Abstract
Protoporphyrinogen IX oxidase is the last enzyme in the common pathway of heme and chlorophyll synthesis and provides precursor for the mitochondrial and plastidic heme synthesis and the predominant chlorophyll synthesis in plastids. We cloned two different, full-length tobacco cDNA sequences by complementation of the protoporphyrin-IX-accumulating Escherichia coli hemG mutant from heme auxotrophy. The two sequences show similarity to the recently published Arabidopsis PPOX, Bacillus subtilis hemY, and to mammalian sequences encoding protoporphyrinogen IX oxidase. One cDNA sequence encodes a 548-amino acid residues protein with a putative transit sequence of 50 amino acid residues, and the second cDNA encodes a protein of 504 amino acid residues. Both deduced protein sequences share 27.2% identical amino acid residues. The first in vitro translated protoporphyrinogen IX oxidase could be translocated to plastids, and the approximately 53-kDa mature protein was detected in stroma and membrane fraction. The second enzyme was targeted to mitochondria without any detectable reduction in size. Localization of both enzymes in subcellular fractions was immunologically confirmed. Steady-state RNA analysis indicates an almost synchronous expression of both genes during tobacco plant development, greening of young seedlings, and diurnal and circadian growth. The mature plastidal and the mitochondrial isoenzyme were overexpressed in E. coli. Bacterial extracts containing the recombinant mitochondrial enzyme exhibit high protoporphyrinogen IX oxidase activity relative to control strains, whereas the plastidal enzyme could only be expressed as an inactive peptide. The data presented confirm a compartmentalized pathway of tetrapyrrole synthesis with protoporphyrinogen IX oxidase in plastids and mitochondria.
In all living organisms the metabolic pathway from 5-aminolevulinate to tetrapyrroles is ubiquitous and highly conserved. In plants, 5-aminolevulinate is synthesized from glutamate in three sequential enzymatic steps, but in animals it is synthesized from succinyl-CoA and glycine. In addition to the general heme formation, plants possess the capacity to synthesize chlorophyll, which is essential for photoautotrophic growth. The animal heme synthesis recruits enzymes sequentially located in the mitochondria, cytoplasm, and finally mitochondria (1), whereas chlorophyll synthesis is exclusively situated in plastids (2). Final steps of plant heme synthesis occur parallel in mitochondria and plastids (3). The last common step of the tetrapyrrolic pathway to heme and chlorophyll is the oxidization of protoporphyrinogen IX to protoporphyrin (4, 5), which is catalyzed by protoporphyrinogen IX oxidase (EC 1.3.3.4). The enzyme was initially characterized from yeast (6) and mammalian liver (7–9), and it catalyzes a six-electron oxidation of protoporphyrinogen IX and contains a flavin as cofactor. The enzyme is associated with the inner mitochondrial membrane facing with its active center, the cytoplasmic side (10). Plant protoporphyrinogen IX oxidase activity has been detected in plastidal and mitochondrial extracts (11). The enzyme was purified from etiolated barley as a 36-kDa protein without detectable differences between the isoenzyme isolated from mitochondria and that from etioplasts (12). The plastidal isoenzyme was found in the envelope and thylakoid membranes, predicting different functions in the dual-subcompartmental localization (13). The plant enzyme is the target of herbicides consisting of various structural classes with a photodynamic mode of action (14–16). The phytotoxicity can be explained by accumulation of excess protoporphyrinogen, which is rapidly oxidized to protoporphyrin in the cytoplasm most likely by nonspecific, membrane-bound peroxidases (17, 18). Protoporphyrin is formed from protoporphyrinogen, known as a potent photosensitizer, which generates singlet oxygen and causes rapid lipid peroxidations and cell death.
Genes involved in protoporphyrinogen IX oxidase activity have been identified first from Escherichia coli (19) and Bacillus subtilis (20) and are designated hemG and hemY. Both genes encode peptides that did not share any sequence similarity. They represent two distinct protoporphyrinogen oxidizing systems, the HemY-type oxygen-dependent and the bacterial multicomponent system. The purified yeast protoporphyrinogen IX oxidase is a 55-kDa protein (21), and its gene was identified by functional complementation of a hem14–1 yeast mutant that is deficient in enzyme activity and resembles the HemY protein (22). Interestingly, the E. coli hemG mutant could be complemented with human, mouse, and Arabidopsis cDNA sequences encoding the HemY-like protein (23–26).
An accurate regulation of tetrapyrrole biosynthesis includes controlled distribution of protoporphyrinogen for plastidal chlorophyll and heme and mitochondrial heme synthesis. Screening for plant cDNA sequences encoding protoporphyrinogen IX oxidase was aimed to provide tools for studies on their expression and the control of metabolic flux in both organelles. Moreover, identification of sequences encoding plant PPX enables genetic and biochemical analysis of the enzyme and the structural significance of the high sensitivity toward the photodynamic herbicides. The present paper shows evidence for two types of nuclear-encoded tobacco protoporphyrinogen IX oxidase with a low overall similarity. Immunological identification of the isoenzymes in subcellular fractions and translocation experiments provided evidence for the compartmental localization of each isoenzyme.
MATERIALS AND METHODS
Plant Material and Growth Conditions.
Tobacco plants (Nicotiana tabacum cv. Samsun NN, Institut für Pflanzengenetik und Kulturpflanzenforschung, Gatersleben) were grown in the greenhouse for 6 weeks as described previously (27). Leaves of 6-week-old plants were harvested from the top to the base. Another set of plants was grown in a growth chamber in a 12-h/12-h light/dark cycle at 24°C for 4 weeks at a light intensity of 75 μmol m−2 s−1. Plantlets were harvested every 4 h over a 24-h time period starting 1 h after onset of the light phase. Another set of plants was transferred from dark/light conditions to continuous light or darkness. Samples were collected starting 48 h after the first harvest. Plant material was immediately frozen in liquid nitrogen and stored at −80°C. Six- to eight-day-old pea seedlings (Pisum sativum L. cv. frühe Harzerin, Saatzucht Quedlinburg, Germany) were used for isolation of mitochondria and plastids.
cDNA Cloning Techniques.
A Lambda-ZAP II tobacco (SR1) cDNA library (Stratagene) from tobacco was amplified, and pBluescript was excised from the phage DNA. E. coli strain R751, which is defective in the hemG gene (28), was electroporated with plasmid DNA containing the library. The cells were plated on Luria–Bertani(LB) agar containing 100 mg/ml ampicillin. Strain R751 grew very poorly in the absence of heme. Complemented E. coli cells were selected on the basis of normal-size colonies. Plasmid DNA was isolated from these colonies and reintroduced into the hemG mutant to confirm the recovery from poor growth. The cDNA fragments were sequenced by the dideoxy chain termination method (29) on both strands using fluorescent-labeled primers for the application to the ALF DNA Analysis System (Pharmacia). Results were analyzed with the pc/gene program (IntelliGenetics).
Southern and Northern Blot Analyses.
DNA extraction and Southern blot hybridization were performed according to standard procedures (30). Total RNA was extracted according to ref. 31. Ten micrograms per sample were separated electrophoretically on 1% formaldehyde-agarose gels followed by vacuum blotting on nylon membranes (Hybond N, Amersham). Equal loading of samples was controlled by hybridization of RNA with an actin cDNA probe. Filters were probed with cDNA inserts that were radiolabeled by random priming, and were washed under high stringency conditions.
Construction of Tobacco Protoporphyrinogen IX Oxidase Expression Vectors.
The synthetic sense primer 5′-GACCCATGGTTGCCAAAGATTACAGTTC and the antisense primer 5′-GACGGATCCTCATTTGTATGCATACCGAGAC were used to amplify by PCR an open reading frame encoding a putative mature protoporphyrinogen IX oxidase beginning with the amino acid valine at position 53 of the entire peptide sequence. The DNA fragment generated was purified, cleaved with NcoI and BamHI, and inserted into NcoI/BamHI-cleaved pQE 60 vector (Qiagen). A DNA fragment encoding a second isoenzyme was amplified by PCR using the sense primer 5′GACGCATGCTTGCTCCTTCTGCCGGAGAAG and the antisense primer 5′GACGGATCCTCAGCAATGTCTTTTGGAGTC. The purified PCR fragment was digested with SphI/BamHI and subcloned into the SphI/BamHI sites of the pDS56-SphI vector (32). The cDNA inserts were inserted in frame behind the ATG initiation codon present in the expression vectors. The E. coli expression strain SG13009 (33) was transformed with the expression vector. Recombinant protein synthesis was induced upon addition of isopropyl β-d-thiogalactopyranoside (1 mM final concentration) during exponential growth at OD600 nm = 0.4 for 4 h at 37°C. Total bacterial cell extracts were separated into soluble and pelletable protein fraction and analyzed on a 10% SDS/polyacrylamide gel. A truncated 41-kDa isoenzyme I without the original C terminus and the complete second isoenzyme were purified and used to immunize rabbits.
Enzyme Assay.
Aliquots of the E. coli cell cultures expressing both recombinant isoenzymes of protoporphyrinogen IX oxidase were centrifuged, lysed in assay buffer (50 mM Tris⋅HCl, pH 7.5/1 mM EDTA/5 mM DTT/0.03% Tween 80) by sonication, and incubated with 2 μM protoporphyrinogen IX at 30°C in the dark for 5 and 10 min according to ref. 34. Protoporphyrinogen IX was always freshly prepared from protoporphyrin IX (Fluka) using sodium amalgam. The formation of protoporphyrin IX was monitored fluorimetrically at 405 nmex and 632 nmem on a LS 50 B Luminescence Spectrophotometer (Perkin-Elmer). Autooxidation of protoporphyrinogen IX was monitored by incubating substrate with heat-inactivated enzyme. Inhibitory effects of herbicides (e.g., acifluorfen) were assayed under the same reaction conditions, using concentrations as indicated.
Production of Radiolabeled Precursor Proteins and Organellar Import Studies.
In vitro transcription and translation of the transcripts in the rabbit reticulocyte lysate in the presence of [35S]methionine were performed according to the manufacturers’ protocols (Stratagene and Promega). Mitochondria were isolated from pea plantlet essentially as described (35). Mitochondrial import assays were described in details elsewhere (36). In vitro transport into pea chloroplasts was carried out according to ref. 37 with minor modifications (38).
RESULTS
Isolation and Analysis of cDNA Clones Encoding Protoporphyrinogen IX Oxidase.
The plasmid pBluescript SK(−) containing the tobacco cDNA library was excised from the amplified λ-ZAP vector. The plasmid library with approximately 1 × 106 individual cDNA sequences was used to transform the E. coli hemG mutant R751 (28). Bacterial clones were selected by colony size and growth rate. Only a few bacterial colonies grew on LB medium without heme supplementation. Retransformation of the hemG mutant R751 with the isolated plasmid DNA confirmed the complementation capacity. The cDNA sequences fell into two groups. The full-length tobacco cDNA sequences were designated PPX I and PPX II (deposited in the GenBank database, accession numbers Y13465 and Y13466). The first cDNA sequence in total was composed of 1,644 nt, including 29 and 216 nt of 5′ and 3′ untranslated region, respectively, and encodes a protein of 548 amino acids, yielding a molecular mass of 59,138 Da. The second cDNA sequence contained an open reading frame of 1,515 nucleotides, a 5′ untranslated region of 38 nt, and a 3′ untranslated region of 386 nt. The cDNA encoded a protein of 505 amino acids with a calculated molecular mass of 55,407 Da. The intactness of the sequences was confirmed by nucleotide sequencing and in vitro transcription/translation experiments. The coding regions of both full-length cDNA sequences were not in frame with the coding sequence of the LacZ, gene indicating that complementation of the hemG mutant is not due to synthesis of the fusion LacZ–PPX translation products. It is proposed that the synthesis of the proteins begins at the first or an internal ATG codon of the cDNA sequences.
The aligned peptide sequences derived from protoporphyrinogen IX oxidases I and II (Fig. 1) showed 27.2% identical amino acid residues. The N-terminal end of both peptides differed in length, and it was not assessable whether the proteins contained a plastidal or mitochondrial target sequence. Alignment of both proteins with known sequences revealed significant similarity to protoporphyrinogen IX oxidases, which were identified by E. coli hemG complementation. The tobacco protoporphyrinogen IX oxidases I and II show sequence identity to the Arabidopsis thaliana PPOX of 71.2%/24.6% (26), Bacillus subtilis HemY of 28.1%/25.1% (20), mouse PPX of 24.6%/24.6% (25), human PPX of 23.5%/21.4% (23), and Saccharomyces cerivisae PPX of 19.3%/19.8% (22). Sequence comparison indicated the close relation of the Arabidopsis protein to protoporphyrinogen IX oxidase I. On the basis of the sequence comparison with other eukaryotic homologous proteins, we could not clearly corroborate which tobacco isoform is the mitochondrial enzyme. An alignment of all known protoporphyrinogen oxidase sequences indicated only a few conserved residues that are most likely involved in structural function and substrate and cofactor binding. A glycine-rich motif G-X-G-X-X-G was previously proposed to be a dinucleotide binding site of well characterized flavoproteins (e.g., monoamine oxidase, putrescine oxidase, dimethylaniline oxidase) (39).
Figure 1.
Sequence alignment of the tobacco protoporphyrinogen oxidases I (NTPPXI) and II (NTPPXII) with the homologous enzymes of the A. thaliana (ATHPPOX), B. subtilis (BACHEMY), and mouse (MUSPO). The glycine-rich motif is underlined. The positions of identical amino acid residues in all sequences are indicated by asterisks, and the positions of well conserved residues are indicated by dots.
Translocation of Protoporphyrinogen IX Oxidase Isoenzymes to Plastids and Mitochondria.
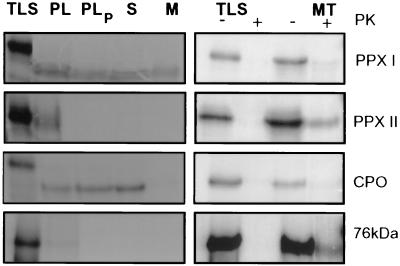
We paid particular attention to the subcellular localization of both isoenzymes. Messenger RNA complementary to the cDNA sequences protoporphyrinogen IX oxidases I and II was synthesized and translated in vitro. The translation products were incubated with intact plastids and mitochondria purified from young green pea plants. Reference precursor proteins whose exclusive import into one of the organelles has been shown before were additionally offered to plastids and mitochondria and served as markers for purity of organelle preparation and selective import. After finishing the translocation experiments, organelles were treated with proteases, and proteins protected from proteolysis were applied to polyacrylamide gels. A processed protoporphyrinogen IX oxidase I with an apparent molecular mass of 53 kDa was equally distributed between stroma and thylakoid fraction (Fig. 2 Left). Coproporphyrinogen oxidase, the preceding enzyme in the pathway (40), was translocated to plastids and exclusively accumulated in the stroma. Protoporphyrinogen IX oxidase II and the 76-kDa NADH:ubiquinone oxidoreductase (41) were attached to plastids but did not resist proteinase treatment of intact plastids, indicating that these proteins were not imported.
Figure 2.
Posttranslational uptake of in vitro synthesized precursor proteins. 35S-labeled precursor of protoporphyrinogen IX oxidases I (PPX I) and II (PPX II) were synthesized by in vitro transcription/translation (TLS) and offered to intact pea chloroplasts (Left) or mitochondria (Right). (Left) Following the import reaction, plastids were either analyzed directly (PL) or after treatment with proteases (PLP). Plastids were reisolated and fractionated into stroma (S) and membrane (M). (Right) Aliquots of the translation products (TLS) and of mitochondria after the import assay were incubated with (+) and without (−) proteinase K (PK) and applied to polyacrylamide gels. In both experiments, coproporphyrinogen oxidase (CPO) as a plastidal marker and a nuclear-encoded mitochondrial subunit of NADH:ubiquinone oxidoreductase (76 kDa) was included as a reference.
The translocation experiment with intact mitochondria was also terminated with protease treatment. Protoporphyrinogen IX oxidase II and NADH:ubiquinone oxidoreductase were protected against protease and integrated in mitochondria (Fig. 2 Right). Processing of both proteins could not be detected assessed from the mobility of both translation and import products on a 10% polyacrylamide gel. Difference in size between precursor and mature NADH:ubiquinone oxidoreductase could not be resolved in the gel system most likely because of the high molecular mass of the protein (ref. 41; L. Grohmann, personal communication). Protoporphyrinogen IX oxidase I and coproporphyrinogen oxidase were bound to mitochondria but obviously not protected from protease treatment. Lack of these radioactive-labeled proteins in the protease-treated mitochondrial fraction additionally indicates that the plastidal contamination in this preparation could be neglected. Immunological detection of the protoporphyrinogen IX oxidase in subcellular fractions confirmed the correct targeting of the in vitro translated and imported isoenzyme I to plastids and II to mitochondria (Fig. 3).
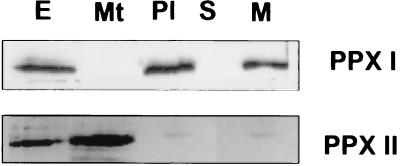
Figure 3.
Immunological determination of the subcellular localization of the protoporphyrinogen IX oxidase isoforms. Proteins from total leaf extract (E), purified mitochondria (Mt), and plastids before (Pl) and after separation into stroma (S) and membrane (M) fraction were subjected to Western blot analysis using monospecific antibodies raised against recombinant plastidal (PPX I) and mitochondrial (PPX II) protoporphyrinogen IX oxidases.
In vivo Expression Studies.
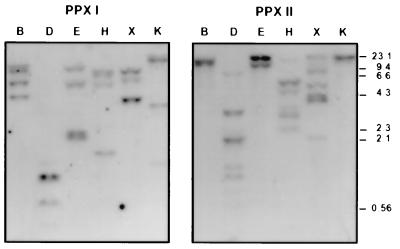
Southern blot hybridization with genomic tobacco DNA (Fig. 4) indicates that genes encoding plastidal and mitochondrial protoporphyrinogen IX oxidase are apparently encoded in small subfamilies. Different patterns of hybridizing DNA bands were obtained with the two cDNA probes. The low similarity of both cDNA sequences excluded cross-hybridization.
Figure 4.
Southern blot analysis of protoporphyrinogen IX oxidases in tobacco. Ten micrograms of genomic DNA was digested with BamHI (B), DraI (D), EcoRV (E), HindIII (H), XbaI (X), and KpnI (K) and subjected to Southern blot hybridization using 32P-labeled cDNA inserts encoding the plastidal (PPX I) and mitochondrial (PPX II) isoenzymes.
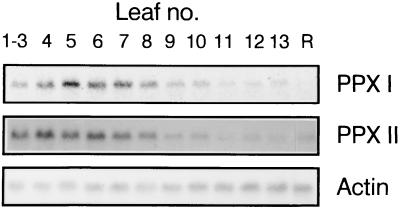
Northern blot analysis revealed protoporphyrinogen IX oxidase transcripts of ca. 1,800 bases. The developmental-dependent expression of genes encoding isoforms I and II were determined in 6-week-old tobacco plants (Fig. 5). These RNA species transiently accumulated in expanding premature leaves (maximum in leaf 5, counting from the top to the bottom of the plant) before the level drastically decreased toward the oldest leaves. In roots, RNA for the mitochondrial protoporphyrinogen IX oxidase was present, whereas the RNA level for the plastidal enzyme was below the detection limit. The amount of transcripts encoding protoporphyrinogen IX oxidases I and II did not alter during greening of 8-day-old etiolated tobacco seedlings (data not shown).
Figure 5.
Accumulation of protoporphyrinogen IX oxidase transcripts during leaf development and in roots. Roots and leaves were harvested from 6-week-old tobacco plants including the youngest leaves (1–3) and the oldest fully expanded leaf (13). Total RNA was extracted, and 10 μg was subjected to Northern blot analysis using 32P-labeled cDNA encoding both isoenzymes (PPX I and PPX II). Actin RNA levels were displayed as control for equal loading of RNA samples.
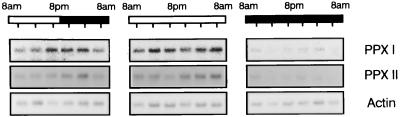
We also analyzed the RNA content within a period of 24 h in tobacco plants grown in a 12-h:12-h light/dark regime and subsequently under constant low light (70 μmol photons m−2 s−1) and dark conditions. RNA content encoding both isoenzymes oscillated under diurnal conditions with a maximum in the dark period (Fig. 6). The RNA displayed an inverse rhythmic periodicity under the light/dark regime in comparison to photosynthetic gene products (42), which accumulate during illumination. Rhythmicity of protoporphyrinogen IX oxidase RNA expression could not be observed during constant growth conditions. This RNA was reduced to a basis level in constant darkness. The RNA expression is apparently not controlled by the endogenous clock. But the light/dark rhythm seems to synchronize the protoporphyrinogen IX oxidase gene activity, leading to an oscillating expression pattern of the RNA species during diurnal growth.
Figure 6.
Steady-state levels of protoporphyrinogen IX oxidase transcripts under diurnal and circadian growth conditions. Tobacco plants were grown for 4 weeks in a 12-h/12-h light/dark cycle. Plants were harvested every 4 h starting 1 h after the onset of the light phase. Another set of plants was subsequently transferred to continuous light or darkness for 48 h. Leaf material was collected for the same 4-h time intervals. Total RNA was isolated and used for Northern blot analysis as described in the legend of Fig. 5.
Expression in E. coli and Activity of Recombinant Protoporphyrinogen IX Oxidases I and II.
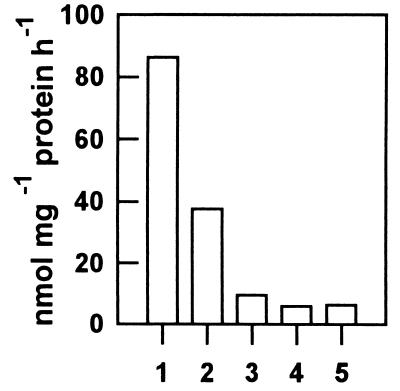
Because purification of protoporphyrinogen IX oxidase activity from plant sources has been found to be rather difficult (12), we attempted to overproduce active enzyme in E. coli. Molecular masses of the recombinant putative mature plastidal (53 kDa) and the mitochondrial isoenzyme (59 kDa) separated by SDS/PAGE were in good agreement with the mass of deduced amino acid sequence (data not shown). Induced synthesis of the mitochondrial enzyme in E. coli strain SG 13009 for 4 h resulted in large amounts of active protein. Synthesis of the plastidal enzyme was immunologically demonstrated but also resulted in accumulation of several other low-molecular-mass peptides (data not shown). Bacterial extracts were collected after different time periods of induced synthesis of the two recombinant protoporphyrinogen IX oxidases. A significant activity of the plastidal enzyme could not be shown relative to the heat-denatured control (data not shown). Protoporphyrinogen IX oxidase activity was determined from bacterial extract containing the recombinant isoenzyme II. This activity was inhibited by acifluorfen at saturating substrate concentration (Fig. 7). Acifluorfen (100 nM) reduced the enzyme activity to less than 50%. This level of inhibition indicates that the tobacco mitochondrial protoporphyrinogen IX oxidase seems to be more sensitive to acifluorfen than the recombinant enzyme of Arabidopsis (26) or the native enzyme from barley etioplasts (18) and human tissues (43, 44), but less prone than the yeast enzyme (21).
Figure 7.
Activity of the recombinant isoform II of protoporphyrinogen IX oxidase. An E. coli clone expressing the mitochondrial protoporphyrinogen IX oxidase (PPX II) was lysed. Enzyme activity of the bacterial homogenate (1) in the presence of 100 nM (2), 1 μM (3), and 10 μM (4) acifluorfen, and endogenous protoporphyrinogen IX oxidase activity of the E. coli strain SG 13009 harboring the plasmid pDS 56 (5) were measured as described in Material and Methods.
DISCUSSION
Two different cDNA sequences have been identified by complementation of the heme auxotrophic E. coli hemG mutant lacking protoporphyrinogen IX oxidase activity. Both deduced peptide sequences contain only 27.2% invariant amino acid residues. The antiserum raised against each of the two isoenzymes did not cross-react with the other (Fig. 3). The two enzymes share only a few identical domains that are also conserved in other bacterial or eukaryotic HemY-type protoporphyrinogen IX oxidase (Fig. 1). The significance of each invariant amino acid residue for structural or functional roles remains to be elucidated. The low homology between both tobacco isoforms encouraged us to search for differences of their gene expression, localization, and biochemical properties.
Early enzymes of tetrapyrrole synthesis are detected exclusively in chloroplasts. Protoporphyrinogen IX oxidase is the first enzyme of the pathway whose activity was determined in plastids as well as in mitochondria (11). In vitro translated protoporphyrinogen IX oxidase I was selectively directed to plastids and accumulated in membrane and stroma fractions (Fig. 2). However, the immunoreactive plastidal enzyme was only detected in the membrane fraction of green plastids (Fig. 3), which is consistent with data on the protoporphyrinogen IX oxidase activity that were found entirely in thylakoid membranes and envelopes of spinach chloroplast (13). The occurrence of the isoenzyme in the stromal fraction might represent an intermediate state of the import or integration process, which has also been observed for other plastid proteins, such as light-harvesting chlorophyll-binding proteins and Rieske iron-sulfur protein (45, 46). In these cases soluble proteinaceous stromal factors have been postulated to promote integration of the processed protein into the thylakoid membrane. The protoporphyrinogen IX oxidase deduced from the Arabidopsis cDNA sequence shares a high similarity with the tobacco isoenzyme I. The assumption that the former protein is localized in mitochondria (26) is in contrast to our import experiment.
The second isoenzyme is specifically recognized by the mitochondrial import machinery and was targeted to and proteolytically protected in isolated mitochondria. The information for mitochondrial import is usually located at the N terminus of the enzyme. The imported mitochondrial enzyme did not show an obvious reduction in size. The enzymes from human and mouse (23, 25) share with tobacco isoform II at the N terminus a rather short stretch of amino acid residues in front of the flavin binding site. The homologous human protein imported in vitro into mitochondria also maintained the molecular size (23), suggesting similar translocation mechanisms of the mammalian and plant mitochondrial enzyme. Compared with other protoporphyrinogen IX oxidase, the yeast enzyme contains an N-terminal extension of 13 amino acid residues that directs the protein to mitochondria and is visibly cleaved off (21). A small transit peptide could be cleaved off from tobacco mitochondrial enzyme, although no clear-cut transit sequence is detectable. Alternatively, this enzyme is inserted into the membranes without any modification, assuming that a targeting signal different from a cleavable peptide extension allows trafficking to the inner membrane. A number of proteins were presented earlier that are localized in the inner membrane space and the outer and inner mitochondrial membrane and do not contain N-terminal targeting signals (47, 48). Apocytochrome c is also initially transferred into the intermembrane space without a processing step. Covalent attachment of heme stabilizes the protein at its target site (49). At the moment, it is speculative whether the substrate protoporphyrinogen IX effects routing of protoporphyrinogen IX oxidase to its target site. Along this line, accumulation of the plastidal protoporphyrinogen IX oxidase in the stroma fraction and interruption of the transfer to the thylakoids could also be explained by the lack of protoporphyrinogen IX.
The mitochondrial heme synthesis requires a transfer system of protoporphyrinogen IX from chloroplasts that protects the substrate from photooxidation. Herbicidal inhibition of PPX and feeding of 5-aminolevulinate caused accumulation of protoporphyrin(ogen) IX in the cytoplasm. This suggests a directed export of protoporphyrinogens from plastids to mitochondria (17). Two pathways of porphyrin transport are suggested. Porphyrins could be actively transported across the cytoplasm. Alternatively, protoporphyrinogen IX is directly channeled from enzyme to enzyme both attached to the membranes of chloroplasts and mitochondria. Additional porphyrin carrier proteins have to be postulated in both cases. The activity of protoporphyrinogen IX oxidase in the plastidal envelopes could be interpreted with a plastidal enzyme involved simultaneously in protoporphyrinogen IX transfer and tetrapyrrole synthesis. However, this idea requires a mechanism that modulates the activity of the plastidic isoenzyme between promotion of substrate channeling and catalytic conversion to protoporphyrin (13).
The mitochondrial form is entirely associated with the capacity for heme synthesis. The plastidic enzyme activity functions in the formation of heme required for apoproteins of the cytochrome b/f complex and, predominantly, of chlorophyll. Compartmentalized isoenzymes with different functions were expected to be differentially regulated in response to cellular and environmental requirements. The developmental and clock-controlled expression of genes encoding both isoforms did not exhibit significant differences between both subfamilies (Figs. 5 and 6). Immune-reactive protoporphyrinogen IX oxidases I and II followed almost the same pattern as their steady-state RNA content (data not shown). We speculate that protoporphyrinogen IX oxidase might have regulatory significance for the distribution of protoporphyrin between both organelles.
The characterization of the two isoenzymes located in the two different cellular compartments will expedite studies both on the interaction between mitochondrial and plastidal pathway, including the regulatory mechanism of substrate distribution, and on the identification of additional porphyrin transport carrier.
Acknowledgments
Prof. Inokuchi, Kyoto University, provided E. coli hemG mutant. We thank Dr. L. Grohmann for making a cDNA clone encoding a subunit of NADH:ubiquinone oxidoreductase available. This work was supported by Deutsche Forschungsgemeinschaft grants to B.G. (Gr 936/3-1 and Gr 936/4-1).
ABBREVIATION
- PPX
protoporphyrinogen IX oxidase
Footnotes
References
- 1.Dailey H A, editor. Biosynthesis of Heme and Chlorophyll. New York: McGraw–Hill; 1990. [Google Scholar]
- 2.Beale S I, Weinstein J D. In: Biosynthesis of Heme and Chlorophyll. Dailey H A, editor. New York: McGraw–Hill; 1990. pp. 287–391. [Google Scholar]
- 3.Smith A G, Marsh O, Elder G H. Biochem J. 1993;292:503–508. doi: 10.1042/bj2920503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sano S, Granick S. J Biol Chem. 1961;236:1173–1180. [PubMed] [Google Scholar]
- 5.Porra R J, Falk J E. Biochem J. 1964;90:69–72. doi: 10.1042/bj0900069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulson R, Polglase W J. J Biol Chem. 1975;250:1269–1274. [PubMed] [Google Scholar]
- 7.Poulson R. J Biol Chem. 1976;251:3730–3733. [PubMed] [Google Scholar]
- 8.Siepker L J, Ford M, de Kock R, Kramer S. Biochim Biophys Acta. 1987;913:349–358. doi: 10.1016/0167-4838(87)90146-4. [DOI] [PubMed] [Google Scholar]
- 9.Dailey H A, Karr S W. Biochemistry. 1987;26:2697–2701. doi: 10.1021/bi00384a007. [DOI] [PubMed] [Google Scholar]
- 10.Deybach J C, da Silva V, Grandchamp B, Nordmann Y. Eur J Biochem. 1985;149:431–435. doi: 10.1111/j.1432-1033.1985.tb08943.x. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs N J, Jacobs J M. Biochem J. 1987;244:219–224. doi: 10.1042/bj2440219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs N J, Borotz S E, Jacobs J M. Biochem Biophys Res Commun. 1989;161:790–796. doi: 10.1016/0006-291x(89)92669-7. [DOI] [PubMed] [Google Scholar]
- 13.Matringe M, Camadro J-M, Block M A, Joyard J, Scalla R, Labbe P, Douce R. J Biol Chem. 1992;267:4646–4651. [PubMed] [Google Scholar]
- 14.Matringe M, Camadro J-M, Labbe P, Scalla R. Biochem J. 1989;260:231–235. doi: 10.1042/bj2600231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witkowski D A, Halling B P. Plant Physiol. 1989;87:1239–1242. doi: 10.1104/pp.90.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duke S O, Lydon J, Becerril J M, Sherman T D, Lehnen L P, Matsumoto H. Weed Sci. 1991;39:465–473. [Google Scholar]
- 17.Jacobs J M, Jacobs N J. Plant Physiol. 1993;101:1181–1187. doi: 10.1104/pp.101.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H J, Duke M V, Duke S O. Plant Physiol. 1993;102:881–889. doi: 10.1104/pp.102.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasarman A, Letowski J, Czaika G, Ramirez V, Nead M A, Jacobs J M, Morais R. Can J Microbiol. 1993;39:1155–1161. doi: 10.1139/m93-174. [DOI] [PubMed] [Google Scholar]
- 20.Hansson M, Hederstedt L. J Bacteriol. 1992;174:8081–8093. doi: 10.1128/jb.174.24.8081-8093.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camadro J-M, Thome F, Brouillet N, Labbe P. J Biol Chem. 1994;269:32085–32091. [PubMed] [Google Scholar]
- 22.Camadro J-M, Labbe P. J Biol Chem. 1996;271:9120–9128. doi: 10.1074/jbc.271.15.9120. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura K, Taketani S, Inokuchi H. J Biol Chem. 1995;270:8076–8080. doi: 10.1074/jbc.270.14.8076. [DOI] [PubMed] [Google Scholar]
- 24.Taketani S, Yoshinaga T, Furukawa T, Kohno H, Tokunaga R, Nishimura K, Inokuchi H. Eur J Biochem. 1995;230:760–765. doi: 10.1111/j.1432-1033.1995.0760h.x. [DOI] [PubMed] [Google Scholar]
- 25.Dailey T A, Dailey H A, Meissner P, Prasad A R K. Arch Biochem Biophys. 1995;324:379–384. doi: 10.1006/abbi.1995.0051. [DOI] [PubMed] [Google Scholar]
- 26.Narita S, Tanaka R, Ito T, Okada K, Taketani S, Inokuchi H. Gene. 1996;182:169–175. doi: 10.1016/s0378-1119(96)00545-8. [DOI] [PubMed] [Google Scholar]
- 27.Kruse E, Mock H-P, Grimm B. EMBO J. 1995;14:3712–3720. doi: 10.1002/j.1460-2075.1995.tb00041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura K, Nakayashiki T, Inokuchi H. DNA Res. 1995;2:1–8. doi: 10.1093/dnares/2.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1979;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Frisch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 32.Stüber D, Ibrahim I, Cutler D, Dobbersten B, Bujard H. EMBO J. 1984;3:3143–3148. doi: 10.1002/j.1460-2075.1984.tb02271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottesmann S, Halpern E, Trisler P. J Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith A G, Griffiths W T. In: Methods in Plant Biochemistry. Dey P M, Harborne J B, editors. London: Academic; 1993. pp. 299–344. [Google Scholar]
- 35.Knorpp C, Hugosson M, Sjöling S, Eriksson A, Glaser E. Plant Mol Biol. 1994;26:571–579. doi: 10.1007/BF00013744. [DOI] [PubMed] [Google Scholar]
- 36.Grohmann L, Rasmusson A G, Heiser V, Thieck O, Brennicke A. Plant J. 1996;10:793–803. doi: 10.1046/j.1365-313x.1996.10050793.x. [DOI] [PubMed] [Google Scholar]
- 37.Grossman A R, Bartlett S G, Schmidt G W, Mullet J E, Chua H-H. J Biol Chem. 1982;257:1558–1563. [PubMed] [Google Scholar]
- 38.Grimm B, Kruse E, Kloppstech K. Plant Mol Biol. 1989;13:583–593. doi: 10.1007/BF00027318. [DOI] [PubMed] [Google Scholar]
- 39.Wierenga R K, Terpstra P, Hol W G J. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 40.Kruse E, Mock H-P, Grimm B. Planta. 1995;196:796–803. [PubMed] [Google Scholar]
- 41.Rasmusson, A. G., Heiser, V., Irrgang, K. D., Brennicke, A. & Grohmann, L. (1997) Planta, in press. [DOI] [PubMed]
- 42.Kloppstech K. Planta. 1986;165:502–506. doi: 10.1007/BF00398095. [DOI] [PubMed] [Google Scholar]
- 43.Corrigall A V, Hift R J, Adams P A, Kirsch R E. Biochem Mol Biol Intern. 1994;34:1283–1289. [PubMed] [Google Scholar]
- 44.Dailey T A, Dailey H A. Protein Sci. 1996;5:98–105. doi: 10.1002/pro.5560050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madueno F, Napier J A, Gray J C. Plant Cell. 1993;5:1865–1876. doi: 10.1105/tpc.5.12.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan J, Henry R, Cline K. Proc Natl Acad Sci USA. 1993;90:8552–8556. doi: 10.1073/pnas.90.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmermann R, Paluch U, Sprinzl M, Neupert W. Eur J Biochem. 1979;99:247–252. doi: 10.1111/j.1432-1033.1979.tb13251.x. [DOI] [PubMed] [Google Scholar]
- 48.Zimmermann R, Paluch U, Neupert W. FEBS Lett. 1979;108:141–146. doi: 10.1016/0014-5793(79)81196-5. [DOI] [PubMed] [Google Scholar]
- 49.Stuart R A, Nicholson D W, Neupert W. Cell. 1990;60:31–43. doi: 10.1016/0092-8674(90)90713-o. [DOI] [PubMed] [Google Scholar]