Abstract
Under nitrogen-limiting conditions Rhizobium meliloti can establish symbiosis with Medicago plants to form nitrogen-fixing root nodules. Nodule organogenesis starts with the dedifferentiation and division of root cortical cells. In these cells the early nodulin gene enod40, which encodes an unusually small peptide (12 or 13 amino acids), is induced from the beginning of this process. Herein we show that enod40 expression evokes root nodule initiation. (i) Nitrogen-deprived transgenic Medicago truncatula plants overexpressing enod40 exhibit extensive cortical cell division in their roots in the absence of Rhizobium. (ii) Bombardment of Medicago roots with an enod40-expressing DNA cassette induces dedifferentiation and division of cortical cells and the expression of another early nodulin gene, Msenod12A. Moreover, transient expression of either the enod40 region spanning the oligopeptide sequence or only the downstream region without this sequence induces these responses. Our results suggest that the cell-specific growth response elicited by enod40 is involved in the initiation of root nodule organogenesis.
Keywords: nodule initiation, plant growth regulator, early nodulin, nitrogen control
Root nodules are symbiotic organs where nitrogen fixation takes place, allowing legume plants to grow in the absence of combined nitrogen. Rhizobium bacteria trigger nodule development through Nod factor production, lipochitooligosaccharide signals (1–3). These signals induce the dedifferentiation and division of root cortical cells where amyloplasts accumulate. Little is known about the molecular mechanisms implicated in nodule organogenesis. The induction of certain genes, the early nodulin (enod) genes, has been demonstrated, although their putative roles have been proposed mainly on the basis of expression. Nevertheless, these genes serve as molecular markers of this developmental process. For example, in alfalfa Msenod12A expression is detected in the initially dividing cortical cells induced either by Rhizobium meliloti or its Nod factor, and transgenic alfalfa carrying an Msenod12A promoter–uidA fusion have, therefore, been used to study the reprogramming of inner cortical cells for nodule initiation (4).
enod40 is one of the first molecular markers expressed at the onset of nodule organogenesis (5–8). Although in various plant species enod40 genes encode highly conserved transcripts of about 0.7 kb, only an unusually small peptide of 12 or 13 amino acids (8, 9) is translated. In addition, computer analyses of the enod40 nucleotide sequences indicate a tendency to form stable secondary structures, a property shared with biologically active RNAs (7). We have previously identified genes, including enod40, expressed in spontaneous nodules (7, 10), nodule-related organs developed in the absence of Rhizobium on certain alfalfa plants during nitrogen limitation (11, 12). This indicates that enod40 expression is associated to the nodule developmental program independently of any infection process. Interestingly, an enod40 homolog was recently found in tobacco whose expression increases auxin tolerance of tobacco protoplasts (9).
To investigate the possible involvement of enod40 in nodule development, we analyzed the effects of the stable and transient expression of enod40 in Medicago roots. In both cases, enod40 expression induced dedifferentiation and division of cortical cells under nitrogen-limiting conditions. Our results suggest that this early nodulin gene is involved in the initiation of nodule development.
MATERIALS AND METHODS
Transformation of Medicago truncatula.
A full-length Mtenod40 cDNA (7) was inserted in the sense orientation in the BamHI site of the binary vector pG3.3 (provided by B. Charrier and P. Ratet, Centre National de la Recherche Scientifique) behind the 35S promoter yielding pGMCSe40. pG3.3 was derived from the binary vector pBin19 (13) and contains a 35S promoter–uidA–35S poly(A) cassette and a second 35S promoter with a polylinker having a unique BamHI site. pGMCSe40 was transferred to Agrobacterium tumefaciens EHA105 by triparental mating (14). M. truncatula 108R plants were transformed as described (15). Segregation of kanamycin resistance, GUS activity (4), and enod40 expression (7) coupled to Southern blot analyses (16) confirmed the transgenic nature of these plants. Inverse PCR (17) indicated the presence of multiple transgene copies (data not shown). Total leaf RNA (10 μg) was used for Northern blot analyses (16). As a positive control, total RNA from young M. truncatula nodules was used and Msc27 served as a RNA loading control (7).
Transient Assay.
Cuttings of transgenic Medicago sativa ssp. varia A2 containing an Msenod12A promoter–uidA fusion (4) were rooted and grown in a medium containing 0.25 mM nitrate. Roots were bombarded on MS medium (ref. 18; 1.5% sucrose/1% Bactoagar) without combined nitrogen, by using a Biolistic PDS 1000/He particle gun [Bio-Rad; rupture pressure, 1350 psi (1 psi = 6.9 kPa); vacuum, 28 in. Hg (1 in. Hg = 3.4 kPa); gap distance, 0.25 in. (1 in. = 2.54 cm); macrocarrier flight distance, 11 mm; microcarrier flight distance, 9 cm; 1.6-μm gold microcarriers]. Constructs used were 35S–Mtenod40 [full-length Mtenod40 (7)], 35S–Mtenod40–Δ7 (nucleotides 31–220), and 35S–Mtenod40–Δ5 (nucleotides 204–611) in the vector pDH51 (19). In each experiment, two to six plants were bombarded. In optimization bombardments, the positive control was pRT103 (20) (35S–uidA). In enod40 bombardments, expression of an irrelevant gene was used as an additional negative control. After 2 days, the roots were checked for β-glucuronidase (GUS) activity.
Histological Analysis and GUS Activity in Medicago.
M. truncatula roots were fixed in FAA as described (21) and cleared for 1 h in commercial bleach (1.75% active chlorine). Prefixation and GUS staining of bombarded M. sativa roots were done according to Bauer et al. (4). Roots were embedded in Spurr resin (22) or paraffin (4), sectioned, and stained with 0.2% toluidine blue. Starch was stained by using Lugol’s solution and nuclei were stained by using 4,6-diamidino-2-phenylindole (23).
RESULTS
M. truncatula Plants Overexpressing enod40 Exhibit Extensive Cortical Cell Divisions in Their Roots Under Nitrogen Deprivation.
Several transgenic plants overexpressing the Mtenod40 gene from the cauliflower mosaic virus 35S promoter were obtained using M. truncatula, a diploid autogamous species, although our initial experiments with M. sativa (alfalfa) showed that the regeneration of somatic embryos was affected by alterations of enod40 expression (7). Two strongly overexpressing M. truncatula plants showed major alterations in growth and development and could not be maintained. Nevertheless, several fertile transgenic plants with relatively high levels of transgene expression were recovered. Their F2 progeny stably inherited the transgene and grew similarly to control plants (Fig. 1a). Northern blot analyses of individual plants indicated variable levels of transgene expression (Fig. 1b). No differences in transgene expression were found between roots and leaves (data not shown). As expected, no (endogenous) enod40 expression could be detected in control leaves (Fig. 1b).
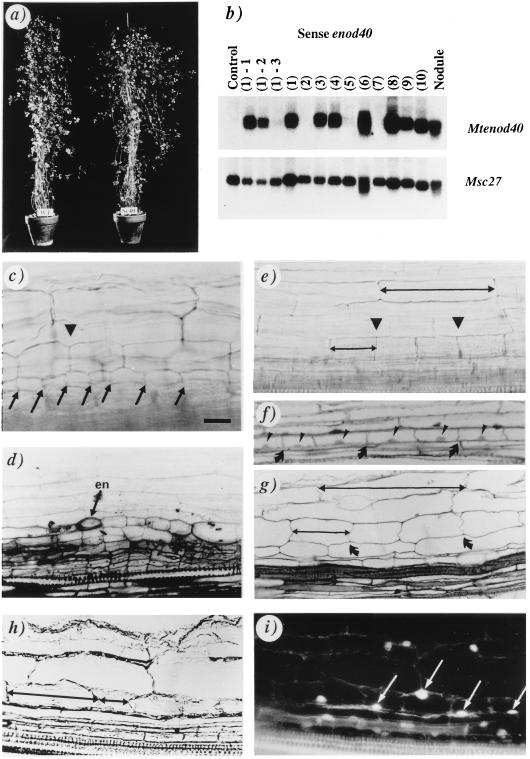
Figure 1.
Analysis of M. truncatula transgenic plants carrying the 35S–Mtenod40 construct. (a) An F2 plant (Se40) overexpressing Mtenod40 compared with a wild-type plant (WT). (b) Northern blot analysis of one control and 10 transgenic enod40 plants with an Mtenod40 probe. Progeny of the enod40 transgenic plant 1 [lanes (1)-1 to (1)-3] are also included. Msc27 was used as RNA-loading control (see ref. 7). (c and d) Lateral root primordia with dividing cortical cells (arrowhead) and divisions in the pericycle (arrows; en, endodermis). (e–i) Mtenod40-induced cortical cell division (e, arrowheads; f and g, arrows). Divisions of pericycle cells cannot be detected. A nucleus is present in each cell (f and i). Compare the size of undivided and divided cortical cells (double arrows in e, g, and h). Dividing cortical cells in a bright-field micrograph (h), showing fluorescent nuclei after 4,6-diamidino-2-phenylindole staining (arrows in i). Whole roots (c and e), 2-μm sections (d, f, and g), 14-μm sections (h and i). [Bar = 20 μm (c, e, h, and i) and 25 μm (d, f, and g).]
The F2 plants were grown under nitrogen-limiting conditions, as required for nodule initiation, and the different patterns of cell division in their roots were analyzed. Divisions occurred either in (i) the pericycle and cortex (putative lateral root primordia; Fig. 1 c and d) or (ii) the cortex only (cortical cell divisions, Fig. 1 e–g). The latter were not accompanied by simultaneous divisions of pericycle cells and endodermis (Fig. 1, compare c and d to e–g). Microscopical analysis of root sections confirmed this localization (Fig. 1 d, f, and g). Division of inner cortical cells was observed at several places in normally developed roots. Shortening of the roots was not detected (data not shown) and the outer cortex contained cells of normal size (Fig. 1e). The presence of nuclei in all divided cells (Fig. 1 f, h, and i) further confirmed that these morphological changes were not due to variations in cell enlargement. Other tissues did not show altered division patterns.
A significant increase in the number of cortical cell divisions per cm of root was found in three independent transgenic plants exhibiting a high level of enod40 expression, when compared with three control plants (Fig. 2a). No differences were found in the number of lateral root primordia between these plants. In the enod40 transgenic plants, cortical cell divisions were more frequent in the upper half of the root, below the hypocotyl. Interestingly, the spontaneous nodules formed on certain alfalfa varieties also appear most frequently in this region (11, 12) and strongly express Msenod40 (7). In the middle part (6–11 cm below the hypocotyl), cortical cell divisions were common in transgenic plants and also detected in the equivalent root region of control plants, albeit at lower frequency.
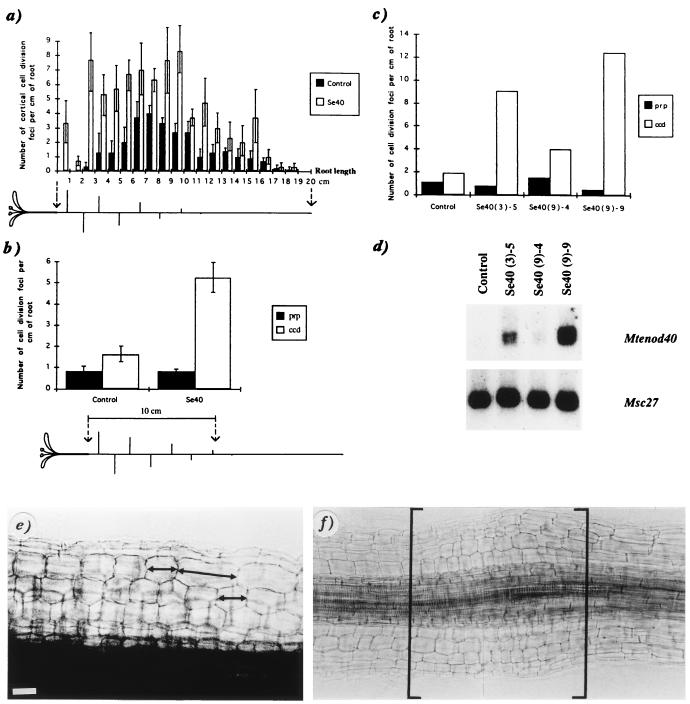
Figure 2.
Correlation between Mtenod40 overexpression and cortical cell division under nitrogen limitation. (a) Number of cortical cell division along the main root for three controls and three independent enod40 plants (Se40) as a function of the distance from the hypocotyl (mean ± SEM). The increase in the number of cortical cell divisions cosegregated 1:1 with the level of transgene expression in the progeny (data not shown). (b) Numbers of cell division foci (ccd, cortical cell divisions, or prp, putative root primordia), in the main root along the first 10 cm below the hypocotyl, for four controls and six independent transgenics (progenies of transgenic lines Se40–3, Se40–9, and Se40–10; mean ± SEM). A plant, with main and lateral roots, is schematically represented below the graph (a and b). (c) Numbers of cell division foci (ccd or prp), along the upper 10 cm of the root, for three transgenic individuals (progeny of Se40–3 and Se40–9) and one control individual. Roots were used for microscopical analysis, and the leaves were used for Northern blot analysis (d) of these individual plants. Root region of an enod40 transgenic plant showing different sizes of cortical cells (e) and an increased diameter due to extensive cortical divisions (in brackets) (f). [Bar = 20 μm (e) and 40 μm (f).]
Further analysis of the upper root region in independent enod40 transgenic plants (progeny of the transgenic lines Se40–3, Se40–9, and Se40–10) revealed cortical cell divisions at high frequency as compared with controls (Fig. 2b), whereas no differences were found in the number of lateral root primordia. Analysis of individual transgenic plants indicated a correlation between the level of transgene expression and the induction of cortical cell division in their roots (Fig. 2 c and d). These divisions were occasionally followed by extensive anticlinal divisions in the entire cortex (Fig. 2e). After 1 month of nitrogen deficiency, certain root portions of enod40 plants even showed an increased diameter (Fig. 2f), forming small bumps. In contrast, no cortical cell divisions were detected in plants grown with combined nitrogen. Control experiments showed that expression of the 35S–enod40 fusion was not affected by this treatment.
Thus, these results indicate that enod40 expression leads to the induction of cortical cell division, which is the initial event in nodule development.
Transient Expression of enod40 Induces Cortical Cell Division and Msenod12A Expression in Alfalfa.
In transgenic plants carrying a Msenod12A promoter–uidA fusion, GUS staining (blue color) of roots marks cortical cell divisions (4). To further analyze the involvement of enod40 in the elicitation of cortical cell division, a transient assay based on particle bombardment of roots from these plants was therefore developed. Control experiments indicated that roots bombarded with vectors or noncoated gold particles did not give blue coloration (Table 1, optimization bombardments; Fig. 3a). Hence, wounding of roots by bombardment did not induce the Msenod12A promoter. Bombardment with a 35S–uidA gene cassette, conferring constitutive GUS expression, yielded several cells transiently expressing GUS in the epidermis and the cortex (Fig. 3b). The blue precipitate was usually confined to single cells and restricted by cell walls. Moreover, a gold particle was frequently observed in the nucleus of the blue cells (Fig. 3c), confirming the transient expression of GUS.
Table 1.
Transient gene expression in transgenic Msenod12A–uidA roots
| Plants, no. | Roots, no. | % + roots | No. + per plant | No. + per root | No. + per + root | |
|---|---|---|---|---|---|---|
| Optimization bombardments | ||||||
| 35S–uidA | 10 | 37 | 97 | 20 | 5.5 ± 0.6 | 5.7 ± 0.6 |
| 35S vector | 16 | 72 | 0 | 0 | 0 ± 0 | 0 ± 0 |
| Au | 10 | 30 | 0 | 0 | 0 ± 0 | 0 ± 0 |
| enod40 bombardments | ||||||
| 35S–Mtenod40 | 28 | 148 | 15 | 1.0 | 0.19 ± 0.04 | 1.3 ± 0.1 |
| 35S–Mtenod40–Δ7 | 18 | 86 | 10 | 0.60 | 0.13 ± 0.04 | 1.1 ± 0.1 |
| 35S–Mtenod40–Δ5 | 25 | 105 | 14 | 1.2 | 0.30 ± 0.1 | 2.1 ± 0.6 |
| 35S–Mtenod40 (+N) | 10 | 21 | 0 | 0 | 0 ± 0 | 0 ± 0 |
| Control DNA | 40 | 142 | 0 | 0 | 0 ± 0 | 0 ± 0 |
| NF + Au | 6 | 38 | 24 | 2.7 | 0.42 ± 0.1 | 1.8 ± 0.3 |
| BAP + Au | 6 | 48 | 17 | 2.5 | 0.31 ± 0.1 | 1.9 ± 0.2 |
| BAP − Au | 4 | 15 | 13 | 0.50 | 0.13 ± 0.09 | 1.00 ± 0 |
Number of positives (+, see below) were counted after particle gun bombardment of roots. Optimization bombardments: positives correspond to nondividing cells transiently expressing the introduced 35S–uidA gene. enod40 bombardments: positives correspond to sites showing cortical cell divisions (ccd) and expression of the endogenous Msenod12A–uidA gene. Numbers of positives are given (mean ± SEM). % + roots is the percentage of roots showing positives per total number of roots. No. + per + root is the number of positives per root showing a response. Au, bombardment with gold particles without bound DNA. +N, cuttings were grown in the presence of combined nitrogen. Control DNA, 35S vector or vector expressing an irrelevant gene. NF, 0.1 μM NodRm-IV (C16:2, AcS) Nod factor (kindly provided by M. Schultze) was added to the roots before bombardment (Au). BAP, 10 μM benzylaminopurine was similarly added, with (+Au) or without (−Au) a following bombardment. Nod factor and BAP treatments induce ccd and Msenod12A (4).
Figure 3.
Transient gene expression in roots of transgenic plants carrying an Msenod12A–uidA fusion after particle bombardment: Optimization bombardments: root bombarded with vector DNA [pDH51 (19)] (a) or with a 35S–uidA construct (20) (b). Blue spots correspond to cells transiently expressing GUS. (c) A cortical cell showing GUS activity with a gold particle (within the ring) in the nucleus. enod40 bombardments: division of cortical cells expressing Msenod12A (blue) in a root bombarded with a 35S–Mtenod40 construct (d). (e) Interference contrast micrograph of a cortical cell division after bombardment as in d. (f and g) Cortical cell division with accumulated amyloplasts (arrows) as seen in 2-μm sections after toluidine blue (f) and Lugol staining (g), respectively. (h) Autofluorescence revealed nuclei (arrows) in dividing cells (8-μm section). (i) Transversal section (8 μm) showing cell divisions (white bracket) outside the endodermis (fluorescent cell walls; en) in front of a xylem pole (x). ph, Phloem tissue. Large and small arrows are as in Fig. 1e. [Bar = 100 μm (a and b), 12 μm (c and i), 25 μm (d–g), and 20 μm (h).]
Subsequently, transient expression of enod40 was assayed on the roots of the transgenic plants carrying the Msenod12A promoter–uidA fusion. After bombardment with a 35S–enod40 DNA construct and then GUS staining, dividing inner cortical cells expressing Msenod12A were detected (Table 1, enod40 bombardments; Fig. 3 d and e). Sectioning of samples confirmed the division of cortical cells with net amyloplast deposition (Fig. 3 f and g) and the central position of nuclei in these cells suggesting recent division (Fig. 3h). Bombardment with control DNA did not induce either cortical cell divisions or Msenod12A expression in the root cortex. Divisions were mainly detected in the inner cortical cells, whereas gold particles were found in neighboring cells of outer layers. This result suggests that enod40 expression resulted in a signal able to reach the inner cortex. Cell divisions related to root primordium development were distinguished from the cortical cell divisions induced by enod40 as described above and were present in both enod40- and control-bombarded roots (data not shown). In lateral roots, expression of Msenod12A was detectable only when the primordium was formed (4), and hence sites of cell divisions in the pericycle normally did not express Msenod12A. These pericycle sites are internal to the typically autofluorescent endodermis (data not shown). In contrast, dividing cortical cells were found external to the endodermis in the enod40-bombarded roots (Fig. 3 g and i).
Treatment of roots with Nod factors or cytokinins prior to bombardment with noncoated particles induced cortical cell division and Msenod12A expression as expected (4) and at a frequency comparable to that observed for transient enod40 expression (Table 1). Plants grown in the presence of combined nitrogen showed no cortical cell divisions after bombardment with enod40 DNA (Table 1). This further supports that enod40-induced cortical cell divisions are connected to nodule initiation.
Thus, these results indicate that enod40 overexpression, either stably or transiently, is able to activate division of certain root cortical cells in legumes.
Both the Oligopeptide-Spanning Region of enod40 and the Region Downstream of This Sequence Are Biologically Active.
Expression of enod40 and its small encoded peptide confers auxin tolerance to tobacco protoplasts (9). Surprisingly, expressing the 3′ region of enod40 (lacking the oligopeptide-coding sequence) also had this effect. In our transient assay, we tested Mtenod40 derivatives spanning the oligopeptide region (Δ7) and a 3′ region lacking this coding sequence (Δ5). Both constructs elicited cortical cell division and Msenod12A expression at similar frequency as the complete construct (Table 1). These results indicate that enod40 contains at least two regions that induce cortical cell division and their mode of action might be connected through a regulatory circuit controlling gene function.
DISCUSSION
Under nitrogen-limiting conditions, R. meliloti and its Nod factor activate the early expression of enod40 in the pericycle and cortex (5–8), followed by the dedifferentiation and division of cortical cells that show amyloplast deposition and Msenod12A expression (2, 4). Herein we show that overexpression of enod40 (stably or transiently) induces the latter responses under similar nitrogen control. This suggests that enod40 action is involved in the reactivation of inner cortical cells, the initial step required for nodule organogenesis. Therefore, we propose that enod40 is a regulatory gene involved in a signal transduction pathway activated during nodule development.
The enod40-induced divisions were mainly detected in inner cortical cells where active nuclei, enod12A expression, and localized amyloplast accumulation were seen. This result confirmed that the observed phenotype is not due to changes in cell enlargement, as can be induced by Rhizobium and Nod factors in Vicia (thick short root or tsr phenotype; ref. 24), an effect mediated by ethylene. Moreover, the tsr phenotype was not observed in the transgenic plants. The inner cortical cells are the main targets of enod40 action because no divisions were detected in other cell types (e.g., pericycle or epidermis) expressing the transgene. Furthermore, the transient assay suggests that the product of the enod40 gene may move in the root symplasmic domain or be secreted (as found for the oligopeptide in the tobacco protoplast system; ref. 9), to reach the inner cortex where its action was detected. As reported (refs. 1, 2, and 4 and references therein), other factors, such as morphogen gradients, carbon allocation, and/or nitrogen status, determine the competence of these cells for division. Furthermore, enod40-induced divisions were under similar nitrogen control although an organized meristem, as induced in alfalfa by Rhizobium (1), was not formed. It is likely that nodule primordium formation is a complex process requiring the action of additional genes and signals. Nevertheless, our work suggests that enod40 participates at least in the beginning of this process by evoking the dedifferentiation and division of cortical cells.
Cytokinins and auxin transport inhibitors mimick Nod factor action in the inner cortex, suggesting that phytohormonal imbalances might occur during nodule initiation (1, 4, 25). Since enod40 is induced both by cytokinins and Nod factors (7, 8, 26), our results suggest that its expression may generate such hormonal imbalances in the root cortex. The reported action of enod40 in tobacco protoplasts (9) might be related to this activity. During nodule organogenesis however, enod40 apparently acts specifically in the cortex because Nod factor derivatives unable to elicit cortical cell division still induce enod40 expression in the root pericycle but not in the cortex (27). Moreover, our enod40 transgenic plants did not show significant alterations in the number or development of lateral roots, which are initiated in the pericycle. The cellular role of enod40 is still unknown; therefore, we cannot exclude more subtle alterations (e.g., in the transport of specific compounds) that might occur in cells of this or other plant tissues.
enod40 function is not nodule-specific because the gene is expressed in other tissues (5–7), and a nonlegume homolog has been identified (9). As mentioned, the enod40 peptide modifies a phytohormonal response in tobacco protoplasts, suggesting that it may be a peptide growth regulator. This together with our results indicating its participation in nodule organogenesis point to an exciting parallel with the well-characterized action of peptide hormones in animal development.
The molecular mechanism of enod40 action seems to be particularly interesting. If enod40 action is only mediated by the oligopeptide, then the capacity of its 3′ region to induce auxin tolerance (9) and cortical cell division (this work) is intriguing. Small ORFs exist within this 3′ region even though they are not conserved in different species. Moreover, to our knowledge, enod40 is the first eukaryotic example (9) of a small oligopeptide acting as the primary translation product. We cannot at present exclude that a second oligopeptide encoded by enod40 is responsible for this activity, but the enod40 genes more probably possess a 3′ untranslated region able to act in trans. Our results indicate that a regulatory process where the enod40 RNA is involved might occur during nodule development. Recently, regulatory 3′ untranslated regions have been implicated in the control in trans of the spatial localization of morphogens in Drosophila embryos (28) and of certain differentiation processes in somatic mammalian cells (29). Therefore, analysis of the molecular mechanism through which the two biologically active regions of enod40 elicit cell-specific growth responses in plants would give novel insights into regulatory processes involved in organogenesis and differentiation.
Acknowledgments
We gratefully acknowledge the help and technical advice of B. Hoffman and H. Trinh in preparing transgenic plants. We thank C. Sousa for the preparation of Δ7; P. Bauer for advice in our initial bombardment experiments; B. Satiat-Jeunemaître, S. Brown, and J. Laporte for suggestions regarding microscopy; S. Poirier for plant tissue culture advice; and B. Gronenborn and J. Schell for careful reading of the manuscript. C.C. and C.J. were supported by the Ministère Français de l’Enseignement Supérieur et de la Recherche and the Swedish Council for Agricultural and Forestry Research, respectively.
ABBREVIATION
- GUS
β-glucuronidase
References
- 1.Hirsch A M. New Phytol. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 2.Mylona P, Pawlowski K, Bisseling T. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé J C, Dénarié J. Nature (London) 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 4.Bauer P, Ratet P, Crespi M D, Schultze M, Kondorosi A. Plant J. 1996;10:91–105. doi: 10.1104/pp.105.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W-C, Katinakis P, Hendriks P, Smolders A, de Vries F, Spee J, van Kammen A, Bisseling T, Franssen H. Plant J. 1993;3:573–585. doi: 10.1046/j.1365-313x.1993.03040573.x. [DOI] [PubMed] [Google Scholar]
- 6.Kouchi H, Hata S. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- 7.Crespi M D, Jurkevitch E, Poiret M, D’Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijn I, Yang W, Pallisgård N, Østergaard Jensen E, van Kammen A, Bisseling T. Plant Mol Biol. 1995;28:1111–1119. doi: 10.1007/BF00032671. [DOI] [PubMed] [Google Scholar]
- 9.Van De Sande K, Pawlowski K, Czaja I, Wieneke U, Schell J, Schmidt J, Walden R, Matvienko M, Wellink J, van Kammen A, Franssen H, Bisseling T. Science. 1996;273:370–373. doi: 10.1126/science.273.5273.370. [DOI] [PubMed] [Google Scholar]
- 10.Coba de la Peña T, Frugier F, McKhann H I, Bauer P, Brown S, Kondorosi A, Crespi M D. Plant J. 1997;11:407–420. doi: 10.1046/j.1365-313x.1997.11030407.x. [DOI] [PubMed] [Google Scholar]
- 11.Truchet G, Barker D G, Camut S, de Billy F, Vasse J, Huguet T. Mol Gen Genet. 1989;219:65–68. [Google Scholar]
- 12.Joshi P A, Caetano-Anollés G, Graham E T, Gresshoff P M. Protoplasma. 1991;162:1–11. [Google Scholar]
- 13.Bevan M. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ooms G, Hooykaas P, van Veen R, van Beelen P, Regensburg-Tunik T, Schilperoort R. Plasmid. 1982;7:15–29. doi: 10.1016/0147-619x(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann B, Trinh T H, Leung J, Kondorosi A, Kondorosi E. Mol Plant–Microbe Interact. 1997;10:307–315. [Google Scholar]
- 16.Sambrook J, Fritsch T, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Does M P, Dekker B M M, de Groot M J A, Offringa R. Plant Mol Biol. 1991;17:151–153. doi: 10.1007/BF00036819. [DOI] [PubMed] [Google Scholar]
- 18.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 19.Bonneville J M, Sanfaçon H, Fütterer J, Hohn T. Cell. 1989;59:1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- 20.Töpfer R, Pröls M, Schell J, Steinbiss H H. Nucleic Acids Res. 1987;15:5890. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKhann H I, Hirsch A M. In: Methods in Plant Molecular Biology and Biotechnology. Glick B R, Thompson J E, editors. Boca Raton, FL: CRC; 1993. pp. 179–205. [Google Scholar]
- 22.Satiat-Jeunemaître B, Hawes C. J Cell Sci. 1992;103:1153–1166. [Google Scholar]
- 23.Gurr E. The Rational Use of Dyes in Biology. London: Leonard Hill; 1965. [Google Scholar]
- 24.van Brussel A A N, Zaat S A J, Canter Cremers H C J, Wijffelman C A, Pees E, Tak T, Lugtenberg B. J Bacteriol. 1986;165:517–522. doi: 10.1128/jb.165.2.517-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper J B, Long S R. Plant Cell. 1994;6:215–225. doi: 10.1105/tpc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch, A. M., Fang, Y., Asad, A. & Kapulnik, Y. (1997) Plant Soil, in press.
- 27.Minami E, Kouchi H, Cohn J R, Ogawa T, Stacey G. Plant J. 1996;9:101–110. doi: 10.1046/j.1365-313x.1996.10010023.x. [DOI] [PubMed] [Google Scholar]
- 28.St. Johnston D. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 29.Rastinejad F, Conboy M J, Rando T A, Blau H M. Cell. 1993;75:1107–1117. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]