Abstract
X-linked adrenoleukodystrophy (X-ALD) is a peroxisomal disorder with impaired β-oxidation of very long chain fatty acids (VLCFAs) and reduced function of peroxisomal very long chain fatty acyl-CoA synthetase (VLCS) that leads to severe and progressive neurological disability. The X-ALD gene, identified by positional cloning, encodes a peroxisomal membrane protein (adrenoleukodystrophy protein; ALDP) that belongs to the ATP binding cassette transporter protein superfamily. Mutational analyses and functional studies of the X-ALD gene confirm that it and not VLCS is the gene responsible for X-ALD. Its role in the β-oxidation of VLCFAs and its effect on the function of VLCS are unclear. The complex pathology of X-ALD and the extreme variability of its clinical phenotypes are also unexplained. To facilitate understanding of X-ALD pathophysiology, we developed an X-ALD mouse model by gene targeting. The X-ALD mouse exhibits reduced β-oxidation of VLCFAs, resulting in significantly elevated levels of saturated VLCFAs in total lipids from all tissues measured and in cholesterol esters from adrenal glands. Lipid cleft inclusions were observed in adrenocortical cells of X-ALD mice under the electron microscope. No neurological involvement has been detected in X-ALD mice up to 6 months. We conclude that X-ALD mice exhibit biochemical defects equivalent to those found in human X-ALD and thus provide an experimental system for testing therapeutic intervention.
X-linked adrenoleukodystrophy (X-ALD) is an X-linked disorder characterized by the accumulation of saturated very long chain fatty acids (VLCFAs) (1) due to an impaired capacity to degrade them. This results from reduced function of the peroxisomal very long chain fatty acyl-CoA synthetase (2, 3) that activates VLCFAs to their CoA derivatives to initiate their degradation. X-ALD affects mainly the nervous system white matter, adrenal cortex, and testis, but the disease phenotype is highly variable (4, 5). The most frequent phenotypes include childhood cerebral X-ALD (6), adrenomyeloneuropathy (AMN) (7), and adrenocortical insufficiency (Addison disease) without nervous system involvement. In childhood cerebral X-ALD, neurological development is normal until at least 4 years of age. The disease involves the cerebral white matter with inflammatory demyelination and rapid progression leading to a vegetative state and death within a few years. AMN, on the other hand, has onset in adulthood (mean age of onset = 27.6 ± 8.7 years old) and progresses slowly. It involves mainly the long tracts of the spinal cord and peripheral nerves, without early involvement of the brain and little or no inflammation. In some cases, adrenocortical insufficiency is the sole clinical finding without any detectable neurological involvement. In a small fraction of cases (about 3%), cerebral X-ALD has onset in adulthood. The various phenotypes frequently occur within the same kindred or nuclear family (8), and identical mutations give rise to all X-ALD phenotypes (5, 9).
In 1993, the X-ALD gene was identified by positional cloning (10), and surprisingly appears to encode an ATP-binding cassette (ABC) transporter protein without homology to any acyl-CoA synthetase. Immunocytochemical studies (11–13) demonstrated that the X-ALD gene product (adrenoleukodystrophy protein; ALDP) is a peroxisomal membrane protein consistent with the observed biochemical abnormality in X-ALD patients. Mutational analyses of the X-ALD gene (9, 13–17) revealed the existence of mutations in all X-ALD patients examined, and complementation studies (18) showed that expression of ALDP restores β-oxidation of VLCFAs in patients’ fibroblasts. While all evidence strongly supports the conclusion that the abnormality in ALDP results in X-ALD, its function is still unknown. Other members of the ABC transporter superfamily (19) have been reported to transport substrates that range from ions to peptides—i.e., cystic fibrosis transmembrane conductance regulator (CFTR) (20, 21), multiple drug resistant protein (MDR) (22), transporters associated with antigen processing (TAP1 and TAP2) (23), a yeast pheromone transporter (STE6) (24, 25), and peroxisomal long chain fatty acid transporters (PXA1 and PXA2) (26). This implies that the function of ALDP might involve the transport of either enzymes, cofactors, or substrates necessary for VLCFA β-oxidation across peroxisomal membranes (11, 27). Based on sequence similarity, there is a small group of peroxisomal membrane proteins that belong to the ABC transporter protein family; these include PMP70 (28, 29), PMP70 related protein (P70R; David Valle, personal communication), and X-ALD related protein (ALDR) (30). Their possible interrelationships and functions are unknown. Thus, the role of ALDP in VLFCA β-oxidation, its relation to very long chain fatty acyl-CoA synthetase and the accumulation of saturated VLCFAs, the role of saturated VLCFAs in dysmyelination/demyelination of the nervous system, as well as the phenotypic variability in X-ALD patients are unresolved. Existing therapeutic methods, including dietary therapy (Lorenzo’s oil) (31, 32) and bone marrow transplantation (33), have had limited success, and gene therapy has been proposed. An animal model will facilitate understanding of the pathophysiology of X-ALD, help elucidate its genetic and phenotypic complexity, and contribute to the evaluation of therapeutic strategies. We have used gene targeting to create an X-ALD mouse model.
MATERIALS AND METHODS
Construction of Targeting Vector.
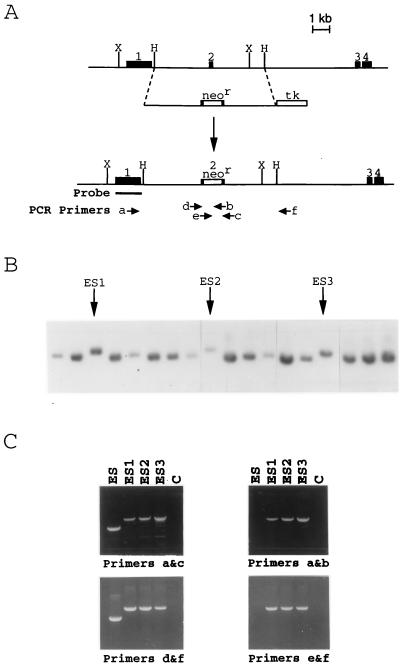
A genomic clone of X-ALD was obtained by probing a mouse strain 129/Sv genomic DNA library (Stratagene) with mouse X-ALD cDNA (34). To obtain full-length mouse X-ALD cDNA, total RNA was prepared from mouse strain 129/Sv fibroblasts and cDNA was generated by reverse transcriptase using random hexamer primers. The first-strand cDNA was amplified by PCR using the following primers: forward primer (5′-TTCAAGGTTTCCAGTTGCCTAGAC-3′) and reverse primer (5′-TAAAAGACACACTGCCCTGAACAG-3′) from the 5′ and 3′ untranslated region of the mouse X-ALD mRNA, respectively. The PCR product was cloned into pCRII vector (Invitrogen). The resulting construct, containing full-length mouse X-ALD cDNA, was then in vitro transcribed by T7 RNA polymerase (Ambion, Austin, TX) in the presence of [α-32p]UTP (Amersham) and used as a probe. A phage clone (λ4-1) with a 13-kb insert containing exons 1–4 of the mouse X-ALD gene (35) was isolated. Fluorescence in situ hybridization was performed to confirm its location on the X chromosome (data not shown). A silent mutation was introduced by site-directed mutagenesis to create an XhoI restriction site in exon 2 (at codon 322). A 6.2-kb HindIII fragment containing the mutated exon 2 (181 nt) of mouse X-ALD gene was isolated from λ4-1 and subcloned into pUC18 vector. A replacement-type targeting vector (36) was constructed by insertion of a neomycin resistant (neor) cassette (37) from pMC1neo poly(A) (Stratagene) into the created XhoI site. This insertional disruption of exon 2 causes a frameshift in the mouse X-ALD gene and results in the premature termination of mouse ALDP (77 codons downstream of the created XhoI site). A thymidine kinase (tk) gene cassette (38) was also incorporated outside of the X-ALD homology region of the targeting vector to allow negative selection by gancyclovir (38–41) against nonhomologous gene targeting events. The final targeting construct (in pBluescript) has a 3-kb 5′ genomic flanking sequence and a 3.2-kb 3′ genomic flanking sequence, with a 1.1-kb neor cassette (in the same orientation as mouse X-ALD gene) in exon 2 and a 1.85-kb tk gene cassette (reverse orientation as mouse X-ALD gene) 3′ to X-ALD sequence (Fig. 1A).
Figure 1.
Targeted disruption of the X-ALD gene in mouse embryonic stem (ES) cells. (A) The genomic structure of the mouse X-ALD gene from 5′ of exons 1–4 is depicted. Gene targeting of the X-ALD locus used a replacement-type vector covering the region between the dashed lines. Following homologous recombination, the targeting vector replaces the wild-type mouse X-ALD gene with the neor cassette-disrupted exon 2. The external probe (from exon 1) used for Southern blot analysis is also indicated. The numbers above the solid boxes indicate exon number. H, HindIII; X, XbaI. (B) Southern blot analysis of targeted mouse ES cells using a probe for exon 1 that flanks the targeted sequence. After digesting DNA with XbaI, the 8-kb targeted allele can be distinguished from the 7-kb wild-type allele. Three targeted ES cell clones (ES1–3) are indicated by arrows. (C) PCR analysis of targeted ES cell clones. Using neor cassette-specific primers (e or b) in combination with the external primers (a or f), the targeted ES cell clones (ES1–3) generated 4-kb PCR products where as wild-type ES cells (ES) did not result in any PCR product (Right). Combining the external primers (a or f) and the internal primers (d or c), targeted ES cell clones gave rise to a 4-kb fragment (including the neor sequence) whereas a 3-kb fragment was generated form wild-type ES cells (Left). Lane C, no template control.
Gene Targeting.
Linearized targeting vector was introduced by electroporation into passage 10 J1 ES cells (42) grown on irradiated G418-resistant primary embryonic fibroblast feeder layers (43, 44). Simultaneous G418 (250 μg/ml) and gancyclovir (2 μM) selection was applied and lasted for 10 days after electroporation. Drug-resistant clones were analyzed by Southern blot analysis. An external probe (Fig. 1A), from exon 1 of the mouse X-ALD gene, was used to screen for homologous recombination events. After digesting mouse genomic DNA with XbaI, this probe distinguished an 8.1-kb targeted allele from a 7-kb nontargeted allele (Fig. 1B). All targeted clones identified by Southern blot analysis were confirmed using PCR analysis to demonstrate the presence of the disrupted X-ALD allele (X-ALD−). As shown in Fig. 1C, X-ALD external primers a (forward primer, 5′-ATGTGGTAGCCTTTGCTG-3′) and f (reverse primer, 5′-GTATTCACCACCACATCTG-3′) were combined either with neor cassette-specific primers e (forward primer, 5′-CATTGGCGAATTCGAACAC-3′) and b (reverse primer, 5′-CCTCAGAAGAACTCGTCAAG-3′), giving rise to a 4-kb fragment from targeted ES cells and no product from nontargeted ES cells; or with internal primers d (forward primer, 5′-CAAGACCTAGCTTCACAGATCAACC-3′) and c (reverse primer, 5′-TAGTCCCTGGAGCCTTG-3′), resulting in a 4-kb PCR product from targeted ES cells or a 3-kb fragment from nontargeted ES cells. A neor cassette probe was also used to demonstrate that only single copy integration occurred in the targeted ES cells (data not shown).
Production of Chimeras.
The targeted ES cells were injected into blastocysts derived from C57BL/6, and the injected blastocysts were transferred into pseudo-pregnant CD-1 female mice to generate chimeras as described (42).
Biochemical Measurements.
Plasma and tissues were harvested from 3-month-old mice and used for fatty acid analyses. Human and mouse skin fibroblasts from normal and X-ALD individuals were derived and cultured in vitro. Human X-ALD postmortem tissues were derived from patients ranging in age from 6 to 50 years old and were processed as described (45, 46). Total lipids in plasma, tissue, fibroblast, and mouse chow were extracted, fractionated, purified by TLC, and subjected to capillary GC analysis as described (47). Fatty acid β-oxidation activity of cultured fibroblasts was determined by measuring their capability to degrade 14C-labeled fatty acid substrates (48). The statistical significance of the measured biochemical differences between normal and X-ALD subjects was determined by one-sided Student’s t test.
Histological Analyses.
Brain and adrenal glands were harvested from 3-month-old X-ALD mice and their normal sibs. Adrenal glands were fixed in 4% glutaraldehyde, postfixed in osmium tetroxide, and embedded in epoxy resin. The embedded tissues were thin sectioned and stained with uranyl acetate–lead citrate and examined under the electron microscope (49, 50). Brain specimens were fixed in 4% paraformaldehyde; coronal brain sections from cerebrum, cerebellum, and brainstem were embedded in paraffin wax and sectioned at 6 micra. Slides were stained with hematoxylin-eosin, Luxol fast blue, and periodic acid–Schiff base, and examined under the light microscope. Other sections of cerebral white matter were postfixed in osmium tetroxide, dehydrated, and embedded in epoxy resin. Then sections were stained with uranyl acetate-lead citrate and examined in an electron microscope (49, 50).
RESULTS
Targeted Disruption of Mouse X-ALD Gene.
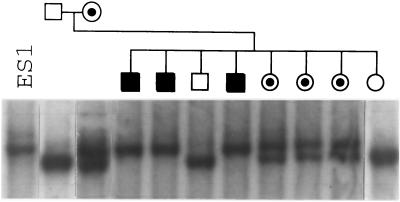
The targeting construct used to disrupt the mouse X-ALD gene is shown in Fig. 1. Among 200 G418 and gancyclovir-resistant ES cell clones screened by Southern blot and PCR analyses (Fig. 1 B and C), six (3%) independent ES cell clones were targeted correctly. Four targeted ES cell clones were injected into blastocysts of C57BL/6 mice and nine chimeras (seven male, two female) were derived, as judged by coat color. Three male chimeras, from two independently targeted ES cell clones, were crossed to C57BL/6 and transmitted the X-ALD− allele to their progeny giving rise to heterozygous female carrier mice. These carrier females were then mated to either 129/Sv or C57BL/6 males to generate X-ALD−/o hemizygous male mice. A normal progeny sex ratio (♂/♀ = 110:102) was observed, indicating that X-ALD−/o hemizygotes are not embryonic lethals. As seen in Fig. 2, the X-ALD− allele segregates with an X-linked inheritance pattern. Among 110 male offspring genotyped, 54 are X-ALD−/o hemizygotes, and 23 of 35 female offspring genotyped are X-ALD−/+ heterozygous carriers. X-ALD−/o hemizygous male mice were backcrossed to X-ALD−/+ heterozygous females to produce X-ALD−/− homozygous carriers. Both hemizygotes and homozygotes are normal in physical appearance and activity at birth and up to 6 months of age. Their fertility is also normal.
Figure 2.
Southern blot analysis of the X-ALD− allele segregation. The X-ALD− allele segregates with an X-linked inheritance pattern as predicted. ES1 is the targeted ES cell clone (Fig. 1) used to derive X-ALD mice.
Accumulation of VLCFAs in X-ALD Mice.
In humans, the diagnosis of X-ALD is usually made by measuring VLCFA levels in plasma (51). In X-ALD mice, surprisingly, there is no detectable elevation of C24:0 or C26:0 (saturated fatty acids with indicated carbon chain length) in plasma (see Table 1). Based on the ratio of C26:0/C22:0, the index commonly used for X-ALD diagnosis (4, 5), there is virtually no difference between normal and X-ALD mice. Because VLCFAs accumulate in tissues of X-ALD patients (1), we measured total VLCFAs in various tissues from X-ALD mice, including brain, adrenal gland, heart, liver, and kidney. As shown in Table 2, the levels of saturated VLCFAs are elevated in all tissues measured in X-ALD mice but the extent of accumulation is variable from tissue to tissue. There is a 5-fold increase of C26:0 level in brain (P < 0.005). In all other tissues, C26:0 levels increases range from 2- to 4-fold (P < 0.025). For comparison, VLCFA levels in various human tissues are also shown in Table 2. In the brain of X-ALD patients, a 2-fold increase in C26:0 level is observed in mildly involved regions (P < 0.005), whereas in demyelinated regions a 7-fold increase is seen (P < 0.005). In addition, a 7-fold elevation is observed in the liver of X-ALD patients (P < 0.005).
Table 1.
Plasma VLCFAs
| C22:0 % | C24:0 % | C26:0 % | C26:0/C22:0 | |
|---|---|---|---|---|
| Mouse | ||||
| Normal (n = 3) | 0.50 ± 0.08 | 0.51 ± 0.04 | 0.018 ± 0.010 | 0.038 ± 0.025 |
| X-ALD (n = 5) | 0.52 ± 0.13 | 0.46 ± 0.15 | 0.023 ± 0.012 | 0.044 ± 0.018 |
| Human | ||||
| Normal (n = 22) | 1.18 ± 0.62 | 0.98 ± 0.49 | 0.014 ± 0.010 | 0.015 ± 0.012 |
| X-ALD (n = 18) | 1.33 ± 0.53 | 1.99 ± 0.76 | 0.081 ± 0.028 | 0.064 ± 0.019 |
Measured values represent means ± SD for fatty acids with the indicated carbon chain length (C22, -24, -26). %, Percentage of total fatty acids. n, Total sample number measured. The differences of C24:0 %, C26:0 % and C26:0/C22:0 values between normal and X-ALD are significant (P < 0.005) in human plasma. Data of human plasma VLCFAs adapted from Moser et al (51).
Table 2.
Tissue total VLCFAs
| C22:0 % | C24:0 % | C26:0 % | C26:0/C22:0 | |
|---|---|---|---|---|
| Mouse tissue, total VLCFAs | ||||
| Brain | ||||
| Normal (n = 4) | 0.701 ± 0.082 | 1.167 ± 0.155 | 0.020 ± 0.004 | 0.029 ± 0.005 |
| X-ALD (n = 4) | 0.902 ± 0.132 | 1.566 ± 0.123 | 0.108 ± 0.013 | 0.123 ± 0.028 |
| Adrenal | ||||
| Normal (n = 6) | 0.124 ± 0.039 | 0.084 ± 0.024 | 0.009 ± 0.003 | 0.074 ± 0.010 |
| X-ALD (n = 7) | 0.093 ± 0.022 | 0.110 ± 0.033 | 0.024 ± 0.009 | 0.254 ± 0.068 |
| Heart | ||||
| Normal (n = 4) | 0.379 ± 0.079 | 0.194 ± 0.030 | 0.006 ± 0.001 | 0.017 ± 0.001 |
| X-ALD (n = 4) | 0.412 ± 0.073 | 0.263 ± 0.031 | 0.025 ± 0.004 | 0.060 ± 0.006 |
| Liver | ||||
| Normal (n = 5) | 0.643 ± 0.398 | 0.378 ± 0.121 | 0.004 ± 0.001 | 0.010 ± 0.007 |
| X-ALD (n = 4) | 0.673 ± 0.066 | 0.596 ± 0.100 | 0.014 ± 0.004 | 0.021 ± 0.005 |
| Kidney | ||||
| Normal (n = 4) | 0.609 ± 0.020 | 1.090 ± 0.115 | 0.013 ± 0.002 | 0.021 ± 0.003 |
| X-ALD (n = 4) | 0.531 ± 0.028 | 1.492 ± 0.191 | 0.052 ± 0.008 | 0.098 ± 0.010 |
| Human tissue, total VLCFAs | ||||
| Brain | ||||
| Normal (n = 19) | 0.65 ± 0.09 | 3.69 ± 0.67 | 0.48 ± 0.09 | 0.74 ± 0.11 |
| X-ALD*(n = 12) | 0.46 ± 0.04 | 2.39 ± 0.46 | 1.09 ± 0.05 | 2.41 ± 1.25 |
| X-ALD†(n = 10) | 0.30 ± 0.09 | 1.99 ± 1.16 | 3.51 ± 2.61 | 10.76 ± 8.05 |
| Liver | ||||
| Normal (n = 4) | 0.63 ± 0.27 | 0.71 ± 0.27 | 0.007 ± 0.004 | 0.009 ± 0.004 |
| X-ALD (n = 7) | 0.43 ± 0.13 | 0.94 ± 0.36 | 0.05 ± 0.02 | 0.12 ± 0.05 |
Measured values represent means ± SD. %, Percentage of total fatty acids. n, Total sample number measured. The differences of C26:0 % and C26:0/C22:0 values between normal and X-ALD are significant (P < 0.025) in all mouse and human tissues.
Brain samples collected from mildly involved regions from X-ALD patients.
Brain samples collected from severely demyelinated regions from X-ALD patients.
We also measured VLCFA content in cholesterol esters. The only significant C26:0 elevation is found in the adrenal glands of X-ALD mice, where the C26:0 level is 10-fold higher than in normal mice (P < 0.005; Table 3). However, significantly elevated levels of C26:0 can be observed from all tissues measured in human X-ALD (P < 0.025; Table 3). All these observations support the conclusion that X-ALD mice have biochemical defects that resemble to those found in hunam X-ALD (4, 5) but to a milder degree.
Table 3.
Tissue cholesterol ester VLCFAs
| C22:0 % | C24:0 % | C26:0 % | |
|---|---|---|---|
| Mouse tissue cholesterol ester VLCFAs | |||
| Brain | |||
| Normal (n = 4) | 1.52 ± 0.32 | 1.10 ± 0.42 | 0.98 ± 0.50 |
| X-ALD (n = 4) | 1.17 ± 0.30 | 0.95 ± 0.22 | 1.21 ± 0.72 |
| Adrenal | |||
| Normal (n = 4) | 0.44 ± 0.05 | 0.46 ± 0.09 | 0.12 ± 0.01 |
| X-ALD (n = 5) | 0.83 ± 0.27 | 2.28 ± 0.24 | 1.19 ± 0.14 |
| Heart | |||
| Normal (n = 2) | 1.27 | 0.71 | 1.34 |
| X-ALD (n = 3) | 0.90 ± 0.12 | 0.60 ± 0.11 | 1.19 ± 0.83 |
| Liver | |||
| Normal (n = 4) | 0.51 ± 0.65 | 0.51 ± 0.68 | 0.25 ± 0.22 |
| X-ALD (n = 4) | 0.24 ± 0.04 | 0.15 ± 0.04 | 0.11 ± 0.02 |
| Kidney | |||
| Normal (n = 3) | 0.77 ± 0.13 | 0.90 ± 0.35 | 0.50 ± 0.17 |
| X-ALD (n = 4) | 0.61 ± 0.20 | 1.04 ± 0.21 | 0.74 ± 0.17 |
| Human tissue cholesterol ester VLCFAs | |||
| Brain | |||
| Normal (n = 12) | 0.74 ± 0.30 | 2.07 ± 0.93 | 1.53 ± 1.08 |
| X-ALD*(n = 5) | 0.81 ± 0.15 | 3.1 ± 1.4 | 3.2 ± 2.0 |
| X-ALD†(n = 14) | 0.46 ± 0.32 | 4.00 ± 0.81 | 11.47 ± 9.37 |
| Adrenal | |||
| Normal (n = 4) | 0.25 ± 0.06 | 0.20 ± 0.12 | 0.03 ± 0.02 |
| X-ALD (n = 6) | 0.40 ± 0.14 | 7.9 ± 3.2 | 27 ± 11 |
| Liver | |||
| Normal (n = 9) | 0.17 ± 0.04 | 0.21 ± 0.06 | 0.07 ± 0.02 |
| X-ALD (n = 6) | 0.25 ± 0.05 | 0.60 ± 0.12 | 1.26 ± 0.27 |
| Kidney | |||
| Normal (n = 4) | 0.91 ± 0.23 | 2.20 ± 0.22 | 1.23 ± 0.99 |
| X-ALD (n = 2) | 0.88 | 8.0 | 5.01 |
Measured values represent means ± SD. %, Percentage of total fatty acids. n, total sample number measured. The differences of C26:0 % values between normal and X-ALD are significant (P < 0.025) in adrenal gland from mouse and brain, adrenal gland and liver from human.
Brain samples collected from mildly involved regions from X-ALD patients.
Brain samples collected from severely demyelinated regions from X-ALD patients.
β-oxidation and VLCFAs in X-ALD Human and Mouse Fibroblasts.
To determine the capability of VLCFA β-oxidation in X-ALD mice relative to human X-ALD patients, skin fibroblasts were derived from X-ALD mice and normal mice as well as from X-ALD and normal humans. Fibroblast VLCFA β-oxidation activity (48) and total VLCFAs (47) were measured. VLCFA β-oxidation in X-ALD mouse fibroblasts is reduced by 60% (P < 0.005) compared with normal mouse fibroblasts (based on C24:0-specific activity) whereas in humans the reduction ranges from 80% (P < 0.005; Table 4) to 60% (52). There is a 5-fold increase of C26:0 level in total VLCFAs of X-ALD mouse fibroblasts (P < 0.025), which is comparable to the result found in patients (53) (Table 5).
Table 4.
Fatty acid β-oxidation by cultured skin fibroblasts
| C16 S.A., nmol/hr per mg protein | C24 S.A., nmol/hr per mg protein | C24/C16 ratio | |
|---|---|---|---|
| Mouse | |||
| Normal (n = 3) | 3.87 ± 0.19 | 0.99 ± 0.15 | 0.26 ± 0.04 |
| X-ALD (n = 4) | 4.35 ± 0.81 | 0.37 ± 0.03 | 0.09 ± 0.02 |
| Human | |||
| Normal (n = 9) | 3.95 ± 0.75 | 1.08 ± 0.21 | 0.28 ± 0.04 |
| X-ALD (n = 8) | 4.21 ± 1.23 | 0.22 ± 0.09 | 0.05 ± 0.02 |
Values reported are mean specific activity (S.A.) ± SD. n, Total sample number measured. The differences of C24 S.A. and C24/C16 ratio values between normal and X-ALD are significant (P < 0.005) in both mouse and human fibroblasts.
Table 5.
VLCFAs in fibroblasts
| C26:0, μg/mg protein | C26:0/C22:0 ratio | |
|---|---|---|
| Mouse | ||
| Normal (n = 3) | 0.062 ± 0.013 | 0.064 ± 0.026 |
| X-ALD (n = 5) | 0.27 ± 0.11 | 0.35 ± 0.18 |
| Human | ||
| Normal (n = 65) | 0.079 ± 0.066 | 0.080 ± 0.029 |
| X-ALD (n = 77) | 0.42 ± 1.15 | 0.67 ± 0.21 |
Measured values represent mean amounts of VLCFAs (μg/mg protein) ± SD. n, Total sample number measured. The differences of C26:0 and C26:0/C22:0 ratio values between normal and X-ALD are significant (P < 0.025) in both mouse and human fibroblasts. Data of VLCFAs in human fibroblasts adapted from Moser et al. (51).
Histological Findings in X-ALD Mice.
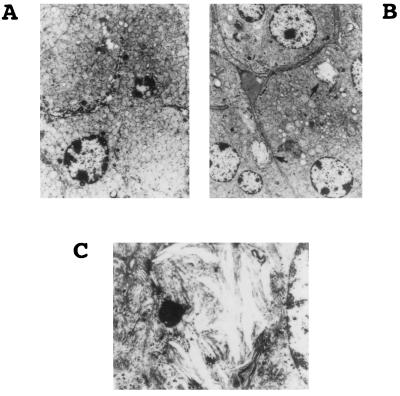
Histological studies were performed on the brains and adrenal glands of 3-month-old X-ALD mice and their normal sibs. Brain samples and adrenal glands were subjected to standard histological examination and electron microscopy, respectively. No light microscopic abnormalities were noted in either sample. Under the electron microscope, adrenocortical cells from normal mice contain abundant mitochondria, scant smooth endoplasmic reticulum, glycogen, and occasional lipofuscin as expected (Fig. 3A). However, in X-ALD mice, clear lipid clefts (indicated by arrows in Fig. 3B) were found in the adrenocortical cells. These lipid clefts were admixed with spicular lamellar material, best seen at high magnification (Fig. 3C). Similar ultrastructural findings have been observed in the adrenal cortex of X-ALD patients. No notable medullary changes were found in adrenal glands of X-ALD mice. No histological abnormalities were observed in the cerebral cortex, cerebellum, or brainstem. Preliminary ultrastructural examination of white matter was unremarkable.
Figure 3.
Histological analysis. (A) Electron micrograph of normal mouse adrenocortical cells with abundant mitochondria, scant smooth endoplasmic reticulum, glycogen, and occasional lipofuscin. (×5,000.) (B) Electron micrograph of X-ALD−/o mouse adrenocortical cells with clear, presumably lipid, clefts consistent with fatty acids (indicated by arrows). (×3,000.) (C) The lipid cleft admixed with spicular lamellar material at high magnification (×15,000.).
VLCFA Analysis of Mouse Chow.
It has been shown that part of the C26:0 accumulation in the brains and plasma of X-ALD patients is of dietary origin (54). As a result, the restriction of saturated VLCFA intake is part of current dietary therapy (Lorenzo’s oil) (31, 32) for X-ALD. To estimate the VLCFA intake of X-ALD mice, we determined the VLCFA content of the mouse chow regularly used as feed for the mice used in this study. We found that the abundance of saturated VLCFAs in mouse chow is relatively low (C22:0, 0.27%; C24:0, 0.20%; C26:0, 0.05%) and the amount of mono-unsaturated fatty acids is high (C18:1ω9, 26.15%; C20:1ω9, 0.46%). It has been shown that mono-unsaturated fatty acids (C18:1ω9 and C22:1ω9), components of Lorenzo’s oil, help normalize VLCFA levels in human X-ALD. Thus, it is possible that exposure to high levels of saturated VLCFAs and low levels of mono-unsaturated VLCFAs could induce the onset of neurological symptoms in X-ALD mice.
DISCUSSION
Biochemically, the X-ALD mouse represents a valid animal model for X-ALD. Most importantly, X-ALD in both mouse and human result in elevated tissue levels of saturated VLCFAs, with the most severe abnormalities localized to the brain and adrenal gland. Ultrastructural lamellar–lipid cleft inclusions in adrenocortical cells are present. Cultured fibroblasts from X-ALD mice show an impairment of VLCFA β-oxidation as do fibroblasts from X-ALD patients. In X-ALD mice, however, the biochemical changes are less severe than in the human disease. For example, human X-ALD has a 5-fold increase of C26:0 level in plasma whereas X-ALD mice do not show any significant elevation (Table 1), and the degree of accumulation of VLCFAs in brain and adrenal is less than that in the human disease (see Tables 2 and 3). Several factors could account for this. (i) The mice may develop more dramatic changes as they get older. (ii) The commonly used mouse chow contains only low amounts of saturated VLCFAs but is rich in mono-unsaturated fatty acids, some of which are known to inhibit saturated VLCFA elongation in human (55). Thus, the X-ALD mice may have been protected from dietary stress. (iii) Partial function redundancy of ALDP in mice may be provided by other peroxisomal ABC transporter proteins, such as ALDR, PMP70, and P70R. (iv) VLCFA metabolism may be influenced by other genes and thus vary in different inbred strains of mice.
At this time, the oldest X-ALD mice are 6 months old and have not shown overt evidence of adrenal insufficiency or nervous system dysfunction. It is still too early to conclude that X-ALD mice will not have neurological involvement equivalent to that seen in human X-ALD. About 50% of human X-ALD hemizygotes remain neurologically uninvolved through adolescence and 3% of human X-ALD commence cerebral involvement in adulthood (4, 5). Powers et al. (50) have demonstrated adrenal inclusions similar to those found in X-ALD mice in X-ALD carriers with normal adrenal function. As a result, the elevation of VLCFAs in cholesterol esters from adrenal glands and the histological findings in X-ALD mice could be the prelude to adrenal insufficiency. The abnormal pattern of VLCFAs in the brains of X-ALD mice is consistent with what is observed in histologically intact brain regions in X-ALD patients. In these regions, unlike the demyelinated areas of brain, the cholesterol ester fraction is normal in respect to total amount and fatty acid composition (45). Thus, it is possible that neurological involvement will appear with time.
In human X-ALD, the degree of VLCFA abnormality, as measured in plasma and fibroblasts, does not correlate with disease severity or age of onset. However, the relative levels of VLCFA in the affected tissues, brain and adrenal gland, at the time of disease onset are unknown. Thus, the lack of adrenal insufficiency and early brain involvement could be related to the VLCFA levels in these tissues in the X-ALD mice. The presence of the biochemical and histological “substratum” for the disease in the X-ALD mouse provides the opportunity to define the pathogenetic mechanisms of the diverse phenotypes of human X-ALD. First, as already noted, the dietary chow commonly supplied to rodents bears some resemblance to the Lorenzo’s oil therapy that is known to normalize the biochemical abnormality in the human disease (31, 32), and thus may have ameliorated the disease process in the mouse mutant. The VLCFAs accumulated in the brain of X-ALD patients has been shown to derive from the diet, at least in part (54). When normal individuals or women heterozygous for X-ALD consumed peanut oil containing 200 mg of C26:0, plasma C26:0 levels increased by a factor of 20 and remained elevated for 12 h in controls and at least 24 h in heterozygotes (A.B.M., unpublished observation). Studies to determine whether dietary stress will accentuate the disease abnormality are now in progress. Second, it has been proposed that a modifier gene contributes to the phenotypic variation found in X-ALD (56–58). Differences in genetic background have been shown to affect the severity of phenotypic presentation of cystic fibrosis in a mouse model leading to the identification of a possible modifier gene (59). Based on the measurable biochemical defect and normal fertility of the X-ALD mouse, crossing X-ALD mice to mice with different genetic backgrounds will provide an excellent opportunity to explore the possible existence of a potential modifier gene. Because the cerebral demyelination in X-ALD is associated with perivascular inflammation, it will be of particular interest to utilize mouse strains that vary in immune competence. For example, different strains of mice vary in their susceptibility to experimental autoimmune encephalomyelitis in response to specific antigenic stimulation (60). Third, there are a number of peroxisomal ABC transporter proteins with significant sequence similarity to ALDP. While the functions of these proteins are unknown, they could potentially provide partial redundancy for the function of ALDP and contribute to the milder biochemical defect found in X-ALD mice. Selective knockouts of these ABC transporter proteins, by targeted homologous recombination or antisense strategies, in X-ALD mice will allow determination of their relation to ALDP and possible role in VLCFA β-oxidation.
Therapies for X-ALD being evaluated in human patients include dietary management, bone marrow transplantation, and anti-inflammatory agents. The evaluation of dietary management is hampered by the fact that a randomized study design was not feasible for ethical reasons (32, 61); now, nearly 10 years after initiation, it is still uncertain whether this intervention is effective. In addition, the saturated VLCFA accumulation in brain persists after its level has been normalized in plasma by Lorenzo’s oil (46, 62), possibly due to the failure of glyceryl trierucate, the active ingredient of Lorenzo’s oil, to cross the blood–brain barrier. Bone marrow transplantation appears promising under carefully defined conditions (33), but the procedure carriers a high risk and evaluation is complicated by the great variability of the natural history of X-ALD. Whereas cerebral demyelination in X-ALD is accompanied by a marked inflammatory reaction (63), the nature of the inflammatory response and its role in demyelination is unclear (64–66) and immunosuppressive approaches that have been used so far have not been successful (67). It is expected that these and other therapeutic approaches, including gene therapy, can be tested rapidly in the animal model where effects on target tissues can be serially measured and then be used to enhance the effectiveness of therapy of the human disease.
Acknowledgments
We thank Dr. Gail Stetten for fluorescence in situ hybridization analysis, Dr. Di Wu for helping with blastocyst injections, Anita Liu for helping with VLCFA measurement, Don Gordon for helping with tissue culture, and Mollie Lange, Mary Blue, and Mary Wilson for helping with sample preparation for histological studies. This work was supported by Grants RP00035, RP00052, RP00072, and PO1HD24605 from the National Institutes of Health and by the John Hirschbeck and the Tommy Pacey Memorial Funds.
ABBREVIATIONS
- X-ALD
X-linked adrenoleukodystrophy
- VLCFA
very long chain fatty acid
- ABC
ATP-binding cassette
- neor
neomycin resistant
- tk
thymidine kinase
- ES cell
embryonic stem cell
- ALDP
adrenleukodystrophy protein
Note Added in Proof
While this manuscript was under review, similar results from an independently derived ALD mouse were published (68).
References
- 1.Igarashi M, Schaumburg H H, Powers J, Kishimoto Y, Kolodny E, Suzuki K. J Neurochem. 1976;26:851–860. doi: 10.1111/j.1471-4159.1976.tb04462.x. [DOI] [PubMed] [Google Scholar]
- 2.Singh I, Moser A E, Moser H W, Kishimoto Y. Pediatr Res. 1984;18:286–290. doi: 10.1203/00006450-198403000-00016. [DOI] [PubMed] [Google Scholar]
- 3.Lazo O, Contreras M, Bhushan A, Stanley W, Singh I. Arch Biochem Biophys. 1989;270:722–728. doi: 10.1016/0003-9861(89)90555-9. [DOI] [PubMed] [Google Scholar]
- 4.Moser H W. In: Advances in Human Genetics. Henry H, Kurt H, editors. Vol. 21. New York: Plenum; 1993. pp. 1–106. [PubMed] [Google Scholar]
- 5.Moser H W, Smith K D, Moser A B. In: The Metabolic Basis of Inherited Disease. Scirver C R, Baeudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 4256–4300. [Google Scholar]
- 6.Schaumburg H H, Powers J M, Raine C S, Suzuki K, Richardson E P., Jr Arch Neurol. 1975;32:577–591. doi: 10.1001/archneur.1975.00490510033001. [DOI] [PubMed] [Google Scholar]
- 7.Schaumburg H H, Powers J M, Raine C S, Spencer P S, Griffin J W, Prineas J W, Boehme D M. Neurology. 1977;27:1114–1119. doi: 10.1212/wnl.27.12.1114. [DOI] [PubMed] [Google Scholar]
- 8.Berger J, Molzer B, Fae I, Bernheimer H. Biochem Biophys Res Commun. 1994;205:1638–1643. doi: 10.1006/bbrc.1994.2855. [DOI] [PubMed] [Google Scholar]
- 9.Kok F, Neumann S, Sarde C O, Zheng S, Wu K H, Wei H M, Bergin J, Watkins P A, Gould S, Sack G, Moser H W, Mandel J L, Smith K D. Hum Mutat. 1995;6:104–115. doi: 10.1002/humu.1380060203. [DOI] [PubMed] [Google Scholar]
- 10.Mosser J, Douar A M, Sarde C O, Kioschis P, Feil R, Moser H, Poustka A M, Mandel J L, Aubourg P. Nature (London) 1993;361:726–730. doi: 10.1038/361726a0. [DOI] [PubMed] [Google Scholar]
- 11.Mosser J, Lutz Y, Stoeckel M E, Sarde C O, Kretz C, Douar A M, Lopez J, Aubourg P, Mandel J L. Hum Mol Genet. 1994;3:265–271. doi: 10.1093/hmg/3.2.265. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T, Yamada T, Yasutake T, Shinnoh N, Goto I, Iwaki T. Biochem Biophys Res Commun. 1994;201:1029–1034. doi: 10.1006/bbrc.1994.1805. [DOI] [PubMed] [Google Scholar]
- 13.Watkins P A, Gould S J, Smith M A, Braiterman L T, Wei H M, Kok F, Moser A B, Moser H W, Smith K D. Am J Hum Genet. 1995;57:292–301. [PMC free article] [PubMed] [Google Scholar]
- 14.Ligtenberg M J, Kemp S, Sarde C O, van Geel B M, Kleijer W J, Barth P G, Mandel J L, van Oost B A, Bolhuis P A. Am J Hum Genet. 1995;56:44–50. [PMC free article] [PubMed] [Google Scholar]
- 15.Krasemann E W, Meier V, Korenke G C, Hunneman D H, Hanefeld F. Hum Genet. 1996;97:194–197. doi: 10.1007/BF02265264. [DOI] [PubMed] [Google Scholar]
- 16.Kemp S, Ligtenberg M J, van Geel B M, Barth P G, Wolterman R A, Schoute F, Sarde C O, Mandel J L, van Oost B A, Bolhuis P A. Biochem Biophys Res Commun. 1994;202:647–653. doi: 10.1006/bbrc.1994.1979. [DOI] [PubMed] [Google Scholar]
- 17.Kemp S, Ligtenberg M J, van Geel B M, Barth P G, Sarde C O, van Oost B A, Bolhuis P A. Hum Mutat. 1995;6:272–273. doi: 10.1002/humu.1380060316. [DOI] [PubMed] [Google Scholar]
- 18.Cartier N, Lopez J, Moullier P, Rocchiccioli F, Rolland M O, Jorge P, Mosser J, Mandel J L, Bougneres P F, Danos O, Aubourg P. Proc Natl Acad Sci USA. 1995;92:1674–1678. doi: 10.1073/pnas.92.5.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 20.Rommens J M, Iannuzzi M C, Kerem B, Drumm M L, Melmer G, Dean M, Rozmahel R, Cole J L, Kennedy D, Hidaka N, Zsiga M, Buchwald M, Riordan J R, Tsui L C, Collins F S. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 21.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J L, Drumm M L, Iannuzzi M C, Collins F S, Tsui L C. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 22.Kane S E, Pastan I, Gottesman M M. J Bioenerg Biomembr. 1990;22:593–618. doi: 10.1007/BF00762963. [DOI] [PubMed] [Google Scholar]
- 23.Androlewicz M J, Anderson K S, Cresswell P. Proc Natl Acad Sci USA. 1993;90:9130–9134. doi: 10.1073/pnas.90.19.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchler K, Sterne R E, Thorner J. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaelis S. Semin Cell Biol. 1993;4:17–27. doi: 10.1006/scel.1993.1003. [DOI] [PubMed] [Google Scholar]
- 26.Shani N, Valle D. Proc Natl Acad Sci USA. 1996;93:11901–11906. doi: 10.1073/pnas.93.21.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Contreras M, Mosser J, Mandel J L, Aubourg P, Singh I. FEBS Lett. 1994;344:211–215. doi: 10.1016/0014-5793(94)00400-5. [DOI] [PubMed] [Google Scholar]
- 28.Kamijo K, Osumi T, Hashimoto T. Nippon Rinsho. 1993;51:2343–2352. [PubMed] [Google Scholar]
- 29.Gartner J, Valle D. Semin Cell Biol. 1993;4:45–52. doi: 10.1006/scel.1993.1006. [DOI] [PubMed] [Google Scholar]
- 30.Lombard-Platet G, Savary S, Sarde C O, Mandel J L, Chimini G. Proc Natl Acad Sci USA. 1996;93:1265–1269. doi: 10.1073/pnas.93.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzo W B, Leshner R T, Odone A, Dammann A L, Craft D A, Jensen M E, Jennings S S, Davis S, Jaitly R, Sgro J A. Neurology. 1989;39:1415–1422. doi: 10.1212/wnl.39.11.1415. [DOI] [PubMed] [Google Scholar]
- 32.Moser H W, Borel J. Annu Rev Nutr. 1995;15:379–397. doi: 10.1146/annurev.nu.15.070195.002115. [DOI] [PubMed] [Google Scholar]
- 33.Aubourg P, Blanche S, Jambaque I, Rocchiccioli F, Kalifa G, Naud-Saudreau C, Rolland M O, Debre M, Chaussain J L, Griscelli C, Fischer A, Bougneres P F. N Engl J Med. 1990;322:1860–1866. doi: 10.1056/NEJM199006283222607. [DOI] [PubMed] [Google Scholar]
- 34.Sarde C O, Thomas J, Sadoulet H, Garnier J M, Mandel J L. Mamm Genome. 1994;5:810–813. doi: 10.1007/BF00292021. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy M A, Rowland S A, Miller A L, Morris C M, Neville L A, Dodd A, Fifield W J, Love D R. Genomics. 1996;32:395–400. doi: 10.1006/geno.1996.0134. [DOI] [PubMed] [Google Scholar]
- 36.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 37.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 38.St Clair M H, Miller W H, Miller R L, Lambe C U, Furman P A. Antimicrob Agents Chemother. 1984;25:191–194. doi: 10.1128/aac.25.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M S, Amico L A, Speelman D J. Antimicrob Agents Chemother. 1984;26:778–780. doi: 10.1128/aac.26.5.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLaren C, Chen M S, Ghazzouli I, Saral R, Burns W H. Antimicrob Agents Chemother. 1985;28:740–744. doi: 10.1128/aac.28.6.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansuri M M, Ghazzouli I, Chen M S, Howell H G, Brodfuehrer P R, Benigni D A, Martin J C. J Med Chem. 1987;30:867–871. doi: 10.1021/jm00388a021. [DOI] [PubMed] [Google Scholar]
- 42.Lee K F, Li E, Huber L J, Landis S C, Sharpe A H, Chao M V, Jaenisch R. Cell. 1992;69:737–749. doi: 10.1016/0092-8674(92)90286-l. [DOI] [PubMed] [Google Scholar]
- 43.Axelrod H R. Dev Biol. 1984;101:225–228. doi: 10.1016/0012-1606(84)90133-7. [DOI] [PubMed] [Google Scholar]
- 44.Doetschman T, Maeda N, Smithies O. Proc Natl Acad Sci USA. 1988;85:8583–8587. doi: 10.1073/pnas.85.22.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theda C, Moser A B, Powers J M, Moser H W. J Neurol. 1992;239:307–310. [Google Scholar]
- 46.Rasmussen M, Moser A B, Borel J, Khangoora S, Moser H W. Neurochem Res. 1994;19:1073–1082. doi: 10.1007/BF00968719. [DOI] [PubMed] [Google Scholar]
- 47.Moser H W, Moser A B. In: Techniques in Diagnostic Biochemical Genetics: A Laboratory Manual. Hommes F A, editor. New York: Wiley–Liss; 1991. pp. 177–191. [Google Scholar]
- 48.Watkins P A, Ferrell E V J, Pedersen J I, Hoefler G. Arch Biochem Biophys. 1991;289:329–336. doi: 10.1016/0003-9861(91)90419-j. [DOI] [PubMed] [Google Scholar]
- 49.Powers J M, Moser H W, Moser A B, Schaumburg H H. Hum Pathol. 1982;13:1013–1019. doi: 10.1016/s0046-8177(82)80093-2. [DOI] [PubMed] [Google Scholar]
- 50.Powers J M, Moser H W, Moser A B, Ma C K, Elias S B, Norum R A. Arch Pathol Lab Med. 1987;111:151–153. [PubMed] [Google Scholar]
- 51.Moser H W, Moser A B, Frayer K K, Chen W, Schulman J D, O’Neill B P, Kishimoto Y. Neurology. 1981;31:1241–1249. doi: 10.1212/wnl.31.10.1241. [DOI] [PubMed] [Google Scholar]
- 52.Rizzo W B, Avigan J, Chemke J, Schulman J O. Neurology. 1984;34:163–169. doi: 10.1212/wnl.34.2.163. [DOI] [PubMed] [Google Scholar]
- 53.Moser H W, Moser A B, Kawamura N, Murphy J, Suzuki K, Schaumburg H, Kishimoto Y. Ann Neurol. 1980;7:542–549. doi: 10.1002/ana.410070607. [DOI] [PubMed] [Google Scholar]
- 54.Kishimoto Y, Moser H W, Kawamura N, Platt M, Pallante S L, Fenselau C. Biochem Biophys Res Commun. 1980;96:69–76. doi: 10.1016/0006-291x(80)91182-1. [DOI] [PubMed] [Google Scholar]
- 55.Rizzo W B, Watkins P A, Phillips M W, Cranin D, Campbell B, Avigan J. Neurology. 1986;36:357–361. doi: 10.1212/wnl.36.3.357. [DOI] [PubMed] [Google Scholar]
- 56.Smith, K. D., Sack, G., Beaty, T., Bergin, A., Naidu, S., Moser, A. B. & Moser, H. W. (1991) Am. J. Hum. Genet. 49 (Suppl), 165.
- 57.Moser H W, Moser A B, Smith K D, Bergin A, Borel J, Shankroff J, Stine O C, Merette C, Ott J, Krivit W, Shapiro E. J Inherited Metab Dis. 1992;15:645–664. doi: 10.1007/BF01799621. [DOI] [PubMed] [Google Scholar]
- 58.Maestri N E, Beaty T H. Am J Hum Genet. 1992;44:576–582. doi: 10.1002/ajmg.1320440509. [DOI] [PubMed] [Google Scholar]
- 59.Rozmahel R, Wilschanski M, Matin A, Plyte S, Oliver M, Auerbach W, Moore A, Forstner J, Durie P, Nadeau J, Bear C, Tsui L C. Nat Genet. 1996;12:280–287. doi: 10.1038/ng0396-280. [DOI] [PubMed] [Google Scholar]
- 60.Brocke S, Gaur A, Piercy C, Gautam A, Gijbels K, Fathman C G, Steinman L. Nature (London) 1993;365:642–644. doi: 10.1038/365642a0. [DOI] [PubMed] [Google Scholar]
- 61.Moser H W. J Inherited Metab Dis. 1995;18:435–447. doi: 10.1007/BF00710055. [DOI] [PubMed] [Google Scholar]
- 62.Poulos A, Gibson R, Sharp P, Beckman K, Grattan-Smith P. Ann Neurol. 1994;36:741–746. doi: 10.1002/ana.410360509. [DOI] [PubMed] [Google Scholar]
- 63.Griffin D E, Moser H W, Mendoza Q, Moench T R, O’Toole S, Moser A B. Ann Neurol. 1985;18:660–664. doi: 10.1002/ana.410180606. [DOI] [PubMed] [Google Scholar]
- 64.Powers J M, Liu Y, Moser A B, Moser H W. J Clin Invest. 1992;90:1864–1870. [Google Scholar]
- 65.McGuinness M C, Griffin D E, Raymond G V, Washington C A, Moser H W, Smith K D. J Neuroimmunol. 1995;61:161–169. doi: 10.1016/0165-5728(95)00084-f. [DOI] [PubMed] [Google Scholar]
- 66.McGuinness M C, Powers J M, Bias W B, Schmeckpeper B J, Segal A H, Gowda V C, Wesselingh S L, Berger J, Griffin D E, Smith K D. J Neuroimmunol. 1997;75:174–182. doi: 10.1016/s0165-5728(97)00020-9. [DOI] [PubMed] [Google Scholar]
- 67.Naidu S, Bresnan M J, Griffin D, O’Toole S, Moser H W. Arch Neurol. 1988;45:846–848. doi: 10.1001/archneur.1988.00520320032011. [DOI] [PubMed] [Google Scholar]
- 68.Kobayashi T, Shinnoh N, Kondo A, Yamada T. Biochem Biophys Res Commun. 1997;232:631–636. doi: 10.1006/bbrc.1997.6340. [DOI] [PubMed] [Google Scholar]