Abstract
Recent studies of mitochondrial DNA (mtDNA) variation among marine turtle populations are consistent with the hypothesis that females return to beaches in their natal region to nest as adults. In contrast, less is known about breeding migrations of male marine turtles and whether they too are philopatric to natal regions. Studies of geographic structuring of restriction fragment and microsatellite polymorphisms at anonymous nuclear loci in green turtle (Chelonia mydas) populations indicate that nuclear gene flow is higher than estimates from mtDNA analyses. Regional populations from the northern and southern Great Barrier Reef were distinct for mtDNA but indistinguishable at nuclear loci, whereas the Gulf of Carpentaria (northern Australia) population was distinct for both types of marker. To assess whether this result was due to reduced philopatry of males across the Great Barrier Reef, we determined the mtDNA haplotypes of breeding males at courtship areas for comparison with breeding females from the same three locations. We used a PCR-restriction fragment length polymorphism approach to determine control region haplotypes and designed mismatch primers for the identification of specific haplotypes. The mtDNA haplotype frequencies were not significantly different between males and females at any of the three areas and estimates of Fst among the regions were similar for males and females (Fst = 0.78 and 0.73, respectively). We conclude that breeding males, like females, are philopatric to courtship areas within their natal region. Nuclear gene flow between populations is most likely occurring through matings during migrations of both males and females through nonnatal courtship areas.
Keywords: population genetics, restriction site, Testudines, natal homing, male-mediated gene flow
Marine turtles are long-lived reptiles whose life histories may encompass entire ocean basins. In most species, posthatchlings presumably drift in open ocean gyres for several years (1) and adults make periodic migrations of up to 2,700 km between feeding and breeding grounds (2, 3). Early tagging studies demonstrated that breeding females returned to nest at specific beaches in subsequent seasons (4, 5) even when they migrated from feeding grounds that overlapped with other breeding populations (5). From these observations arose two behavioral models to explain female site fidelity, known as the natal homing and social facilitation models. In the natal homing model (6), first time nesters return to their natal region to breed and remain faithful to that region throughout their lives. In the social facilitation model (4, 7), first time nesters follow experienced breeders away from shared feeding grounds to established breeding grounds and nesting beaches to which they subsequently return. Recently, studies of mitochondrial DNA (mtDNA) variability in marine turtles have revealed that regional populations are genetically disjunct, even when they share feeding grounds, a finding that supports the natal homing hypothesis for females (see, for example, refs. 8–13; but also see ref. 14). In contrast, no studies have examined natal homing in male marine turtles, and it is possible that males could display greater flexibility in their choice of breeding grounds, perhaps through social facilitation.
Our knowledge of male marine turtles is limited (but see refs. 15–18) because unlike females, they rarely come ashore; thus most studies require the capture of males in the water at breeding or feeding grounds. Tagging studies of male green turtles (Chelonia mydas) in the southern Great Barrier Reef found that breeding males display fidelity to specific courtship areas near nesting beaches chosen by females (17) and that they return to specific feeding grounds after mating (C.J.L., unpublished data). Tagging and telemetry studies of male C. mydas in the Hawaiian archipelago lend further support for a hypothesis that adult males, like females, are faithful to particular feeding and breeding regions (15). A better understanding of the behavior of male marine turtles and male-mediated gene flow should provide insights into the evolution of mating systems, the dynamics of rookery colonization, and the nature of population divergence and subdivision.
Early studies of sea turtle population structure used allozyme variation to investigate nuclear gene flow (incorporating the effects of both males and females), but obtained mixed results (19, 20). In the first test of male-mediated gene flow in marine turtles, Karl et al. (21) analyzed populations of C. mydas with anonymous single copy nuclear loci (ascnDNA) for comparison with the mtDNA divergence found in the same populations (22). On a global scale, only 28% of pairwise tests (n = 455 tests) of population divergence at nuclear loci were significant, whereas 93% of pairwise tests with mtDNA (n = 91 tests) indicated significant divergence between populations (21). Hypotheses to explain these results included the smaller effective size, and higher mutation rate of mtDNA relative to ascnDNA, but focused on life history differences between males and females (21). A moderate level of male-mediated gene flow was suspected to occur when different breeding populations overlapped, either at feeding grounds or during migrations, whether or not males migrated to natal regions for breeding. However, because many of the populations compared by Karl et al. (21) did not overlap on feeding grounds, we interpret these results as suggesting that males are less philopatric than females.
To pursue these questions in more detail, FitzSimmons et al. (23) investigated population divergence at nuclear loci in Australian C. mydas populations. Four regional groupings, each with overlapping feeding grounds, had already been identified by tagging data (3, 24, 25): Western Australia, Gulf of Carpentaria (GoC), northern Great Barrier Reef (nGBR), and the southern Great Barrier Reef (sGBR) (see Fig. 1). Previous mtDNA analyses (26, 27) detected strong divergence between each of these regions, but not within regions, supporting a hypothesis of natal homing by C. mydas females to specific regions, but not to specific nesting beaches. Nuclear, and thus male-mediated, gene flow between regions was assessed (23) at the same ascnDNA loci analyzed by Karl et al. (21), and at four highly variable microsatellite loci developed for marine turtles (28). At ascnDNA loci, pairwise tests indicated a lack of divergence between regions, even though population structure was evident among all regions combined, suggesting at least moderate male-mediated gene flow between regions. Further testing of genetic structure at highly variable microsatellite loci demonstrated that all regions were statistically distinct with the exception of the nGBR and sGBR regions. Moderate levels of nuclear gene flow occurred between most regions, by contrast, nuclear gene flow between the nGBR and sGBR appeared to be extensive (23).
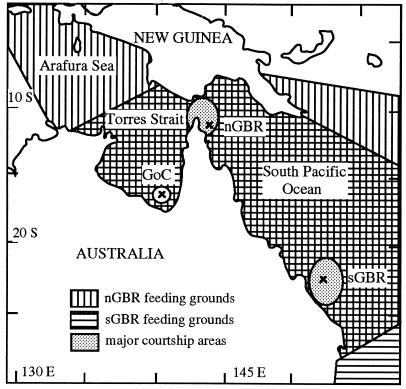
Figure 1.
Sample locations of C. mydas populations in Australian waters. GoC, Bountiful Island, Gulf of Carpentaria; nGBR, Raine Island, northern Great Barrier Reef; sGBR, Heron Island, southern Great Barrier Reef. The feeding ground distributions of the nGBR and sGBR populations are depicted. The extent of feeding ground locations for the GoC is unknown.
Variation in nuclear gene flow between Australian populations may indicate differences in the degree of natal homing displayed by males in different populations. Behavioral differences in natal homing could be a response to the geographic location of rookeries, given that the Western Australia, GoC, and the GBR populations breed in biogeographically distinct regions that are influenced by different oceanic currents (29). In contrast, the majority of nesting sites in the nGBR and sGBR are within the same barrier reef system on widely separated coral cays. Within the GBR, natal homing by males may be relaxed relative to females, and social facilitation (7) may operate. Alternatively, variation in the extent of nuclear gene flow may simply reflect variation in opportunities for interpopulation matings that correspond to the extent of population mixing at feeding grounds and on migratory routes, this being extensive for the nGBR and sGBR populations.
Given the different patterns observed for nuclear gene flow between regions (23) and on a broader geographic scale (21), it is unknown whether male-mediated gene flow is a common and perhaps extensive phenomenon in marine turtles, and how strongly nuclear gene flow reflects a lack of natal homing behavior in males. To address the extent of male philopatry directly we determined the mtDNA haplotypes of breeding males to infer their natal origins. However, because the mtDNA haplotypes of nesting populations are not unique (26), we could not positively identify individual breeding males as originating from particular populations. Therefore, we compared the frequencies of mtDNA haplotypes between breeding males and nesting females in the same areas. If males are as strongly philopatric as females, then the pattern and magnitude of mtDNA differentiation among populations should be similar for both males and females. Conversely, if males exhibit less philopatry, then less mtDNA divergence is expected among breeding males from different populations, relative to that among breeding females. For these analyses we selected three regions, GoC, nGBR, and sGBR (Fig. 1), which vary in the extent of interpopulation nuclear gene flow (23). Given the high level of nuclear gene flow between the nGBR and sGBR populations (23), we expected to see relatively little divergence in mtDNA allele frequencies between males of these two populations relative to that in females, but similar mtDNA divergence for males and females from the GoC population, if this phenomenon is due to relaxed philopatry in males.
MATERIALS AND METHODS
Breeding males (n = 82) were captured in the Heron Island lagoon (sGBR) in 1993 and 1994 during the peak of the mating season (mid-October to mid-November) by the turtle rodeo method (30) or by catching animals in shallow water off the beach. At Raine Island (nGBR), males (n = 13) were captured in 1994 as they patrolled the nearby waters during the peak of the nesting season (December). The sample size from Raine Island was small due to the limited numbers of breeding males present when nesting surveys were conducted. Breeding males (n = 38) at Bountiful Island (GoC) were captured while mounted on females in shallow water during the peak of the nesting season (late July 1993; ref. 31). A male was considered to be of breeding status only if (i) he was observed mounted or courting a female turtle, (ii) if he was an attendant male, or (iii) if he had recent and extensive bite scars on his rear flippers or tail inflicted by competing males (17). The breeding condition of several (n = 21) males at Heron Island was also confirmed by laparoscopy (32) in 1994. Samples for genetic analysis were obtained by collecting blood (≈0.5 ml) from the dorsal cervical sinus (33) or by removing a small (<1 cm2) skin biopsy from the shoulder region with a sharp knife. Blood was preserved in a lysis buffer, and genomic DNA was extracted by a salting out procedure (28). Skin samples were placed in a preservative solution (20% dimethyl sulfoxide, saturated with NaCl), and DNA was extracted using Chelex (Bio-Rad) beads. With this method a very small (≈0.5 mm) piece of tissue was placed in 1 ml of a 5% Chelex solution for 1–4 hr at 55–60°C (with occasional inversion), heated at 95°C for 5 min, and spun for 5 min at 13,000 rpm, leaving the DNA dissolved in the supernatant.
A portion (≈385 bp) of the mtDNA control region was amplified in all samples by PCR using the TCR-5 and TCR-6 (TCR, turtle control region) primers designed by Norman et al. (26). PCR amplifications were performed with 25 μl volumes containing 5–50 ng of template DNA, 0.5 μM of each primer, 200 μM of dNTPs, 3 mM MgCl2, 50 mM KCl, 10 mM Tris⋅HCl, 0.01% Tween 20, 0.01% Nonidet P-40, and 0.4 unit of Taq polymerase. Amplifications were carried out at 93°C for 1 min, followed by 30 cycles at 93°C for 40 sec, 55°C for 50 sec, and 72°C for 40 sec, and a final extension at 72°C for 2 min. Negative controls were used for all sets of reactions, and 5 μl of each PCR product was run through 1.2% agarose gels and visualized using ethidium bromide to ensure proper amplification. To distinguish the three major haplotype classes (A, B, and C; Table 1) previously identified in these populations (26), PCR products were digested with the restriction enzyme MseI, separated on small 8% polyacrylamide gels, and visualized with ethidium bromide following the procedure of Norman et al. (26). Samples identified as haplotype C were further tested to identify a variant haplotype (F) previously observed in samples from Java, by digesting PCR products with EcoRI (26).
Table 1.
Diagnostic restriction sites of mtDNA haplotypes found in Australian C. mydas breeding populations as identified by Norman et al. (26)
| Haplotype | Location of variable restriction sites*
|
||||||
|---|---|---|---|---|---|---|---|
|
MseI
|
EcoRI, 60 bp | AseI,† 78 bp | HphI,‡ 164 bp | MaeI,§ 269 bp | |||
| 56 bp | 75–76 bp | 374 bp | |||||
| Aa, Ac | − | + | − | NA | + | − | + |
| Ab | − | + | − | NA | + | + | + |
| B | + | + | + | NA | NA | NA | NA |
| Ca-1 | − | + | + | − | + | + | − |
| Ca-2 | − | + | + | − | + | + | + |
| Cb-1 | − | + | + | − | + | − | − |
| Cb-2 | − | + | + | − | − | − | − |
| D | − | − | + | − | NA | NA | NA |
| E¶ | − | + | − | − | NA | NA | NA |
| F | − | + | + | + | NA | NA | NA |
Presence of the restriction site is denoted by +, absence by −; NA, not applicable.
Restriction site created by mismatch primer TCR-Ase, 5′-ATTGAATCCACATAAATATATTA-3′, amplified in conjunction with TCR-Hph.
Restriction site created by mismatch primer TCR-Hph, 5′-TTTAAGAAATAACCAATCAC-3′, amplified in conjunction with TCR5 or TCR-Ase.
Restriction site created by mismatch primer TCR-Mae, 5′-CCCATTTAGTTTATAGCGTACCTA-3′ amplified in conjunction with TCR-6.
Identified by a 10-bp insertion (ref. 26).
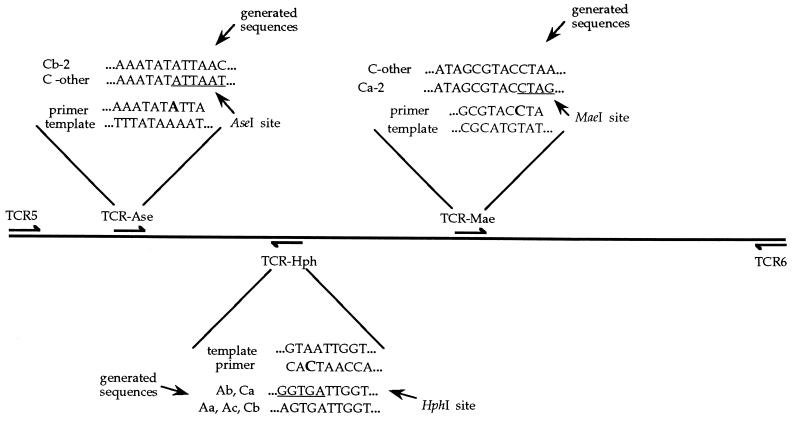
To identify variants within the A and C haplotype classes (Aa, Ab, Ac, Ca-1[LAC], Ca-2 [NWC], Cb-1[GoC], and Cb-2-[SWK]; ref. 26) that differ in sequence but not available restriction sites, mismatch primers (34) were designed to produce new restriction sites that incorporated the definitive variable sites (Fig. 2, Table 1). To distinguish the Ab haplotype from Aa and Ac and also the Ca from the Cb haplotype, a mismatch primer was designed that produced a HphI site in the Ab and Ca haplotypes, but not in the Aa, Ac, or Cb haplotypes. To separate the Ca-2 haplotype from all other C haplotypes, a mismatch primer was designed that incorporated a MaeI restriction site into the Ca-2 sequence. The Cb-2 haplotype was identified from other C haplotypes with the use of a mismatch primer that incorporated an AseI restriction site into all C haplotypes except Cb-2. PCR conditions for the use of mismatch primers were modified as follows: 93°C for 1 min, followed by 5 cycles at 93°C for 40 sec, 42°C for 60 sec, and 72°C for 45 sec, followed by 30 cycles with the annealing temperature increased to 50°C, and a final extension at 72°C for 2 min. Known controls were digested at the same time as the samples to ensure the effectiveness of the enzyme digests and the digested mismatch PCR products were run on 8–12% polyacrylamide gels. Any PCR product that displayed a new restriction fragment pattern in any digest was sequenced by cycle sequencing using primers end labeled with [γ-33P]ATP, separated on 6% denaturing polyacrylamide gels, and visualized by autoradiography. Additional samples previously taken from breeding females (or their hatchlings) by Norman et al. (26) were further analyzed using the mismatch primers to assign haplotype class for each of the three regions investigated (C.M., unpublished data).
Figure 2.
Location and design of mismatch primers to generate diagnostic restriction sites in the mtDNA control region of C. mydas mtDNA. The TCR-5 or TCR-6 primers (26) were typically paired with the appropriate mismatch primer, though TCR-Ase and TCR-Hph were also used together. Shown are the template DNA sequence, the portion of the primer sequence containing the mismatch primer, and the sequences generated through PCR amplifications that indicate the presence (or absence) of the new restriction site.
Differences in haplotype frequencies between breeding males and females were tested for each of the three geographic areas by χ2 randomizations (10,000 each) of the data sets in the Monte Carlo routine of reap (35). Measures of population subdivision (Fst; ref. 36) and gene flow (Nm) among regions were estimated for the male and female breeding components to assess qualitative similarity. Fst was estimated in fstat (37) with significance levels tested by permutations (n = 3000), and Nm was calculated as Nm = (1/2)[(1/Fst) − 1].
RESULTS
Restriction digests of the TCR-5/TCR-6 PCR products using MseI gave the same pattern of bands as observed for the A, B, and C haplotypes reported by Norman et al. (26). PCR products that were identified as C haplotypes were also digested with EcoRI, but no F haplotypes were found. One sample from Raine Island had a unique MseI fragment pattern and was sequenced to identify unique site changes. No novel variants were found in the sGBR or GoC samples. These results are qualitatively similar to those observed for breeding females where two variant haplotypes (D and E) were found (out of 66 samples) in the nGBR breeding population, and none in the sGBR or GoC (26). Sequencing of the nGBR male that had the variant haplotype revealed that a single mutation (T to C) at 179 bp (26) had eliminated an MseI site. Construction of a phylogeny (neighbor joining, Kimura 2-parameter) using mega (38) indicated that the variant haplotype was most closely affiliated with a French Polynesian sample (FPa) sequenced by Norman et al. (26). Even so, the two sequences differed by 2.6%; a relatively high value given a maximum divergence of 7.0% observed among Indo-Pacific rookery haplotypes (26).
Mismatch primers were successful in amplifying shorter segments of the control region and creating diagnostic restriction sites. All A haplotypes showed two bands of 198 and 8 bp after amplification with the TCR-5/TCR-Hph primer combination and HphI digestion; only controls without the restriction site revealed a single uncut 206-bp band. C haplotypes showed either a single uncut 131-bp band, or two bands of 123 and 8 bp after amplification with TCR-Ase/TCR-Hph and HphI digestion. Once identified as Cb haplotypes, the products were also digested with AseI and showed either the single 131-bp band or two bands of 110 and 21 bp in length. All Ca haplotypes showed a single uncut 169-bp band after amplification with the TCR-Mae/TCR-6 primers and digestion with MaeI digestion. No novel haplotypes were observed in the digest profiles from the mismatch PCR products.
In each of the three regions studied, breeding males and females had statistically indistinguishable mtDNA haplotype frequencies (Table 2). χ2 tests of randomized data sets indicated that haplotype frequencies of breeding males and females were not different from each other in any of the areas: P = 0.78, 0.84, and 0.26 for the sGBR, nGBR, and GoC, respectively. As observed for females, a large (90%) shift in allele frequencies was observed between the nGBR and sGBR breeding males, and there was a fixed difference between these and the GoC.
Table 2.
Frequencies of mtDNA haplotypes in breeding male and female C. mydas at three locations: Bountiful Island (GoC), Raine Island (nGBR), and Heron Island (sGBR)
| Haplotype* | Location
|
|||||
|---|---|---|---|---|---|---|
| GoC
|
nGBR
|
sGBR
|
||||
| F | M | F | M | F | M | |
| Aa, Ac | 0 | 0 | 5 | 1 | 93 | 76 |
| Ab | 0 | 0 | 0 | 0 | 0 | 0 |
| B | 0 | 0 | 46 | 11 | 7 | 6 |
| Ca-1 | 11 | 12 | 0 | 0 | 0 | 0 |
| Ca-2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cb-1 | 7 | 16 | 0 | 0 | 0 | 0 |
| Cb-2 | 3 | 10 | 0 | 0 | 0 | 0 |
| D | 0 | 0 | 1 | 0 | 0 | 0 |
| E | 0 | 0 | 1 | 0 | 0 | 0 |
| F | 0 | 0 | 0 | 0 | 0 | 0 |
| Variants | 0 | 0 | 0 | 1 | 0 | 0 |
M, male; F, female.
See ref. 26.
Estimates of Fst showed a high degree of structure across the three regions and were qualitatively similar for males and females (Fst = 0.78 and 0.73, respectively; P < 0.001 each). Pairwise estimates of Fst indicated that males and females share the same qualitative patterns in genetic subdivision between regions: nGBR–sGBR, Fst = 0.79 and 0.80; nGBR–GoC, Fst = 0.52 and 0.57; sGBR–GoC, Fst = 0.71 and 0.68, for males and females, respectively. Estimates of migration were low (Nm < 1) between all regions for both the male and female breeding components: nGBR–sGBR, Nm = 0.13 and 0.14; nGBR–GoC, Nm = 0.38 and 0.47; and sGBR–GoC, Nm = 0.23 and 0.20, respectively.
DISCUSSION
Regardless of the extent of male-mediated gene flow between regions, our data indicate that males, like females, are philopatric to natal regions in their choice of breeding grounds. How well do these data agree with our knowledge about male C. mydas and parallel female behavior? Tagging data of adult male C. mydas suggest fidelity to specific feeding and breeding grounds (15, 17), as is known for females (3, 39) and long distance breeding migrations have been observed in males (16, 17, 40). Additionally, mtDNA haplotype frequencies of males and females were similar at feeding grounds within the GBR (Princess Charlotte Bay, nGBR; and Shoalwater Bay, sGBR), suggesting parallel behaviors (27). In contrast, however, males appear to migrate more frequently to breeding grounds (17, 40) than do females and a greater proportion of males may breed in close proximity to their feeding grounds in comparison to females (C.J.L., unpublished data). These varied results suggest that male behavior has strong parallels to that of females regarding natal philopatry, and to a somewhat lesser extent regarding choice of feeding grounds and migratory behavior.
Our results suggest that the large variation in nuclear gene flow previously observed in these populations does not result from variation in the degree of male philopatry to natal regions. In particular, male philopatry appears to be as strong between the nGBR and sGBR as between the GBR and GoC rookeries, despite evidence for extensive nuclear gene flow between the former. Instead, variation in nuclear gene flow may depend upon the geographic positioning of feeding grounds relative to mating grounds. As suggested by Karl et al. (21), nuclear gene flow could occur even if males were philopatric to natal sites through interpopulation matings when populations overlapped at feeding grounds or along migration corridors. If so, then the extent of male-mediated gene flow would be influenced by the degree to which mating grounds overlap with mixed feeding grounds or migration routes, and also the extent to which the timing of migration varies among populations. The timing of migration is important because it is linked with hormonal changes that dictate breeding behavior which is of a relatively short duration (1–2 weeks for females, 1–2 months for males; refs. 15 and 41; C.J.L., unpublished data).
Females from all of the three populations we studied occur together at feeding grounds located in the GoC and nGBR regions (3) (Fig. 1). Ostensibly, the greatest overlap occurs between the nGBR and sGBR populations that share many feeding grounds throughout the GBR, resulting in shared migratory pathways through the Torres Strait and along the GBR. sGBR turtles who utilize feeding grounds in the GoC and adjacent areas must migrate through Torres Strait, one of the main courtship areas for nGBR turtles, thus increasing the likelihood of gene flow between these populations. Within Torres Strait, male-mediated gene flow would occur when breeding sGBR males migrate through nonnatal regions if they mate in transit with receptive nGBR females. Conversely, breeding nGBR males may be able to mate with migrating sGBR females that pass through the nGBR mating grounds. This latter scenario depends upon whether or not migrating females are receptive, and if not, whether males can successfully mate with unreceptive females. This scenario contrasts with other C. mydas populations in which the breeding areas are geographically distant from shared feeding grounds, or where migratory breeders do not pass through nonnatal breeding grounds (e.g., Hawaiian archipelago).
Given the diverse opportunities for breeding throughout the region, it seems curious that males would be philopatric to natal regions in their choice of mating grounds. If mating areas coincide with feeding grounds, males could forgo breeding migrations and mate instead at their chosen feeding sites, whether or not these were within their natal region. If feeding grounds were distant from mating areas, then first-time breeding males could simply follow experienced breeders to diverse mating grounds, irrespective of natal homing. Such behavior seems plausible and was originally proposed for first-time breeding females under the social facilitation model (4, 7). It is probable that selective pressures on females to return to their natal region to locate successful nesting beaches would be linked to similar behavior in males as well to insure their co-occurrence at mating grounds. A proximate mechanism driving this selection is the local environmental conditions which dictate the timing of nesting seasonality. Near equatorial regions, the timing of mating and nesting can vary substantially over relatively short geographic distances (4, 42). In Australia, turtles breeding at GBR rookeries or along the west coast do so in mid-summer, whereas the centrally located GoC population breeds primarily during the winter. Winter nesting on Bountiful Island has presumably arisen because hot sand temperatures in the summer months may diminish embryo survival. This bimodality within Australian C. mydas populations indicates relatively strong selective pressures that would couple natal homing and the timing of reproduction in both sexes. This is particularly true for the GoC population which nests in an area that was not available for nesting prior to 9,500 years BP (43), only 190–270 turtle generations ago, given maturity of 35–50 years (44). Similar variation in the timing of nesting occurs within Indonesia (42), perhaps indicating that environmental barriers (via sand temperature) to gene flow may be a general pattern where populations overlap in equatorial regions.
Our results, in combination with tagging and tracking data on male C. mydas, suggest that male behavior is similar to that of females in many respects. However, male behavior and mating systems can vary across species, or populations, and generalizations would be inappropriate at this time. For example, less genetic structure is observed among populations of leatherback turtles (Dermochelys coriacea) (14), and mating in olive ridley turtles (Lepidochelys olivacea) may occur opportunistically over broad geographic areas (18). It may be possible that there are other geographic hot spots for male-mediated gene flow, such as we believe occurs in the restricted migratory corridor of Torres Strait where several C. mydas populations overlap. To gain further insight into how much breeding occurs outside of natal areas, future studies should sample breeding pairs at the advent of the mating season, or look for mating animals in migratory corridors who have not completed migrations to natal areas. Given the apparent geographic specificity of our results, studies of male behavior in other regions, and other species, are necessary for a better understanding of this under studied half of the turtle world.
Acknowledgments
We thank T. Tucker and B. Bowen for useful comments on the manuscript. We thank the Queensland Turtle Research Project of the Queensland Department of Environment and the Heron Island Research Station for supporting the field work. This research was funded with a grant from the Australian Research Council and a scholarship to N.N.F. from the University of Queensland.
ABBREVIATIONS
- GBR
Great Barrier Reef
- sGBR
southern GBR
- nGBR
northern GBR
- ascnDNA
anonymous single copy nuclear loci
- GoC
Gulf of Carpentaria
References
- 1.Carr A. Conserv Biol. 1987;1:103–121. [Google Scholar]
- 2.Carr A. Copeia. 1975;1975:547–555. [Google Scholar]
- 3.Limpus C J, Miller J D, Parmenter C J, Reimer D, McLachlan N, Webb R. Wildl Res. 1992;19:347–358. [Google Scholar]
- 4.Hendrickson J R. Proc Zool Soc London. 1958;130:455–535. [Google Scholar]
- 5.Carr A, Ogren L. Bull Am Mus Nat Hist. 1960;121:7–48. [Google Scholar]
- 6.Carr A. So Excellente a Fishe. New York: Natural History Press; 1967. [Google Scholar]
- 7.Owens D W, Grasman M A, Hendrickson J R. Herpetologica. 1982;38:124–135. [Google Scholar]
- 8.Meylan A B, Bowen B W, Avise J C. Science. 1990;248:724–727. doi: 10.1126/science.2333522. [DOI] [PubMed] [Google Scholar]
- 9.Bowen B W, Avise J C, Richardson J I, Meylan A B, Margaritoulis D, Hopkins-Murphy S R. Conserv Biol. 1993;7:834–844. [Google Scholar]
- 10.Broderick D, Moritz C, Miller J D, Guinea M, Prince R J, Limpus C J. Pac Conserv Biol. 1994;1:123–131. [Google Scholar]
- 11.Bowen B W, Avise J C. In: Conservation Genetics: Case Histories from Nature. Avise J C, Hamrick J L, editors. New York: Chapman & Hall; 1995. pp. 190–237. [Google Scholar]
- 12.Bass A L, Good D A, Bjorndal K A, Richardson J I, Hillis Z M, Horrocks J, Bowen B W. Mol Ecol. 1996;5:321–328. [PubMed] [Google Scholar]
- 13.Encalada S E, Lahanas P N, Bjorndal K A, Bolten A B, Miyamoto M M, Bowen B W. Mol Ecol. 1996;5:473–483. [PubMed] [Google Scholar]
- 14.Dutton, P. H. (1995) Ph.D. dissertation (Texas A&M Univ., College Station).
- 15.Dizon A E, Balazs G H. Mar Fish Rev. 1982;44:13–20. [Google Scholar]
- 16.Green D. J Herpetol. 1984;18:212–130. [Google Scholar]
- 17.Limpus C J. Wildl Res. 1993;20:513–523. [Google Scholar]
- 18.Plotkin P T, Owens D W, Byles R A, Patterson R. Herpetologica. 1996;52:1–7. [Google Scholar]
- 19.Bonhomme F, Salvidio S, Lebeau A, Pasteur G. Genetica. 1987;74:89–94. [Google Scholar]
- 20.Coates D J, Carstairs S A, Prince R I T. Proceedings of the Australian Marine Turtle Conservation Workshop. Queensland Department of Environment and Heritage and Australian Nature Conservation Agency; 1994. pp. 163–166. [Google Scholar]
- 21.Karl S A, Bowen B W, Avise J C. Genetics. 1992;131:163–173. doi: 10.1093/genetics/131.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowen B W, Meylan A B, Ross J P, Limpus C J, Balazs G H, Avise J C. Evolution. 1992;46:865–881. doi: 10.1111/j.1558-5646.1992.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 23.FitzSimmons, N. N., Moritz, C., Pope, L. & Limpus, C. J. (1997) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 24.Limpus C J, Fleay A, Guinea M. In: The Capricornia Section of the Great Barrier Reef: Past, Present, and Future. Ward W T, Saenger P, editors. R. Soc. of Queensland and Australian Coral Reef Soc.; 1984. pp. 61–78. [Google Scholar]
- 25.Prince R. In: Proceedings of the Australian Marine Turtle Conservation Workshop. James R, editor. Queensland Department of Environment and Heritage & Australian Nature Conservation Agency; 1994. pp. 1–14. [Google Scholar]
- 26.Norman J A, Moritz C, Limpus C J. Mol Ecol. 1994;3:363–373. doi: 10.1111/j.1365-294x.1994.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 27.Norman, J. A. (1996) Ph.D. dissertation (Univ. of Queensland, Brisbane).
- 28.FitzSimmons N N, Moritz C, Moore S S. Mol Biol Evol. 1995;12:432–440. doi: 10.1093/oxfordjournals.molbev.a040218. [DOI] [PubMed] [Google Scholar]
- 29.Wilson B R, Allen G R. In: Fauna of Australia. Dyne G R, Walton D W, editors. 1A. Canberra: Australian Government Publishing Service; 1987. pp. 43–68. [Google Scholar]
- 30.Limpus C J. In: Exploration North. Lavery H J, editor. Melbourne, Australia: Richmond Hill; 1978. pp. 187–220. [Google Scholar]
- 31.Limpus C J, Miller K D, Preece N. In: Proceedings of 14th Annual Sea Turtle Symposium: Sea Turtle Biology and Conservation. Bjorndal K A, Bolten A B, Johnson D A, Eliazer P J, editors. Washington, DC: Natl. Oceanic and Atmospheric Administration; 1994. NOAA Tech. Mem. NMFS-SEFSC-351, pp. 76–77. [Google Scholar]
- 32.Limpus C J, Reed P C. In: Biology of Australasian Frogs and Reptiles. Grigg G, Shine R, Ehmann H, editors. Sydney: Surrey Beatty; 1985. pp. 47–52. [Google Scholar]
- 33.Owens D W, Ruiz G J. Herpetologica. 1980;36:17–20. [Google Scholar]
- 34.Dowling T E, Moritz C, Palmer J D, Rieseberg L H. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 249–320. [Google Scholar]
- 35.McElroy D, Moran P, Bermingham E, Kornfield I. J Hered. 1992;83:157–158. doi: 10.1093/oxfordjournals.jhered.a111180. [DOI] [PubMed] [Google Scholar]
- 36.Weir B S, Cockerham C. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 37.Goudet J. fstat: A Program for IBM PC Compatibles to Calculate Weir and Cockerham’s Estimators of F-Statistics. Lausanne, Switzerland: Lausanne Univ.; 1994. [Google Scholar]
- 38.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis. University Park: Pennsylvania State Univ.; 1993. , Version 1.0. [Google Scholar]
- 39.Carr A F, Carr M, Meylan A. Bull Am Mus Nat Hist. 1978;162:1–46. [Google Scholar]
- 40.Balazs G. Synopsis of Biological Data on the Green Turtle in the Hawaiian Islands. Washington, DC: Natl. Oceanic and Atmospheric Administration; 1980. , NOAA Tech. Mem. NMFS-SWFC-7. [Google Scholar]
- 41.Comuzzie D K C, Owens D W. Herpetologica. 1990;46:195–202. [Google Scholar]
- 42.Nuitja N S, Lazell J D. Copeia. 1982;1982:708–710. [Google Scholar]
- 43.Jones M R, Torgersen T. Aust J Earth Sci. 1988;35:313–324. [Google Scholar]
- 44.Limpus C J, Walter D G. Herpetologica. 1980;36:162–165. [Google Scholar]