Abstract
Among the four subtypes of Hodgkin disease (HD), lymphocyte-predominant (LP) HD is now generally considered as a separate entity. The B cell nature of the typical Hodgkin and Reed–Sternberg (HRS) cells and their variants (L and H, lymphocytic and histiocytic cells) in LP HD has long been suspected, but the question of whether these cells represent a true tumor clone is unclear. We previously demonstrated clonal Ig gene rearrangements in one case of LP HD. In the present study, five cases of LP HD were analyzed by micromanipulation of single HRS cells from frozen tissue sections and DNA amplification of rearranged Ig heavy chain genes from those cells. Clonal V gene rearrangements harboring somatic mutations were detected in each case. In three cases ongoing somatic mutation was evident. This shows that HRS cells in LP HD are a clonal tumor population derived from germinal center B cells. The pattern of somatic mutation indicates that HRS cells in LP HD are selected for antibody expression. This, and the presence of ongoing mutation discriminates LP from classical HD.
Hodgkin disease (HD) is one of the most common malignant lymphomas. It is characterized by a small number of putative tumor cells, the Hodgkin and Reed–Sternberg (HRS) cells, which are surrounded by a large population of lymphocytes and histiocytes (1). On the basis of morphological, immunohistochemical, and clinical criteria, HD can be differentiated in at least two distinct entities: the more common classical HD (accounting for about 95% of cases) and the rare lymphocyte-predominant (LP) HD (2, 3).
Recently, we established a method to micromanipulate single cells from frozen tissue sections and analyze those cells for rearranged Ig variable (V) region genes (4). Because Ig gene rearrangements are specific for B cells and are highly diverse, this approach is not only suitable to determine whether cells analyzed belong to the B cell lineage, but also to reveal the clonal relationship of different cells isolated from the tissue. In addition, sequence analysis of such rearrangements can reveal further characteristics of a B cell carrying the rearrangement. Whereas naive B cells express unmutated V region genes (5–7), antigen-activated B cells acquire somatic mutations within their V region genes in the course of the germinal center (GC) reaction (4, 8, 9). Thus, somatically mutated V region genes are a hallmark of GC B cells and their descendants—i.e., memory B cells and GC-derived plasma cells (4, 6, 7). An analysis of variable domain of heavy chain (VH) genes for somatic mutation can now reliably be carried out because the human VH locus has recently been cloned and extensively analyzed (10). Furthermore, sequence analysis of VH genes amplified from single human IgD+ B cells revealed that the vast majority of rearranged VH genes of this population can be assigned to the respective germ-line genes with 100% homology (unpublished observation).
Using amplification of rearranged V region genes from micromanipulated single cells, we investigated 13 cases of classical HD for a potential B-lineage derivation and clonality of the HRS cells (11–13). The results showed that HRS cells in classical HD represent clonal populations of tumor cells derived from GC B cells, at least some of which had acquired “crippling” mutations (13).
In the case of LP HD, a B cell origin has been suspected for many years on the basis of immunohistochemical and in situ hybridization data. Thus, several B cell-specific markers were found to be expressed by HRS cells and their variants, including L and H (lymphocytic and histiocytic) cells, in LP HD (and usually not found in classical HD): the pan B cell marker CD20 (14, 15), the Ig-associated molecules mb-1 (16, 17) and J chain (18), and mRNA for Ig κ or λ light chains (19–21). Immunhistological studies further indicated that LP HD might represent a GC lymphoma (14). Because of its indolent clinical course (22), LP HD might not represent a true neoplasm but merely a peculiar reactive or premalignant condition (23, 24).
In one case of LP HD that we had previously analyzed, the tumor was apparently derived from a GC B cell, because clonal V region genes harboring somatic mutations and showing evidence for ongoing mutation were amplified from single HRS cells (11). However, in another study, in which HRS cells were isolated from suspensions prepared from fixed tissues, four cases of LP HD were described in which the HRS cells appeared to be a polyclonal population (25). Because of the small number of cases so far investigated, it is unclear whether these apparently contradictory results reflect true variability of the disease or merely technical matters (26).
The present investigation was performed to confirm the clonality of HRS cells in LP HD and their GC derivation, and to evaluate potential structural differences of V gene rearrangements carried by HRS cells in LP and classical HD, respectively.
MATERIALS AND METHODS
Tissues and Clinical Data.
Clinical features of the HD patients are summarized in Table 1. Patients 1–4 represent primary presentation of disease, and patient 5 is a relapse, 11 years after initial manifestation.
Table 1.
Case description of LP HD patients whose HRS cells were analyzed for Ig heavy chain rearrangements
| Patient | Age | Sex | Localization | HRS cells |
|---|---|---|---|---|
| 1 | 43 | Female | Axillary LN | CD20+ |
| 2 | 58 | Male | Axillary LN | CD20+ |
| 3 | 7 | Male | Inguinal LN | CD20+ |
| 4 | 18 | Male | Cervical LN | CD20+ |
| 5 | 37 | Male | Cervical LN | CD20+ |
LN, lymph node
Immunostaining and Micromanipulation.
Five- to 10-μm-thick frozen sections were stained with anti-CD20 (L26; Dako) or anti-CD3 (OKT3; Ortho Diagnostic) monoclonal antibodies. The staining procedure was as described (4). Bound alkaline phosphatase was visualized with Fast Red. Stained sections were overlaid with TBS, and single cells were mobilized and aspirated with the help of micropipettes fixed to a hydraulic micromanipulator. Only CD20+ large cells with the morphology of HRS (L and H) cells located solitarily outside remnants of GCs were micromanipulated as HRS cells. Isolated cells were stored in 20 μl of PCR buffer at −20°C.
Single Cell PCR.
Rearranged Ig genes were amplified from single cells using family specific VH leader primers (27) together with sets of nested joining domain of heavy chain (JH) primers as described (13). The first round of amplification consisted of 1 cycle of 2 min at 95°C, 4 min at 65°C, and 1 min at 72°C, followed by 34 cycles of 30 sec at 95°C, 45 sec at 61°C, and 45 sec at 72°C in 1× Expand buffer (Boehringer Mannheim) with 2 mM MgCl2, 200 μM for each dNTP, 0.25 μM of each of the VH and JH primers, and 7.25 units Expand enzyme mix (Boehringer Mannheim). The second round of PCR was carried out in separate reactions for each of the VH leader primers. The amplification consisted of 1 cycle of 2 min at 95°C, 4 min at 67°C, and 1 min at 72°C, followed by 39 cycles of 30 sec at 95°C, 45 sec at 63°C, and 45 sec at 72°C in 1× PCR buffer with 2 mM MgCl2, 200 μM each dNTP, 2.5 μM of one of the VH leader primers, 2.5 μM of nested JH primer mix, and 0.7 unit of Taq DNA polymerase (Boehringer Mannheim).
PCR products were analyzed on agarose gels and purified using a gel extraction kit (QiaExII; Qiagen, Hilden, Germany). PCR products were directly sequenced using the Ready Reaction DyeDeoxyTerminator cycle sequencing kit (Perkin–Elmer) and an automated sequencer (model ABI377; Applied Biosystems). Sequences were compared with the GenBank data library (release 98) using gene works (IntelliGenetics) software.
Analysis of Mutation Pattern.
Ratios of replacement to silent (R/S) mutations and frequencies of mutations creating stop codons on the basis of random mutagenesis were calculated as described (28).
RESULTS
PCR Analysis of Ig Gene Rearrangements in Single HRS Cells.
Single CD20+ HRS cells (29–67 cells/patient) were micromanipulated from frozen lymph node sections of five cases of LP HD (Tables 1 and 2). In two of the cases, a second micromanipulation was performed (Table 2). During all micromanipulations CD3+ cells from adjacent sections or CD20− small cells from the same sections and aliquots of the buffer covering the sections during the procedure were included in a blinded fashion as negative controls. In one case an Ig gene rearrangement was obtained from a buffer control. It was clonally related to those amplified from the corresponding HRS cells. All buffer controls from a second micromanipulation of this case were negative (Table 2). Occasionally, single IgD+ peripheral blood B cells sorted by flow cytometry or batches of two to three micromanipulated small CD20+ cells were included as positive controls. About 60–70% of the positive controls yielded at least one PCR product of expected size.
Table 2.
Summary of single cell analysis of five cases of LP HD for Ig VH gene rearrangements
| Patient | Exp. | Cells positive | PCR products
|
Rearrangements
|
||
|---|---|---|---|---|---|---|
| Total | Sequenced | Repeated | Unique | |||
| 1 | 1a | 2/17 | 2 VH3 | 2 | 2 VH3 | |
| 1b | 26/50 | 26 VH3 | 26 | 25 VH3 | 1 VH3 | |
| 2 | 2 | 33/60 | 33 VH5 | 19 | 19 VH5 | |
| 3 | 3a | 6/38 | 6 VH4 | 5 | 5 VH4 | |
| 3b | 6/9 | 6 VH4 | 6 | 6 VH4 | ||
| 4 | 4 | 4/29 | 4 VH3 | 4 | 4 VH3 | |
| 5 | 5 | 9/32 | 9 VH3 | 9 | 9 VH3 | |
| Controls | 1a | 0/8 T cells | — | |||
| 1b | 0/15 T cells | — | ||||
| 0/8 buffer | — | |||||
| 2 | 0/24 buffer | — | ||||
| 3a | 1/12 buffer | 1 VH4 | 1 | 1 VH4 | ||
| 3b | 0/9 buffer | — | ||||
| 4 | 0/5 L26− cells | — | ||||
| 0/6 buffer | — | |||||
| 5 | 0/8 T cells | — | ||||
| 0/8 buffer | — | |||||
As in our previous work the micromanipulated single HRS cells were analyzed for Ig heavy chain gene rearrangement by PCR and subsequent direct sequencing of the PCR products. However, in the present study we used a set of VH family-specific primers hybridizing to sequences within the leader region instead of primers hybridizing to framework region (FR) I. Because somatic mutations are rarely observed within the leader region, this reduces the risk of missing a rearrangement because of mutations present at primer binding sites. Indeed, in all five cases analyzed rearrangements could be amplified with efficiencies ranging from 11 to 68% in different experiments (Table 2). Because of the high efficiency of VH amplification, we did not perform additional studies of light chain rearrangements in the present work.
HRS Cells from LP HD Harbor Mutated In-Frame Ig Gene Rearrangements That Are Clonal.
Whereas 1 of 28 sequenced PCR products from patient 1 contained a unique VH3-30 rearrangement, the remaining 27 sequences represented clonally related rearrangements involving the VH3-23 and the JH1b gene segments (Tables 2 and 3; ref. 30 is for JH genes). The clonal rearrangement is potentially functional because the correct reading frame was preserved in the recombination process. A 3-bp deletion (codon 54) was found in all sequences and may be due to either somatic mutation or VH polymorphism. Compared with the most homologous germ-line V gene segment, VH3-23, 45 mutations (corresponding to a mutation frequency of 14.5%) were observed with 9 additional mutations in the leader intron. The unique VH3-30 rearrangement contains three mutations and may derive from cellular contamination in the micromanipulation procedure.
Table 3.
Sequence analysis of clonal Ig heavy chain gene rearrangements
| Patient | VH family | VH gene | % mutation | In-frame | Ongoing mutation detected |
|---|---|---|---|---|---|
| 1 | 3 | 3-23 | 14.5 | + | Yes |
| 2 | 5 | 5-51 | 4.0 | + | Yes |
| 3 | 4 | 4-34 | 9.9 | + | Yes |
| 4 | 3 | 3-11 | 8.8 | + | No |
| 5 | 3 | 3-11 | 4.8 | ? | No |
Reference for VH germ-line genes is ref 29. In the three cases with ongoing mutation, sequences with greatest homologies to the germ-line VH genes were considered for the calculation of the mutation frequencies.
Nineteen of 33 PCR products from patient 2 were sequenced and all consisted of a potentially functional in-frame rearrangement using the VH5-51 and the JH5b segment (Tables 2 and 3). As members of the small VH5 family are only rarely used in B cells, the remaining 14 unsequenced VH5 PCR products most likely also belong to the same clone. The clonal sequence contains 12 nucleotide substitutions (4.0%) compared with the germ-line gene, and one further mutation in the leader region.
For patient 3, 11 of 12 PCR products obtained with the primer for the VH4 family were sequenced. All represented the same in-frame rearrangement of VH4-34 to JH6b, containing 29 mutations (9.9%) in the V segment and 1 further mutation in the leader intron (Tables 2 and 3). In the first experiment on this case the same clonal rearrangement was also amplified from 1 of 12 buffer controls (Table 2). In a second micromanipulation of this case, 9 of 9 buffer controls were negative, and the clonal rearrangement was amplified from 6 of 9 HRS cells. The contamination of the buffer control in the first micromanipulation may be due to fragments of HRS cells generated during attempts to mobilize and aspirate HRS cells from the slide.
In patient 4, all of four sequenced PCR products contained the same in-frame rearrangement of VH3-11 to JH6b and the same 26 mutations (8.8% mutation frequency) compared with the germ-line V gene segment (Tables 2 and 3). The leader and leader intron harbored 7 mutations.
For patient 5 another clonal VH3-11 rearrangement was amplified and sequenced from 9 HRS cells (Tables 2 and 3). In this rearrangement a stretch of at least 11 guanidines within complementarity determining region III prevented further sequencing in the 3′ direction. Therefore, we were unable to determine whether this rearrangement is functional. The rearrangement harbored 14 mutations (4.8% mutation frequency) compared with the VH3-11 germ-line V segment and two additional mutations in the leader and leader intron.
All splice donor and acceptor sites of the leader introns examined were not affected by mutations, except that in one case a cytosine was replaced by a thymidine in the pyrimidine-rich sequence 5′ to the splice acceptor.
Taking the present five cases together with the one case of LP HD analyzed previously (11), in which we identified a clonal in-frame as well as out-of-frame VH rearrangement (with 14 and 15% mutation, respectively), all six cases of LP HD so far analyzed show clonal Ig heavy chain gene rearrangements in the HRS cells. These rearrangements are mutated with frequencies ranging from 4 to 15%. With the possible exception of patient 5, where the reading frame is unclear, all cases harbored potentially functional rearrangements.
Analysis of Mutation Patterns.
In the FR of functional V region genes replacement mutations are usually counterselected to preserve the structure of the antibody V domain. This results in R/S ratios below those expected on the basis of random mutagenesis. For nonfunctional rearrangements, however, which are not under selective pressure for antigen receptor expression, the R/S ratios should correspond to those expected assuming random mutagenesis.
Indeed, as previously shown (28), the R/S ratio observed in out-of-frame rearrangements from diffuse large cell lymphomas and HRS cells does not differ significantly from the calculated value (3.3 versus 3.0; Table 4). For in-frame rearrangements of nodular sclerosis and mixed cellularity HD we observed a R/S ratio of 1.9, whereas the calculated ratio is 3.1 (Table 4). For the four cases of LP HD analyzed here (patient 5 was excluded from this analysis as the functionality of the rearrangement is unclear) and the one analyzed previously (11), the calculated R/S ratio is 2.9, whereas a ratio of 1.3 is observed. This indicates counterselection of replacement mutations in the FRs of these rearrangements as it is seen in IgM only and class switched memory B cells, which are selected for expression of functional antibody (Table 4).
Table 4.
R/S analysis of mutated V genes
| Cell origin* | Rearrangement | No. of mutations | R/S expected | R/S found |
|---|---|---|---|---|
| LP HD | In-frame | 77 | 2.9 | 1.3 |
| NS and MC HD | In-frame | 301 | 3.1 | 1.9 |
| DLL and HD | Out-of-frame | 141 | 3.0 | 3.3 |
| IgM memory B cells | In-frame | 260 | ND | 1.5 |
| Class-switched | In-frame | 188 | ND | 1.0 |
| memory B cells |
Another indication for selection of a B cell for antigen receptor expression is the absence of mutations creating stop codons within functional V gene rearrangements (28). Accordingly, the “intrinsic” frequency of mutations creating stop codons was calculated for the germ-line V gene segments rearranged in the HRS cells from the LP HD patients and was found to be 5% (Table 5). In the absence of selection one would therefore expect to find about 7 mutations creating stop codons among the 141 mutations present in the five in-frame VH region genes [four cases analyzed here and the one analyzed previously (11)]. However, with the exception of one unique mutation in patient 2 (see Discussion), stop codons were not observed in the present analysis, clearly indicating that they were counterselected.
Table 5.
Analysis of V genes for mutations creating stop codons
| Cell origin* | Rearrangement | No. of mutations | % stop codons expected | No. of stop codons
|
|
|---|---|---|---|---|---|
| Expected | Found | ||||
| LP HD | In-frame | 141† | 5.0 | 7 | 0 |
| NS and MC HD | In-frame | 307 | 4.6 | 14 | 5 |
| DLL and HD | Out-of-frame | 155 | 4.8 | 7 | 7‡ |
DLL, diffuse large cell lymphomas; MC, mixed cellularity; NS, nodular sclerosis.
Mutations due to ongoing mutation were not considered.
Three more stop codons were caused by two point mutations within a codon.
Taken together, the analysis of the mutation pattern in the V gene segments as to R/S ratios in FRs and the frequency of mutations causing stop codons demonstrates that in LP HD the tumor cell or its precursor was selected for expression of functional antibodies while accumulating somatic mutations.
Ongoing Mutation in LP HD.
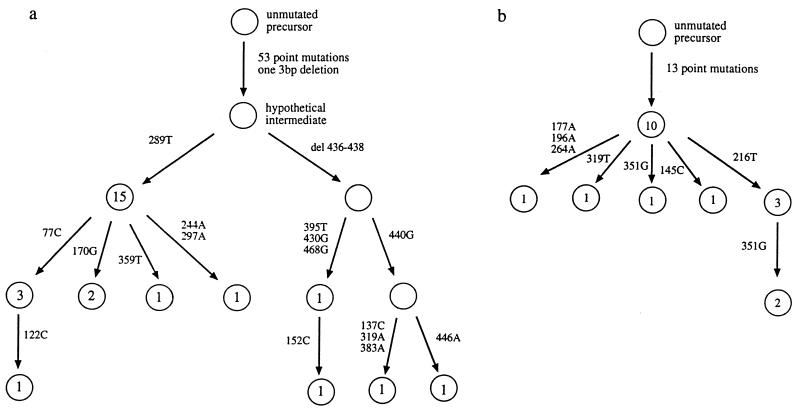
In four of the six cases of LP HD analyzed (including the one from ref. 11), intraclonal diversity of Ig gene rearrangements was observed. In patient 1, 10 different, but clonally related, sequences were found with various frequencies. One sequence was amplified from 15 cells, whereas the remaining sequences were found one to three times. Using the germ-line VH sequence as reference, the different sequences can be ordered in a genealogical tree, showing that the mutations accumulated in a stepwise manner (Fig. 1). While most of the sequences differ from the sequence with the greatest homology to the germ-line sequence by one or two nucleotide exchanges, four sequences also harbor the same 3-bp deletion. Seven different, clonally related sequences were observed in patient 2, again allowing the construction of a genealogical tree (Fig. 1). One sequence was obtained 10 times, the others 1 to 3 times. One unique mutation creates a stop codon (see Discussion). In patient 3, nine sequences are identical, while two sequences each contain a different further point mutation. Ongoing mutation had been likewise identified in the case of LP HD analyzed previously (11).
Figure 1.
Genealogical trees explaining the relationship between clonally related sequences for LP HD patient 1 (a) and patient 2 (b). Each circle represents a different sequence. Numbers of single HRS cells with a specific sequence is given within the circles. Numbers and bases beside the arrows indicate the nucleotide position and type of exchange, respectively, defining a sequence. For intraclonal diversity within complementarity-determining region III (CDRIII), the most frequently occurring nucleotide was assumed to represent the unmutated variant.
There is thus clear evidence for ongoing mutation in LP HD. The failure to detect sequence variations in patients 4 and 5 may be due to a lower rate of ongoing mutation and/or the small number of sequences analyzed (three for patient 4, and nine for patient 5).
DISCUSSION
HRS Cells in LP HD Represent a Clonal Population of Tumor Cells.
In the present study, single HRS cells were micromanipulated from frozen tissue sections of five cases of LP HD and analyzed for rearranged Ig heavy chain genes. In each case clonal VH region genes were repeatedly amplified from single HRS cells (Tables 2 and 3). This shows that the HRS cells in all cases represent a clonal population of B lineage-derived cells. Clonality of HRS cells in LP HD is supported by earlier studies, in which mRNA for either κ or λ light chain was detected in HRS cells in the majority of cases by in situ hybridization (19–21).
The polyclonality of HRS cells in the analysis of LP HD by Delabie et al. (25) may thus reflect technical problems associated with the approach of preparing single cell suspensions from fixed tissues (26).
HRS Cells in LP HD Are Derived from GC B Cells.
Sequence analysis of the amplified VH gene rearrangements revealed the presence of somatic mutations in all cases, with mutation frequencies in the V segment ranging from 4.0 to 14.5% (Table 3). Of particular importance is the finding of ongoing mutation within the tumor clone in three of the cases (Fig. 1), which is in accord with what we had already observed in a further patient earlier (11). Because the process of somatic hypermutation is active in and specific for GC B cells (4, 9), this identifies a GC B cell as the precursor of the tumor clone in these cases. In the two other cases, ongoing mutation may not have been detected because of the relatively small number of sequences analyzed (Table 2). A relationship of LP HD to GC B cell lymphoma had previously been suspected on the basis of a varity of immunohistochemical findings: (i) the presence of follicular dendritic cells, a cell type restricted to B cell follicles and GCs is a characteristic feature of LP HD (32); (ii) CD57+ T cells, specific for GC, are present in LP HD (14, 33); and (iii) HRS cells in LP HD strongly express the BCL-6 protein, which is predominantly expressed by GC B cells (34).
HRS Cells in LP HD Show Evidence for Antigen Selection.
The low R/S value within the FRs as well as the absence of mutations causing stop codons in the amplified in-frame rearrangements both indicate that the tumor clones of the LP HD cases were selected for antigen receptor expression (Tables 4 and 5). This interpretation, which is based on the sequence analysis of the heavy chain rearrangements, implies that the corresponding light chain gene rearrangements, known to be expressed in the HRS cells from LP HD (19–21), show a similar mutation pattern because antibodies are selected as molecules consisting of heavy and light chains. A single stop codon was detected in a single sequence of patient 2. As this mutation was thus found only in one single HRS cell, it may have occurred recently and could potentially interfere with a further expansion of that cell. On the other hand, patient 2 was the one with the highest R/S value (four R mutations/one S mutation). Although the number of mutations within the FR in this case is small, both findings together might also be taken as an indication that in this tumor clone, antibody expression was no longer required for cellular growth.
The Mutation Pattern of Rearranged V Genes Discriminates LP from Classical HD, and also from Follicular Lymphoma.
In classical HD, selection against R mutations and mutations generating stop codons within FRs of in-frame V region gene rearrangements is considerably weaker than in memory B cells (ref. 28; Tables 4 and 5). This suggests that the tumor clone in classical HD acquired somatic mutations at least in part independent of selection for antigen receptor expression. This contrasts with the selection for antibody expression in LP HD (see above). Whether functional antibody is still needed at later stages of the latter disease, however, remains to be clarified.
A second discriminating feature between LP and classical HD is the observed ongoing mutation in the former, which was not observed in the latter. Taking these findings together, it appears that HRS cells in classical HD are derived from GC B cells that have lost the ability to recognize antigen because of “crippling” somatic mutations (13), whereas in LP HD the HRS cell is a GC B cell which is still under selection by antigen and undergoing somatic hypermutation.
Ongoing mutation and selection for antigen receptor expression is also well established for follicular lymphoma (35, 36), which is regarded as the typical GC B cell lymphoma (2). Interestingly, the presence of follicular dendritic cells and CD57+ T cells, which are both constituents of normal GCs, is a characteristic feature of follicular lymphoma as well as LP HD (2, 14, 32, 37). These cell types, which are believed to play an important role in the establishment of GCs and the selection of GC B cells (9), may thus be required for ongoing mutation and cellular selection of antibody mutants in follicular lymphoma and LP HD.
However, LP HD and follicular lymphoma can be clearly discriminated on the basis of histological, morphological and clinical criteria (2, 37). Furthermore, intraclonal diversity seems to be more pronounced in follicular lymphoma (38–40). The finding that in each of the four cases of LP HD with detectable ongoing mutation the majority of HRS cells harbor the same sequence could have one of several reasons. First, the rate of somatic mutation may be lower in the tumor cells of LP HD compared with follicular lymphoma, the latter representing a subset of GC B cells with a more active hypermutation machinery. Second, in LP HD a subclone of HRS cells, that down-regulated somatic hypermutation, might have acquired a growth advantage over the other ones. Third, if the major subclone of HRS cells represents a selected cell producing high-affinity antibodies for the unknown and hypothetical antigen driving tumor expansion, then further mutations (most of which would reduce affinity) might be counterselected.
Because follicular lymphoma as well as LP HD originate from a mutating GC B cell, one may speculate that it is not the differentiation stage of the precursor of the lymphoma, but rather the nature of the transforming events that determine which of the two diseases will arise. Whereas it is well known that a chromosomal translocation involving the bcl-2 protooncogene and one of the Ig loci is a characteristic feature of follicular lymphoma (41), transforming events involved in LP HD remain to be uncovered.
Acknowledgments
We thank Christiane Gerhardt, Tanja Schaffer, and Julia Jesdinsky for excellent technical assistance and Holger Kanzler for help throughout the work. This work was supported by the Deutsche Forschungsgemeinschaft through SFB502 and the Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung.
ABBREVIATIONS
- FR
framework region
- GC
germinal center
- HD
Hodgkin disease
- HRS
Hodgkin and Reed–Sternberg
- LP
lymphocyte predominant
- NS
nodular sclerosis
- MC
mixed cellularity
- R/S
replacement to silent ratio.
Footnotes
References
- 1.Burke J S. In: Neoplastic Hematopathology. Knowles D M, editor. Baltimore: Williams & Wilkins; 1992. pp. 497–533. [Google Scholar]
- 2.Harris N L, Jaffe E S, Stein H, Banks P M, Chan J K C, Cleary M L, Delsol G, de Wolf-Peeters C, Falini B, Gatter K C, Grogan T M, Isaacson P G, Knowles D M, Mason D Y, Müller-Hermelink H K, Pileri S A, Piris M A, Rafkiaer E, Warnke R A. Blood. 1994;84:1361–1392. [PubMed] [Google Scholar]
- 3.Mason D Y, Banks P M, Chan J, Cleary M L, Delsol G, de Wolf-Peeters C, Falini B, Gatter K, Grogan T M, Harris N L, Isaacson P G, Jaffe E S, Knowles D M, Müller-Hermelink K, Pileri S, Ralfkiaer E, Stein H, Warnke R. Am J Surg Pathol. 1994;18:526–530. doi: 10.1097/00000478-199405000-00014. [DOI] [PubMed] [Google Scholar]
- 4.Küppers R, Zhao M, Hansmann M L, Rajewsky K. EMBO J. 1993;12:4955–4967. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein U, Küppers R, Rajewsky K. Eur J Immunol. 1993;23:3272–3277. doi: 10.1002/eji.1830231232. [DOI] [PubMed] [Google Scholar]
- 6.Klein U, Küppers R, Rajewsky K. J Exp Med. 1994;180:1383–1393. doi: 10.1084/jem.180.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascual V, Liu Y J, Magalski A, de Bouteiller O, Banchereau J, Capra J D. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Nature (London) 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 9.Kelsoe G. Adv Immunol. 1995;60:267–288. doi: 10.1016/s0065-2776(08)60587-8. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda F, Honjo T. Adv Immunol. 1996;62:1–29. doi: 10.1016/s0065-2776(08)60426-5. [DOI] [PubMed] [Google Scholar]
- 11.Küppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R, Hansmann M L. Proc Natl Acad Sci USA. 1994;91:10962–10966. doi: 10.1073/pnas.91.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanzler H, Hansmann M L, Kapp U, Wolf J, Diehl V, Rajewsky K, Küppers R. Blood. 1996;87:3429–3436. [PubMed] [Google Scholar]
- 13.Kanzler H, Küppers R, Hansmann M L, Rajewsky K. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timens W, Visser L, Poppema S. Lab Invest. 1986;54:457–461. [PubMed] [Google Scholar]
- 15.Coles F B, Cartun R W, Pastuszak W T. Mod Pathol. 1986;1:274–278. [PubMed] [Google Scholar]
- 16.Kuzu I, Jones M, Gatter K C, Mason D Y. Histopathology. 1993;22:141–144. doi: 10.1111/j.1365-2559.1993.tb00092.x. [DOI] [PubMed] [Google Scholar]
- 17.Korkolopoulou P, Cordell J, Jones M, Kakamanis L, Tsenga A, Gatter K C, Mason D Y. Histopathology. 1994;24:511–515. doi: 10.1111/j.1365-2559.1994.tb00568.x. [DOI] [PubMed] [Google Scholar]
- 18.Stein H, Hansmann M L, Lennert K, Brandtzaeg P, Gatter K C, Mason D Y. Am J Clin Pathol. 1986;86:292–297. doi: 10.1093/ajcp/86.3.292. [DOI] [PubMed] [Google Scholar]
- 19.Hell K, Pringle J H, Hansmann M L, Lorenzen J, Colloby P, Lauder I, Fischer R. J Pathol. 1993;171:137–143. doi: 10.1002/path.1711710211. [DOI] [PubMed] [Google Scholar]
- 20.Stoler M H, Nichols G E, Symbula M, Weiss L M. Am J Pathol. 1995;146:812–818. [PMC free article] [PubMed] [Google Scholar]
- 21.van Wasielewski R, Wilkens L, Nolte M, Werner M, Georii A. Mod Pathol. 1996;9:334–338. [PubMed] [Google Scholar]
- 22.Hansmann M L, Zwingers T, Böske A, Löffler H, Lennert K. J Cancer Res Clin Oncol. 1984;108:321–330. doi: 10.1007/BF00390466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regula D P, Hoppe R T, Weiss L M. N Engl J Med. 1988;318:214–219. doi: 10.1056/NEJM198801283180404. [DOI] [PubMed] [Google Scholar]
- 24.Pan L X, Diss T C, Peng H Z, Norton A J, Isaacson P G. Blood. 1996;87:2428–2434. [PubMed] [Google Scholar]
- 25.Delabie J, Tierens A, Wu G, Weisenburger D D, Chan W C. Blood. 1994;84:3291–3298. [PubMed] [Google Scholar]
- 26.Küppers, R., Kanzler, H., Hansmann, M. L. & Rajewsky, K. (1996) Ann. Oncol. 7 (Suppl. 4), S27–S30. [DOI] [PubMed]
- 27.Klein U, Klein G, Ehlin-Henriksson B, Rajewsky K, Küppers R. Mol Med. 1995;1:495–505. [PMC free article] [PubMed] [Google Scholar]
- 28.Küppers R, Rajewsky K, Hansmann M L. Eur J Immunol. 1997;27:1398–1405. doi: 10.1002/eji.1830270616. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda F, Shin E K, Nagaoka H, Matsumura R, Haino M, Fukita Y, Takaishi S, Imai T, Riley J H, Anand R, Soeda E, Honjo T. Nat Genet. 1993;3:88–94. doi: 10.1038/ng0193-88. [DOI] [PubMed] [Google Scholar]
- 30.Yamada M, Wasserman R, Reichard B A, Shane S, Caton A J, Rovera G. J Exp Med. 1991;173:395–407. doi: 10.1084/jem.173.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein U, Küppers R, Rajewsky K. Blood. 1997;89:1288–1298. [PubMed] [Google Scholar]
- 32.Alavaikko M, Hansmann M L, Nebendahl C, Parwaresch M R, Lennert K. Am J Clin Pathol. 1991;95:194–200. doi: 10.1093/ajcp/95.2.194. [DOI] [PubMed] [Google Scholar]
- 33.Hansmann M L, Fellbaum C H, Hui P K, Zwingers T. J Cancer Res Clin Oncol. 1988;114:405–410. doi: 10.1007/BF02128186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falini B, Bigerna B, Pasqualucci L, Fizzotti M, Martelli M F, Pileri S, Pinto A, Carbone A, Venturi S, Pacini R, Cattoretti G, Pescarmona E, Lo Coco F, Pelicci P G, Anagnastopoulos I, Dalla-Favera R, Flenghi L. Blood. 1996;87:465–471. [PubMed] [Google Scholar]
- 35.Cleary M L, Meeker T C, Levy S, Lee E, Trela M, Sklar J, Levy R. Cell. 1986;44:97–106. doi: 10.1016/0092-8674(86)90488-5. [DOI] [PubMed] [Google Scholar]
- 36.Bahler D W, Levy R. Proc Natl Acad Sci USA. 1992;89:6770–6774. doi: 10.1073/pnas.89.15.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poppema S, Kaleta J, Hugh J, Visser L. Immunol Rev. 1992;126:163–178. doi: 10.1111/j.1600-065x.1992.tb00636.x. [DOI] [PubMed] [Google Scholar]
- 38.Zelenetz A D, Chen T T, Levy A. J Exp Med. 1991;173:197–207. doi: 10.1084/jem.173.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu D, Hawkins R E, Hamblin T J, Stevenson F K. Br J Haematol. 1994;86:505–512. doi: 10.1111/j.1365-2141.1994.tb04780.x. [DOI] [PubMed] [Google Scholar]
- 40.Wu H J, Kaartinen M. Scand J Immunol. 1995;42:52–59. doi: 10.1111/j.1365-3083.1995.tb03625.x. [DOI] [PubMed] [Google Scholar]
- 41.Korsmeyer S J. Blood. 1992;80:879–886. [PubMed] [Google Scholar]