). The β-chemokines macrophage inflammatory protein 1β (MIP-1β) and RANTES (regulated upon activation, normal T cell expressed and secreted), which are potent agonists of CCR-5, competitively inhibit infection of target cells by primary HIV-1 isolates at the virus entry stage, and can block cell-to-cell fusion (8, 9, 11). Although Env-mediated cell-to-cell fusion has now been demonstrated for macrophage-tropic nonsyncytium-inducing primary isolates, little is known as yet concerning the requirements for this phenomenon, particularly with respect to its sensitivity to different anti-Env antibodies.
To study the requirements for HIV-1 Env-mediated fusion, we have developed an assay system based on the use of Semliki Forest virus (SFV) recombinants that express the Env gp120/gp41 either from the TCLA HIV-1LAI strain or from a primary macrophage-tropic, isolate HIV-1BX08 (12). The abilities of the Env proteins expressed by the recombinants to induce syncytium formation in HeLa CD4+, or HeLa CD4+/CCR-5+ cells were examined in the presence and absence of a variety of potential inhibitors of HIV infection. The results of this study are presented below.
MATERIALS AND METHODS
Sources of Reagents.
Recombinant human β-chemokines and anti-β-chemokine mAbs were purchased from R & D Systems. Soluble CD4 was obtained from American Biotechnologies, Cambridge, MA). The mAbs that were used were provided by the indicated sources or colleagues, or came from the reagent repository of the Medical Research Council (MRC), or were purchased from commercial suppliers. Primary citations for these mAbs and/or references to previous characterizations of them are as follows: CRA-1 [MRC (13)]; IgG1b12 [D. Burton (14–16)]; CD4-IgG2 [Progenics Pharmaceuticals, Tarrytown, NY (17, 18)]; 12.22.F5.C4 anti-CD4 antibody [MRC (19)]; 2G12 and 2F5 [H. Katinger (20)]; 447-D (16–18), 697-D (21, 22), 670-D (21) and 694/98D (23) (Cellular Products); F105 and F240 [L. Cavacini (24, 25)]; 62C [MRC (26)]; 1121 (Agmed, Bedford, MA); and K24, F5-5, and 41A (Hybridolab, Institut Pasteur). D7324 is a polyclonal sheep antibody from Aalto-Bio Reagents (Dublin) (27).
CCR-5 Retroviral Vector.
The CCR-5 gene was inserted into the Moloney murine leukemia virus (MoMLV)-derived retroviral vector LXSH (28) to generate plasmid LR5. The LXSH vector encodes the gene for hygromycine resistance and allows expression of transgenes under the control of the MoMLV LTR. The CCR-5 insert was obtained by PCR amplification using Pfu DNA polymerase (Stratagene) (40 thermal cycles at 92°C for 15 sec, 50°C for 1 min, 75°C for 2 min). Primers used were as follows: 5X-CCR5, 5′-CTAGCGCTCGAGATGGATTATCAAGTGTC-3′; and 3B-CCR5, 5′-CGGGATCCGTCACAAGCCCACAGATA-3′.
XbaI and BamHI restriction sites were included in the primers to allow cloning of the PCR product into LXSH. Retroviral particles carrying the LR5 vector were obtained by electroporation of the CRIP packaging cell line (29). Because Hela cells are poorly susceptible to infection by amphotropic murine leukemia virus-derived particles, vector particles were pseudotyped with the highly fusogenic vesicular stomatitis virus (VSV) Env. Fifteen micrograms of both the CCR-5 vector plasmid and pCMV–VSV–G was used to electroporate 107 CRIP cells with a single pulse of 280 V and 960 μF in serum-free DMEM. CCR-5 vector particles were collected 48 h postelectroporation.
Cell Lines.
P4 indicator cells are HeLa–CD4+ cells that carry the lacZ gene under the control of the HIV-1 LTR (30). Because the HIV-1 coreceptor CXCR-4 is expressed on the surface of HeLa cells, P4 cells may be infected by T cell-adapted HIV-1 strains and by syncytium-inducing HIV-1 primary isolates. P4 cells were grown in DMEM (GIBCO) supplemented with 10% fetal calf serum and 500 μg/ml of G418 (GIBCO).
P4 cells were infected with CCR-5 vector particles in the presence of 15 μg/ml of DEAE-dextran. After hygromycine selection, the transduced P4P cell population was tested for its permissivity to infection with the CCR-5-dependent macrophage-tropic molecular clone NLAD8 (a gift from E. O. Freed, National Institutes of Health). P4P cells were grown in DMEM (GIBCO) supplemented with 10% fetal calf serum, 500 μg/ml of G418, and 150 μg/ml of hygromycine B (Sigma). All fusion assays were performed on P4P cells that had been passaged less than four times.
SFV Recombinants.
The Env-encoding gene from HIV-1BX08 or HIV-1LAI was subcloned from plasmids 133-3 (a gift from B. Rovinski; Willowdale, ON, Canada) and pBru respectively into the pSFV1 plasmid (31) as described by Paul et al. (32) to give recombinants pSFV–BX08 and pSFV–LAI. Recombinant pSFV DNA was linearized by SpeI and purified using the Wizard PCR Kit (Promega). Transcription reactions were carried out as described (31, 32). Electroporations were performed on BHK-21 cells (107 cells/ml) in PBS. To synthesize recombinant virus, 800 μl of cells were mixed with the transcribed RNA [20 μl (1 μg) of SFV–Helper 2 RNA transcripts plus 20 μl (1 μg) of SFV–LAI, SFV–BX08, or SFV–LacZ RNA transcripts]. Electroporation was carried out at room temperature in a 0.4 cm electroporation cuvette using a Bio-Rad Gene Pulser, with two pulses of 830V/25 μF at maximum resistance. Cells were diluted 20-fold in complete DMEM and transferred to tissue culture plates. At 24 h posttransfection the supernatant was harvested and clarified by centrifugation for 20 min at 4°C, and the viruses were pelleted by ultracentrifugation (SW28 rotor, 1.5 hr at 26 000 rpm, 4°C). The viral pellet was resuspended in 50 mM Tris⋅HCl (pH 7.4), 100 mM NaCl, and 0.5 mM EDTA and frozen at −80°C. Before infection, particles were activated with chymotrypsin as described (33). Virus stocks were titered on BHK cells, infection was detected by immunohistostaining with mAbs 41A and F5.5 (1 μg/ml) as primary antibodies, followed by biotinylated anti-mouse IgG then avidin and biotinylated horseradish peroxidase H (Vectastain ABC kit; Vector Laboratories).
Syncytium Assays.
P4 (CCR-5−) or P4P (CCR-5+) HeLa cells, at a density of 2 × 105 cells per well in a 24-multiwell plate, were washed with DMEM without serum and infected with defective recombinant SFV–LAI, SFV–BX08, or SFV–LacZ at a multiplicity of infection of 3.6 infectious particles per cell (as measured on BHK cells). The SFV inoculum was removed 30 min after infection by aspiration and the cells covered with DMEM supplemented with 5% fetal calf serum. After 12 h incubation at 37°C, cells were fixed with 0.5% (vol/vol) glutaraldehyde and stained with Giemsa. The syncytia present were counted from triplicate experiments using a grid composed of 8 squares (1/7th of the total well surface) distributed evenly throughout the well. Total number of syncytia counted in control wells was between 400 and 1,000. Inhibitors were added 30 min after infection. The percent inhibition, compared with control wells, was calculated as a function of the concentration of the specific inhibitors. Each figure for inhibition of syncytium formation was calculated from at least three independent assays.
RESULTS AND DISCUSSION
Syncytium Induction by a Macrophage-Tropic Primary HIV-1 Env Protein Requires CD4 and CCR-5.
The SFV vector was used to express HIV gp160 because this system offers the major advantage of allowing studies without manipulation of infectious HIV. An additional safety feature is that the recombinant SFV is defective, so only a single round of infection can be initiated in target cells. Thus, defective SFV recombinants (31–33) were constructed to carry the unmodified, full-length env gene from either the TCLA syncytium-inducing HIV-1LAI strain, or from a primary HIV-1BX08 isolate in place of the SFV structural proteins (Fig. 1; see Materials and Methods). HIV-1BX08 was isolated in 1992 from a French seropositive individual (BX08) who had been infected for less than 8 months. The amino acid sequence of the V3 loop region in the gp120 of this primary isolate is homologous to the consensus sequence found amongst French clade B HIV-1 isolates (12). The BX08 virus grows in primary peripheral blood cells, but not T cell lines (12). As target cells for fusion studies, we used the previously characterized P4 HeLa-CD4+ cells (CD4+ CXCR-4+, HIV-1 LTR–LacZ) (30). For these studies, we also constructed the P4P cell line by transduction of P4 cells with a retroviral expression vector (L.CCR5.5H) carrying the cDNA of human CCR-5 (see Materials and Methods). The P4P cell line thus expresses CD4 and both known coreceptors of HIV and is readily susceptible to infection by both T cell and macrophage-tropic HIV-1 strains.
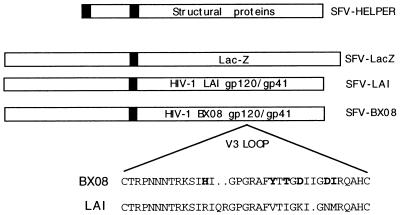
Figure 1.
Defective SFV recombinants were constructed by substitution of the sequence of the SFV structural proteins with those of the Escherichia coli β-galactosidase gene (lacZ) or those of the env gene from either HIV-1LAI or HIV-1BX08. Stocks of recombinant viruses were obtained by cotransfection of the hybrid constructs with a helper SFV mutant bearing a mutation in the virus spike gene, which is incapable of encapsidation (33). The amino acid sequence of the V3 loops of HIV-1BX08 and HIV-1LAI are indicated in the Inset.
The capacity of the LAI and BX08 Env glycoproteins to induce cell-to-cell fusion was examined by infecting P4, or P4P cells with each of the SFV recombinants (Fig. 2). Although both recombinant SFV preparations synthesized gp120/gp41 as judged by Western blot analysis (data not shown), or by immunohistostaining with mAb 2G12 (Fig. 2), only the glycoprotein from the HIV-1LAI strain induced significant cell-to-cell fusion in P4 cells. By contrast, in P4P (CCR-5+) cells, the formation of syncytia was as pronounced in the SFV–BX08-infected cells as in the SFV–LAI-infected cells (Fig. 2). These data confirm that the gp120/gp41 from a macrophage-tropic primary HIV-1 isolate, which behaves as an nonsyncytium-inducing virus in T cell lines, can be as fusogenic as a virus representing an syncytium-inducing TCLA strain in cells that express the appropriate coreceptor (5, 34). The multiplicity of infection (moi) used for these preliminary experiments resulted in the productive infection of 1 cell in 5, as judged by immunohistostaining (Fig. 2). Interestingly, while the number and size of syncytia observed per well increased markedly with higher moi, syncytium formation with either SFV–LAI or SFV–BX08 was abrogated in experiments where all the target cells were infected (moi exceeding 10 infectious SFV particles per cell, data not shown). These results suggest that down-regulation of cell surface receptor expression, which is typical of heavily HIV-1-infected cell populations ex vivo also occurred in a virus-free system that uses CD4+ HeLa cells and Env proteins expressed by SFV recombinants.
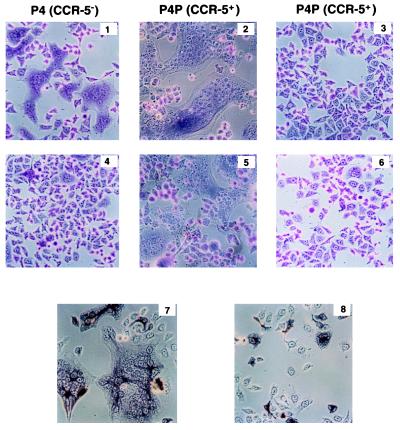
Figure 2.
Formation of syncytia in CD4+ HeLa cells expressing CCR-5 and gp120/gp41 from a macrophage-tropic primary HIV-1 isolate. P4 (CCR-5−) (1, 4, 7, and 8) or P4P (CCR-5+) (2, 3, 5, and 6) HeLa cells (as indicated) were infected with defective recombinant SFV-LAI (1, 2, and 7), SFV-BX08 (4, 5, and 8) or SFV-LacZ (3). Control cells were left uninfected (6). After 12 h incubation at 37°C, cells were fixed with 0.5% (vol/vol) glutaraldehyde and stained with Giemsa (1-6) or reacted with mAb 2G12 (7 and 8) as primary antibody, followed by biotinylated anti-mouse IgG then avidin and biotinylated horseradish peroxidase H (Vectastain ABC kit). The cells were examined by microscopy. (×125.)
Sensitivity of Fusion to Soluble CD4 (sCD4) and the Natural Ligands of CCR-5.
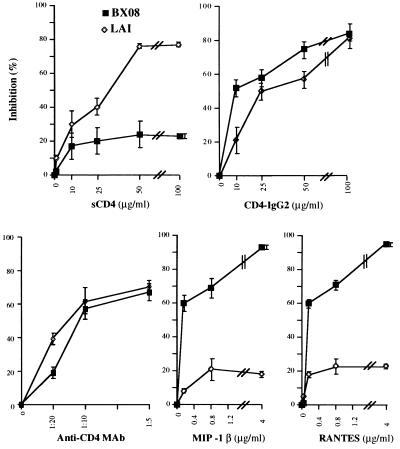
To characterize further the requirements for syncytium formation, we examined the capacities of the β-chemokines RANTES and MIP-1β (natural ligands of CCR-5), and of sCD4, to inhibit cell-to-cell fusion mediated by the two HIV-1 Env proteins. Toward this end, a range of concentrations of the potential inhibitors was included in the culture medium of SFV-infected P4 or P4P cells. As expected from recent reports demonstrating that β-chemokines block entry of macrophage-tropic-HIV-1 (8, 35, 36). BX08 Env-, but not LAI Env-mediated fusion was almost completely blocked by MIP-1β or RANTES and in a dose-dependent manner (Fig. 3). Antibodies to CD4 reduced the fusion mediated by both Env proteins, as expected if the interaction of a primary Env with CCR-5 was facilitated by its interaction with CD4, as suggested recently (37, 38). The addition of sCD4 dramatically reduced gp120/gp41 LAI-mediated fusion. It had, however, relatively little effect on fusion induced by gp120/gp41 BX08. This observation correlates well with earlier studies showing that infection by primary HIV-1 isolates is relatively insensitive to sCD4 (39). However, it was recently demonstrated that sCD4 efficiently blocks primary isolate gp120-mediated inhibition of MIP-1β binding to activated T cells (38). This apparent discrepancy in the reported effect of sCD4 may reflect the different affinities for sCD4 of cell surface-expressed gp120/gp41 (as used in the present study), and the purified monomeric gp120 used by Moore and colleagues (38). Inhibition of BX08 Env-mediated formation of syncytia by tetrameric CD4–IgG2 at equivalent concentrations confirmed that BX08 Env-mediated fusion in P4P cells was indeed CD4-dependent (Fig. 3) and reflects the greater avidity of tetravalent CD4–IgG2 (as compared with sCD4) for surface expressed gp120/gp41 oligomers (17, 18).
Figure 3.
Syncytium formation inhibition assay. P4P HeLa cells were infected with either SFV–LAI or SFV–BX08 as described. At 30 min after infection, the SFV inoculum was removed and the cells were incubated in medium containing the indicated β-chemokines, anti-CD4 mAb (12.22.F5.C4), sCD4, or CD4–IgG2 at increasing concentrations (final concentrations in the medium are indicated in μg/ml, except for mAb anti-CD4 where numbers refer to dilution of ascites fluids).
Inhibition of Syncytium Formation by Antibodies to gp120/gp41.
The domains in gp120/gp41 involved in the interaction with CD4, and/or CCR-5, were probed using antibodies that recognize linear or conformation-dependent epitopes of Env. Previous studies on antibody neutalization of HIV-1 have shown that neutralizing antibodies may be separated into several classes. One class reacts with epitopes corresponding to linear amino acid sequences in the V3 loop of gp120. These antibodies neutralize TCLA strains very efficiently in a type-specific manner. Studies with other mAbs have shown that the V1/V2 domain of gp120 contains multiple neutralization epitopes, including linear, conformational, or glycan-dependent species. A third class of neutralizing antibodies reacts with conformational structures of the CD4 binding site on gp120. These latter antibodies appear late in the course of disease and show broad, group-specific neutralizing activities. A fourth, potentially important, group-specific antibody, is that reported by Muster et al. (40) which is directed against the ecto-domain of gp41 and which neutralizes many type B isolates of HIV-1.
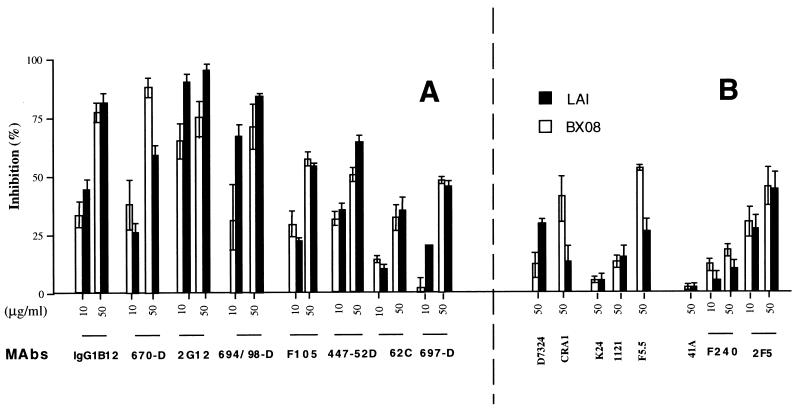
To gain an insight into the topology of the BX08 gp120/gp41–CD4/CCR-5 interactions that lead to cell-to-cell fusion, we tested a panel of mAbs whose epitopes have been previously defined, and monitored their abilities to inhibit the formation of syncytia in P4P cells infected with either SFV–LAI, or SFV–BX08 recombinants. As shown in Fig. 4, the greatest levels of inhibition of both LAI and BX08 Env-mediated fusion were observed with antibodies that recognize discontinuous, conformation-dependent epitopes (Fig. 4A). For example, an inhibition of syncytium formation of greater than 70% was observed for either gp120/gp41 LAI, or gp120/gp41 BX08, using human mAb 2G12, which is known to recognize a conformation-dependent epitope in the C3–V4 region of gp120. This site is destroyed by amino acid substitutions affecting N-linked glycosylation sites near the bases of the V3 and the V4 loops (38). mAb 2G12 has also been shown to reverse gp120-induced blocking of 125I-labeled MIP-1β/CCR-5 interactions (38). Similarly, an inhibition of syncytium formation of more than 70% was observed with mAbs 670-D (anti-gp120 C-terminus), 694/98-D (anti-V3 loop) (21), and IgG1b12 (14–16). mAb 670-D recognizes gp120 on the surface of cells infected with syncytium-inducing and nonsyncytium-inducing primary isolate HIV-1 clades A through E, whereas mAb 694/98-D is specific for the GRAF motif at the tip of the V3 loop in clade B viruses (15, 21, 23). IgG1b12 recognizes discontinuous epitopes that overlap the CD4-binding site on gp120 (15, 18) and neutralizes most primary isolates at a concentration of 1 μg/ml (15). Inhibition of fusion with mAb F105 (anti-CD4 binding site) and 447-52D (anti-V3 loop) was less dramatic than with the other mAbs, but was reproducibly in excess of 50% (Fig. 4). mAbs IgGb12, 2G12, F105, and 447-52D have previously all been shown to neutralize primary clade B HIV-1 isolates (40, 41). Interestingly, significant effects were also observed with mAb 697-D (anti-V2 loop) on both LAI and BX08 Env-induced fusions, although 50% fusion inhibition was not reproducibly attained with this antibody. mAb 697-D has previously been reported to neutralize primary but not laboratory isolates HIV-1 (21, 22), but has no competitive effect against a primary monomeric gp120 in the 125I-labeled MIP-1β competition assay (38). It is noteworthy that most of the conformation-dependent mAbs are known to recognize epitopes that have been mapped to the neutralizing face of monomeric gp120 (42). These inhibited gp120/gp41 BX08-mediated cell-to-cell fusion efficiently. One exception was mAb 62c (26), which reacts with the V2 domain. This antibody has previously been shown to possess no neutralizing activity against either laboratory or wild HIV-1 strains (26).
Figure 4.
Inhibition of syncytium formation by antibodies to gp120/gp41. P4P HeLa cells were infected with either SFV–LAI or SFV–BX08 as described. At 30 min after infection, the SFV inoculum was removed and the cells incubated in medium containing the indicated antibodies. (A) mAbs targeted to conformational epitopes. (B) mAbs targeted to linear epitopes (final concentrations in μg/ml, as indicated).
Most anti-HIV-1 mAbs that recognize linear epitopes on gp160 that were tested failed to significantly block fusion induced by either HIV-1LAI, or HIV-1BX08 gp120/gp41 at the concentrations used (Fig. 4B) (examples were mAb 1121, an anti-V3 MN mAb that neutralizes laboratory strains of HIV-1 at a concentration of 10 μg/ml; mAb K24, an anti-V3 SC mAb that neutralizes many laboratory strains including HIV-1MN, HIV-1LAI, and HIV-1SF2 at concentrations of 100 μg/ml; mAb 41A, an anti-gp41 LAI mAb that has no neutralizing activity against HIV-1LAI; and F240, an anti-gp41 mAb). Similarly, a sheep hyperimmune serum (D7324), which contains antibodies to the C5 region of gp120, was inefficient in the fusion inhibition assay. D7324 has previously also been shown to be ineffective in the 125I-labeled MIP-1β competition assay (38) and appears to be specific to the nonneutralizing face of gp120 (42). Although some antibodies targeted to linear epitopes exhibited a detectable inhibitory activity in our assay (Fig. 4B), such reactions never reached the levels observed with the best conformation-dependent mAbs. Examples include mAb 2F5, a human neutralizing mAb, which has previously been shown to recognize a conserved immunodominant epitope in gp41 (40), mAb F5.5 (an anti-V3 MN mAb that neutralizes HIV-1MN and HIV-1LAI) and mAb CRA-1 (an anti-gp120-C terminus mAb). Interestingly, mAbs F5.5 and CRA-1 inhibited BX08 Env-mediated fusion to a greater extent than LAI Env-mediated fusion. It is possible that the higher activity observed for mAb F5.5 is due to the unusually high affinity of this antibody for the relevant V3 peptide (KD of 4 × 10−11 M for the GPGRAFYT HIV-1MN peptide, as opposed to 3 × 10−9 M for the equivalent sequence of HIV-1LAI (François Traincard, personal communication). This differential affinity might explain why mAb F5.5 had no effect on LAI gp120/gp41-mediated fusion (see Conclusion). We cannot explain the moderate inhibition of fusion observed with mAb CRA-1. This mAb exhibited no detectable activity in the 125I-labeled MIP-1β competition assay using gp120 from HIV-1JRFL (38). The epitope recognized by mAb CRA-1 has been mapped to the nonneutralizing face of gp120 (42). However it is possible that this epitope is exposed on gp120/gp41 expressed on the cell surface but not on the monomeric gp120 used in the previous studies, or is exposed following interaction of gp120/gp41 with CD4 and/or CCR-5.
Detection of Two Fusion-Inhibitory Activities in Human Serum Samples.
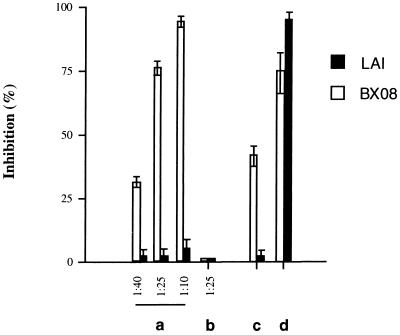
We also examined the effect of a serum sample obtained from the patient from whom the BX08 virus strain was isolated. Serum obtained 8 months after seroconversion inhibited gp120/gp41 BX08-induced fusion in a dose-dependent fashion, but was without effect in assays using gp120/gp41 LAI (Fig. 5). No detectable inhibition was observed in control experiments using standard HIV-1 seronegative serum samples.
Figure 5.
Syncytium inhibition assay with serum from patient BX08. P4P HeLa cells were infected with either SFV–LAI or SFV–BX08 as described. At 30 min after infection, the SFV inoculum was removed and the cells were incubated in medium containing the serum from patient BX08 diluted as indicated (lane a); the serum from patient BX08 diluted at 1:25 plus 10 μg/ml each of anti-RANTES, anti-MIP-1α, and anti-MIP-1β mAbs (lane c); or 10 μg/ml each of anti-RANTES, anti-MIP-1α, anti-MIP-1β mAbs plus 50 μg/ml mAb 2G12 (lane d). (Lane b) Control seronegative human serum.
Because the β-chemokines RANTES, MIP-1α, and MIP-1β have been identified as HIV-suppressive factors that are secreted by CD8+ T lymphocytes in HIV-positive individuals (11), their possible roles in syncytium inhibition in BX08 serum were determined by repeating the fusion-inhibition experiment in the presence or absence of a mixture of antibodies to RANTES, MIP-1α, and MIP-1β. Antibody concentrations of 10 μg/ml reduced BX08 serum inhibitory activity by about 50% (Fig. 5). However, their presence in the fusion assay did not affect fusion inhibition with human mAb 2G12, suggesting that their effects were specific. The concentration of anti-chemokine antibodies used was sufficient to totally reverse β-chemokine-mediated inhibition of BX08 fusion using 160 ng/ml of MIP-1β and RANTES (data not shown). The concentration of RANTES in the BX08 serum, as measured by ELISA, was 50 ng/ml, a concentration that correlates well with the level of anti-chemokine antibody-sensitive inhibition observed in this experiment and with those reported in several previous studies (11, 43, 44). Thus, the inhibitory activity of the patient’s serum on BX08 env-mediated cell-to-cell fusion was most likely due to the mixed effect of the presence of β-chemokines and antibody.
CONCLUSIONS
Here we have described a sensitive assay system for the analysis of CCR-5-dependent, CD4+-dependent cell-to-cell fusion mediated by Env proteins from a primary HIV-1 isolate. Because HeLa cells naturally express CXCR-4, the ability of Env proteins from primary isolates to induce fusion in P4 or P4P cells allows their rapid characterization as T, macrophage-, or dual-tropic. It is worth noting that the fusion assay described here mimics the entry of HIV-1 particles into target cells. Syncytium formation in our system was inhibited to the greatest extent by mAbs to conformation-dependent epitopes, rather than by the mAbs to linear epitopes that were tested in this study. Because several of these antibodies to linear epitopes have previously been shown to neutralize some HIV-1 isolates, the apparent lack of fusion inhibitory activity observed here might imply that they act at a step subsequent to fusion. However, several other antibodies to linear determinants (especially in the V3 loop) have previously been proven also to be powerful inhibitors of fusion (see, for example, ref. 45). Thus it is likely that the lack of activity of certain of our mAbs results from a restricted-type specificity, and that inhibition of fusion would indeed be possible with isolate-specific, high affinity antibodies to linear epitopes. Nevertheless, it has recently been shown that conformation-dependent neutralization epitopes recognized by antibodies such as IgG1b12 are major epitopes on virions and infected cells, in contrast to recombinant gp120 on which weakly neutralizing epitopes predominate (46). Interestingly, we detected significant neutralizing activity with certain mAbs (e.g., CRA-1 and 697-D) that had no activity in the 125I-labeled MIP-1β competition assay (46). We believe that these differences highlight the importance of using native oligomeric surface-expressed gp160 from primary isolates as neutralization targets rather than the monomeric, soluble forms of gp160 or gp120 that have been used predominantly to date. In this respect it is interesting to note that all conformation-dependent mAbs capable of cross-clade recognition of surface expressed gp120/gp41 (21) that we tested were efficacious in the fusion inhibition assay.
In addition, the fusion inhibition assay is sufficiently sensitive not only to detect but also to differentiate between Ab- and β-chemokine mediated inhibition with a human serum sample. The finding that almost 50% of fusion inhibitory activity with the BX08 serum could be attributed to β-chemokines underscores the risk that serum neutralization titers determined on primary virus isolates grown on peripheral blood mononuclear cell by classical infection inhibition assays might be artificially elevated. We suggest that IgG purified from serum samples might provide a more reliable means of determining Ab-mediated inhibition of fusion with primary HIV-1 isolates.
Finally, given the sensitivity of the assay with human sera, it will be interesting to compare the syncytium inhibition activity with the neutralizing activity of reference serum samples and to screen sera from vaccinated volunteers and animals. Toward this end, we are currently expanding the approach described in this paper by expressing a number of Env proteins of primary isolates from other HIV-1 clades. The ability to distinguish between antibody- and β-chemokine-mediated inhibition in such samples (using anti-β-chemokine antibodies) should prove useful in the quest for correlates of vaccine-induced protection and of HIV-1 disease progression.
Acknowledgments
We are grateful to Katherine M. Kean and David Bishop for their interest and advice during this work and for critical comments regarding the manuscript. We also thank François Clavel for his interest in the project and Vesna Mimic for excellent technical assistance. We are also grateful to H. Katinger, L. Cavacini, G. P. Allaway, P. Maddon, and D. Burton for donation of mAbs. F.C.V. was supported by the Fondation Merieux; A.M.B. was supported by a Bourse Roux fellowship from the Institut Pasteur. This work was supported by grants from the Agence Nationale de Recherches sur le SIDA, the Centre National de la Recherche Scientifique, and the Institut Pasteur.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: Env, envelope; TCLA, T cell line-adapted; RANTES, regulated upon activation, normal T cell expressed and secreted; MIP-1, macrophage inflammatory proteins 1; SFV, Semliki Forest virus; sCD4, soluble CD4.
References
- 1.Robey E, Axel R. Cell. 1990;60:697–700. doi: 10.1016/0092-8674(90)90082-p. [DOI] [PubMed] [Google Scholar]
- 2.Sattentau Q J, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 5.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 8.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 9.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 10.Kozak S L, Platt E J, Madani N, Ferro F E, Peden K, Kabat D. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 12.Moog C, Spenlehauer C, Fleury H, Heshmati F, Saragosti S, Letourneur F, Kirn A, Aubertin A M. AIDS Res Hum Retroviruses. 1997;13:19–27. doi: 10.1089/aid.1997.13.19. [DOI] [PubMed] [Google Scholar]
- 13.Page M, Vella C, Corcoran T, Dilger P, Ling C, Heath A, Thorpe R. AIDS. 1992;6:441–446. doi: 10.1097/00002030-199205000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W, Sawyer L S, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Cao Y, Moore J P, Ho D D, Barbas C F., III Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 15.Roben P, Moore J P, Thali M, Sodroski J, Barbas C F r, Burton D R. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton D R, Barbas C F d, Persson M A, Koenig S, Chanock R M, Lerner R A. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore J P, McKeating J A, Huang Y X, Ashkenazi A, Ho D D. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas C F r, Burton D R, Ho D D. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilks D, Walker L, O’Brien J, Habeshaw J, Dalgleish A. Immunology. 1990;71:10–15. [PMC free article] [PubMed] [Google Scholar]
- 20.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinper H. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 21.Zolla-Pazner S, O’Leary J, Burda S, Gorny M K, Kim M, Mascola J, McCutchan F. J Virol. 1995;69:3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorny M K, Moore J P, Conley A J, Karwowska S, Sodroski J, Williams C, Burda S, Boots L J, Zolla-Pazner S. J Virol. 1994;68:8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorny M K, Conley A J, Karwowska S, Buchbinder A, Xu J Y, Emini E A, Koenig S, Zolla-Pazner S. J Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavacini L A, Emes C L, Power J, Underdahl J, Goldstein R, Mayer K, Posner M R. J AIDS. 1993;6:1093–1102. [PubMed] [Google Scholar]
- 25.Cavacini L A, Emes C L, Power J, Desharnais F D, Duval M, Montefiori D, Posner M R. J Immunol. 1995;155:3638–3644. [PubMed] [Google Scholar]
- 26.Shotton C, Arnold C, Sattentau Q, Sodroski J, McKeating J A. J Virol. 1995;69:222–230. doi: 10.1128/jvi.69.1.222-230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, et al. Nature (London) 1985;313:277–84. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 28.Miller A D, Rosman G J. BioTechniques. 1989;7:980–982. , 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 29.Danos O, Mulligan R C. Proc Natl Acad Sci USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charneau P, Mirambeau G, Roux P, Paulous S, Buc H, Clavel F. J Mol Biol. 1994;241:651–662. doi: 10.1006/jmbi.1994.1542. [DOI] [PubMed] [Google Scholar]
- 31.Liljestrom P, Lusa S, Huylebroeck D, Garoff H. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul N L, Marsh M, McKeating J A, Schulz T F, Liljestrom P, Garoff H, Weiss R A. AIDS Res Hum Retroviruses. 1993;9:963–970. doi: 10.1089/aid.1993.9.963. [DOI] [PubMed] [Google Scholar]
- 33.Berglund P, Sjoberg M, Garoff H, Atkins G J, Sheahan B J, Liljestrom P. Bio/Technology. 1993;11:916–920. doi: 10.1038/nbt0893-916. [DOI] [PubMed] [Google Scholar]
- 34.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arenzanaseisdedos F, Virelizier J L, Rousset D, Clarklewis I, Loetscher P, Moser B, Baggiolini M. Nature (London) 1996;383:400. doi: 10.1038/383400a0. [DOI] [PubMed] [Google Scholar]
- 36.Schmidtmayerova H, Sherry B, Bukrinsky M. Nature (London) 1996;382:767. doi: 10.1038/382767a0. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A, Desjardin E, Newman W, Gerard C, Sodroski J. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 38.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Chengmayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 39.Daar E S, Li X L, Moudgil T, Ho D D. Proc Natl Acad Sci USA. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posner M R, Cavacini L A, Emes C L, Power J, Byrn R. J AIDS. 1993;6:7–14. [PubMed] [Google Scholar]
- 42.Moore J P, Sodroski J. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cocchi F, Devico A L, Garzinodemo A, Cara A, Gallo R C, Lusso P. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 44.Paxton W A, Martin S R, Tse D, O’Brien T R, Skurnick J, VanDevanter N L, Padian N, Braun J F, Kotler D P, Wolinsky S M, Koup R A. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 45.Scott C F, Silver S, Profy A T, Putney S D, Langlois A, Weinhold K, Robinson J E. Proc Natl Acad Sci USA. 1990;87:8597–8601. doi: 10.1073/pnas.87.21.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parren P, Fisicaro P, Labrijn A F, Binley J M, Yang W P, Ditzel H J, Barbas C F, Burton D R. J Virol. 1996;70:9046–9050. doi: 10.1128/jvi.70.12.9046-9050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]