Abstract
As previously reported, Listeria monocytogenes infection of P388D1 macrophages results in a rapid induction of NF-κB DNA-binding activity. Here we show that this induction of NF-κB activity occurs in a biphasic mode: first, a transient, IκBα degradation-dependent phase of activity, also induced by the nonvirulent species Listeria innocua, which is mediated by binding of the bacteria to the macrophage, or by adding Listeria-derived lipoteichoic acid to the macrophage; the second persistent phase of activation is only markedly induced when the bacteria enter the cytoplasm of the host cell and express the virulence genes plcA and plcB, encoding two phospholipases. We suggest that products of the enzymatic activity of phospholipases directly interfere with host cell signal transduction pathways, thus leading to persistent NF-κB activation via persistent IκBβ degradation.
The transcription factor NF-κB, a pleiotropic mediator of inducible and tissue-specific gene control, is involved in the transcription of a variety of genes, many of which are activated during the immune response. NF-κB belongs to the Rel family of transcriptional activator proteins. The major form of this transcription factor is a heterodimer composed of a 50-kDa (NF-κB1 or p50) protein and a 65-kDa RelA DNA-binding protein, which is sequestered in the cytoplasm by association with inhibitory proteins of the IκB family (1, 2). The most intensively studied inhibitor of the Rel protein dimers is IκBα. Stimulation of cells with different inducers lead to hyperphosphorylation of IκBα, which in turn results in polyubiquitination. Subsequent degradation of IκBα by the proteasome allows NF-κB to be released from this inactive complex (3–6). Recently it was shown that IκBα and IκBβ are mediators of either transient (IκBα) or persistent (IκBβ) NF-κB activation in response to stimulators like tumor necrosis factor α and lipopolysaccharide, respectively (7). Free NF-κB enters the nucleus, where it binds to its target sequences and activates transcription (1).
Listeria monocytogenes, a Gram-positive bacterium and a causative agent of food-borne septicemia and meningitis, has been widely used as a model system of facultative intracellular bacteria (8, 9). L. monocytogenes can invade, survive, and replicate within nonprofessional phagocytic cells as well as in macrophages of different origin. It has been conclusively shown that the extracellular protein listeriolysin O (LLO) is absolutely required for intracellular survival and replication (10, 11). The gene encoding LLO (12) is part of a gene cluster which includes genes encoding a phosphatidylinositol-specific phospholipase C (plcA; PI-PLC) (13, 14), a metalloprotease (mpl) (15, 16), a protein necessary for induction of actin polymerization (actA) (17, 18), and a broad spectrum specificity phospholipase called phosphatidylcholine-specific phospholipase C (plcB; PC-PLC) (19). These genes, as well as the internalin family genes (inlA, inlB, and inlC) (20, 21), which are not part of the virulence gene cluster are under the control of the transcriptional activator PrfA encoded by the prfA gene (22).
Recently it was shown that heat-killed Staphylococcus aureus, invasive Shigella flexneri, and pneumococcal cell walls induce NF-κB (RelA/p50)-like activities in murine macrophages, HeLa cells, and human macrophages, respectively (23–25). We have reported on the rapid generation of nuclear NF-κB complexes in the macrophage-like cell line P388D1 after infection with a virulent L. monocytogenes strain (26). In the present study we show that L. monocytogenes infection of P388D1 macrophages results in a biphasic NF-κB activation in which the first transient phase is induced by lipoteichoic acid (LTA) and paralleled by IκBα decrease. The second persistent phase is mediated by the expression of the PrfA-dependent listerial phospholipases PI-PLC and PC-PLC and is paralleled by IκBβ degradation.
MATERIALS AND METHODS
Bacteria, Mammalian Cell Culture, and Infection.
Listeria used for cell culture infections were grown in brain heart infusion broth (Difco) at 37°C with aeration. Mid-log phase cultures were washed twice in PBS and stored at −80°C. The bacterial strains used for infection are shown in Table 1. P388D1 macrophages were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum and 2 mM l-glutamine (all from GIBCO). Macrophages were seeded 48 h prior to infection in tissue culture plates (Greiner, Nurtingen, Germany) at 5 × 106 cells/plate. Twenty-four hours before infection the medium containing only 0.5% fetal calf serum was added. The macrophages were infected with Listeria to give a multiplicity of infection of 50 bacteria per eukaryotic cell and incubated for 40 min. They were washed with PBS and incubated further in medium containing gentamicin (50 μg/ml) to kill extracellular bacteria and prevent reinfection.
Table 1.
Listeria strains used
| Strain | Genotype | Characteristics | Source or ref. |
|---|---|---|---|
| L. monocytogenes | |||
| Sv 1/2a EGD | WT | S. H. E. Kaufmann | |
| Sv 4b ATCC 19115 | WT | ATCC | |
| DP-L2161 | Δhly | In-frame deletion | D. A. Portnoy (56) |
| A42 | ΔprfA | In-frame deletion | B. Middendorf (57) |
| E5 | Δmpl | In-frame deletion | R. Böckmann (58) |
| A49 | ΔactA | In-frame deletion | This study |
| WL-103 | ΔplcB | In-frame deletion | This study |
| WL-105 | ΔplcA | In-frame deletion | This study |
| WL-106 | Δ(plcA,plcB) | In-frame deletion | This study |
| Δ2 | Δ(mpl,actA,plcB) | In-frame deletion | This study |
| A76 | ΔinlA | In-frame deletion | R. Böckmann (58) |
| PKP-1 | Δ(plcA,hly,mpl,actA,plcB) | In-frame deletion and insertion of kanR cassette | C. Ochs (21) |
| L. innocua | |||
| Sv 6a NCTC 11288 | WT | NCTC | |
| Sv 6a ATCC 33090 | ATCC | ||
| INN-LLO | prfA+,hly+ | LLO+ | This study |
ATCC; American Type Culture Collection; NCTC; National Cancer Tissue Cultures, National Institutes of Health; WT, wild type.
Preparation of Cytoplasmic and Nuclear Protein Extracts.
At the indicated time points following infection, cytoplasmic and nuclear protein extracts were prepared essentially as described (26, 27). The washed macrophages were scraped from the culture plate and collected by centrifugation. Pellets were resuspended in 0.4 ml of hypotonic lysis buffer containing 10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). After 15 min on ice, Nonidet P-40 was added to give a final concentration of 0.5%, and the cells were vortex mixed for 10 sec. The cells were centrifuged (30 sec, 4°C, 12,000 × g). The supernatant (=cytoplasmic extract) was frozen immediately and the nuclear pellets were resuspended in 130 μl buffer containing 20 mM Hepes (pH 7.9), 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF and vortex mixed for 30 min at 4°C. After centrifugation (5 min, 4°C, 12,000 × g) the supernatant (= nuclear extract) was frozen at −80°C.
Electrophoretic Mobility Shift Assay (EMSA).
The oligonucleotides 5′-AGCTTCAGAGGGGACTTTCCGAGAGG-3′ and 5′-TCGACCTCTCGGAAAGTCCCCTCTGA-3′ (28) were hybridized to form a double-stranded probe, and the overhanging ends were 32P-labeled with Klenow polymerase. The binding reactions (27) were performed on ice in a volume of 15 μl and contained 5 μl 3× binding buffer (60 mM Hepes, pH 7.9/3 mM DTT/3 mM EDTA/150 mM KCl/12% Ficoll), 20,000 cpm of 32P-labeled DNA probe, 2 μg nuclear proteins, and 1.6 μg poly(dI-dC) (Boehringer Mannheim). After 30 min on ice, the complexes were separated on native 5% polyacrylamide gels. Densitometric analysis of DNA–protein complexes from EMSA was performed with an ELSCRIPT 400 densitometer (Hirschmann Instruments, Unterhaching, Germany).
Detection of IκB Proteins.
For Western blot analysis 4–40 μg of cytoplasmic protein extract was separated on 12% SDS/PAGE, transferred to nitrocellulose, probed with the IκBα- or IκBβ-specific antibodies sc-203 and sc-945 (Santa Cruz Biotechnology), respectively, and developed using an enhanced chemoluminiscense kit (ECL, Amersham).
Construction of the In-Frame Deletion Mutants.
A 358-bp fragment from the 5′ end of the plcB gene (17) was amplified from L. monocytogenes chromosomal DNA by PCR with the primers PLCB-IN1 (5′-GGTGAATGATATGGAATTCAAAAATGTGG-3′) and PLCB-IN2 (5′-TGTACTGGTACCATAATATGG-3′). The primers were designed to generate sticky end restriction sites for EcoRI and KpnI, respectively. A second 226-bp fragment from the 3′ end of the plcB gene was amplified with the primers PLCB-IC3 (5′-TACGTAGGTACCATTAAACAC-3′) and PLCB-IC4 (5′-GCTGTGGATCCCTTAGTCTAGCTCCAG-3′). These primers were designed to generate restriction sites for KpnI and BamHI, respectively. The inserts were then ligated into pUC18 in a two-step ligation resulting in plasmid pUC18ΔplcB. A 372-bp fragment from the 5′ end of the plcA gene (13) was amplified from L. monocytogenes chromosomal DNA by PCR with the primers PLCA-IN1 (5′-TGGCGGAATTCGCTTCTAAAGATGAAACGC-3′) and PLCA-IN2 (5′-GCTCATGGTACCATGTGTACCTGG-3′). The primers were designed to generate sticky end restriction sites for EcoRI and KpnI, respectively. A second 281-bp fragment from the 3′ end of the plcA gene was amplified with the primers PLCA-IC3 (5′-ATCAAGGTACCAAAGCGGAC-3′) and PLCA-IC4 (5′-GTATATGGATCCGAGGTTGCTCGG-3′). These primers were designed to generate restriction sites for KpnI and BamHI, respectively. The inserts were then ligated into pUC18 in a two-step ligation resulting in plasmid pUC18ΔplcA. A 352-bp fragment from the 5′ end of the mpl gene (16) was amplified with the primers MSZ1 (5′-GTTACGAATTCTACGCTCGCGC-3′) and MSZ2 (5′-GCGCCGGTACCTACGTCCACTTG-3′) to generate a fragment with sticky ends after EcoRI and KpnI digestion. This fragment and the fragment obtained with the primers PLCB-IC3 and PLCB-IC4 from the plcB gene, were then ligated into pUC18 in a two-step ligation resulting in plasmid pUC18Δ(mplactAplcB). The EcoRI–BamHI inserts from pUC18ΔplcB, pUC18ΔplcA, and pUC18Δ(mplactAplcB) were then independently ligated into the shuttle vector pLSV-1 (29) resulting in plasmids pLSV-BNH1, pLSV-ANH1, and pSZZ-1, respectively. A 327-bp fragment (A) from the 3′ end of the actA gene (17) was amplified with the primers ACT1 (5′-ACACTGCAGACCTAATAGCAATGT-3′) and ACT2 (5′-GCATACTAGTATCTAAGTCACT-3′) to generate sticky ends after digestion with PstI and SpeI. A second fragment (B) from the 3′ end of the actA gene was amplified with the primers ACT3 (5′-GCTCTGACTAGTGACATAACTA-3′) and ACT4 (5′-GCTGATTCGCTTTCCTCTACCAT-3′) to generate a SpeI site at the 5′ end of the fragment. Fragment B was ligated into SmaI-treated pUC18 by blunt end ligation. The resulting plasmid was PstI and SpeI digested and ligated with fragment A to yield plasmid pUC18ΔactA. A BamHI–EcoRI fragment from pUC18ΔactA harboring fragments A and B was ligated into pLSV-1 (29), resulting in plasmid pSZZ-2. By using the plasmids pLSV-BNH1, pLSV-ANH1, pSZZ-1, and pSZZ-2, allelic exchange was performed as described (30) to generate the plcB deletion strain L. monocytogenes WL-103, the plcA deletion strain L. monocytogenes WL-105, the strain L. monocytogenes Δ2 with a deletion ranging from the mpl to the plcB gene, and the actA deletion strain L. monocytogenes A49. The strain L. monocytogenes WL-106 with deletions in plcA and plcB was constructed using plasmid pLSV-ANH1 and performing allelic exchange with strain L. monocytogenes WL-103 already harboring a deletion in plcB. The correct deletions on the chromosome of the respective strains was confirmed by PCR amplification of the deleted alleles and by sequencing the deletion sites.
Construction of the LLO-Expressing L. innocua Strain.
Plasmid pERL3–503 (31) harboring the L. monocytogenes genes prfA and hly was introduced by electroporation into strain L. innocua Sv 6a (NCTC 11288) to yield the LLO-producing L. innocua strain INN-LLO.
Isolation of LTAs.
LTAs (32, 33) from heat inactivated and mechanically disintegrated L. monocytogenes cells were extracted with 40% (vol/vol) phenol at 65°C for 45 min and the phases separated by centrifugation. The aqueous phase containing LTA was dialyzed against 0.05 M sodium acetate (pH 4.0). LTA was then purified by separation on an octyl-Sepharose CL-4b column (Pharmacia) with a linear gradient [0–66% (vol/vol) propanol in 0.05 M sodium acetate, pH 4.0]. Phosphate containing fractions eluted within the propanol gradient were pooled, dialyzed against double distilled water, and lyophilized.
RESULTS
Listeria Infection Results in a Biphasic Mode of NF-κB Activation.
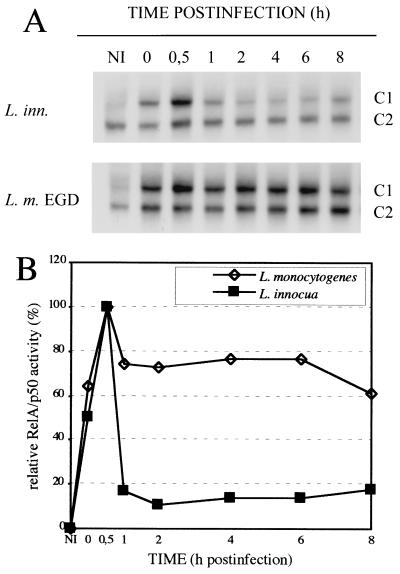
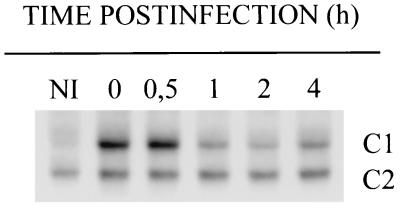
We have recently reported on the rapid and persistent induction of NF-κB DNA-binding activity in L. monocytogenes infected macrophages of the cell line P388D1 (26). This is confirmed here and a densitometric analysis of the complex C1 (RelA/p50) formed upon infection shows that L. monocytogenes infection indeed results in a peak of NF-κB activity at 0.5 h postinfection (pi) and a persistent activity at a slightly lower level for several hours thereafter (Fig. 1). The avirulent species L. innocua, although closely related with L. monocytogenes, is also able to induce a strong, rapid, and mainly transient activation of NF-κB, which, after the initial peak stays at a very low level for several hours (Fig. 1). These types of kinetics suggest that there might be two signals involved: a first one, which is also present in L. innocua, resulting in rapid, but transient activation, and the second being largely L. monocytogenes specific and responsible for the high level persistent NF-κB activation.
Figure 1.
(A) Kinetics of NF-κB DNA-binding activity in L. monocytogenes and L. innocua infected macrophages as detected with EMSAs. P388D1 macrophages were infected with Listeria as described, and NF-κB activity was determined with EMSAs at time points ranging from 0 to 8 h pi. Complex C1 (RelA/p50) is heavily induced, whereas complex C2 (p50/p50) remains largely unaffected. Only the relevant part of the EMSAs are shown. NI, noninfected control. (B) Densitometric analysis of the RelA/p50 containing complexes C1 from A.
LTA Induces a Rapid and Transient NF-κB Activation.
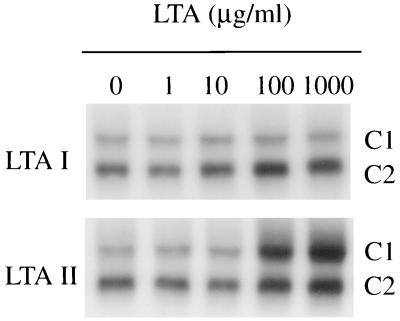
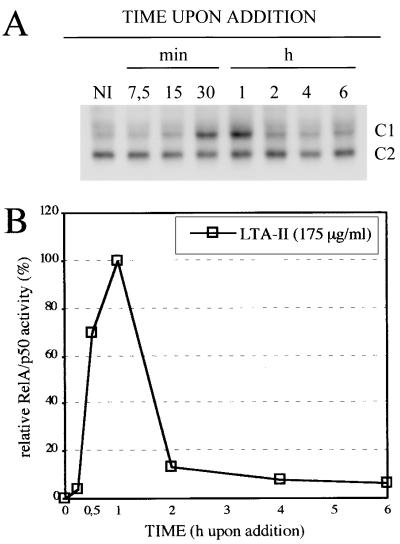
Binding of L. monocytogenes to P388D1 macrophages alone leads to NF-κB activation as described (26). Together with the fast kinetics of NF-κB activation upon infection it is reasonable to assume that bacterial surface structures activate the transcription factor after binding to host cell receptors. Bacterial surface structures like lipopolysaccharide (7) or lipoarabinomannan (34) are well-known activators of NF-κB, but they are both absent in Listeria spp. LTA can be phenol-extracted and purified from Listeria (33). Hydrophobic chromatography on octyl-Sepharose yielded, in the case of L. monocytogenes Sv 4b, and in L. innocua (ATCC 33090), two fractions, LTA I and LTA II. The latter was found to contain a glycolipid anchor substituted with a phosphatidyl residue that is absent in LTA I (35). The fractions of LTA from a L. monocytogenes Sv4b were assayed for their ability to enhance NF-κB DNA-binding activity in P388D1 macrophages. As shown in Fig. 2, LTA II gave a significant dose-dependent activation of NF-κB (in a range of 100–1,000 μg/ml) within 30 min after addition, whereas LTA I had no effect on NF-κB activity. The analogous preparations from L. innocua resulted in a similar NF-κB activation when added to P388D1 macrophages. Again, fraction I was inactive, but fraction II resulted in a significant activation of NF-κB (data not shown). In contrast to LTAs, Listeria-derived teichoic acids also present on the bacterial surface (35) do not induce NF-κB in a concentration of 200 μg/ml (data not shown). The kinetics of L. monocytogenes-derived LTA II induced NF-κB DNA-binding activity is, as outlined in Fig. 3, clearly transient. It follows roughly the pattern of the first phase of the biphasic NF-κB activation pattern postulated for L. monocytogenes. The rapid decrease of NF-κB DNA-binding activity between 1 and 2 h after addition of LTA II closely resembles the situation observed after infection with L. innocua (Fig. 1B), suggesting that listerial LTA II is the mediator of the rapid first phase of NF-κB activation observed after L. monocytogenes and L. innocua infection and may be also responsible for the low level persistent NF-κB activation found upon L. innocua infection (Fig. 1B).
Figure 2.
LTA II but not LTA I from L. monocytogenes induces a dose-dependent induction of NF-κB DNA-binding activity. P388D1 macrophages were treated with LTAs in concentrations ranging from 1 to 1,000 μg/ml for 30 min, and NF-κB activity was determined with EMSA.
Figure 3.
(A) LTA II from L. monocytogenes induces a rapid but transient NF-κB DNA-binding activity. P388D1 macrophages were treated with 175 μg/ml L. monocytogenes-derived LTA II for the indicated times before nuclear extracts were prepared and NF-κB activity determined with EMSA. NI, noninfected control. (B) Densitometric analysis of the RelA/p50 containing complexes C1 from A.
Release of L. monocytogenes into the Cytoplasm Is a Prerequisite for PrfA-Dependent Proteins to Induce Persistent NF-κB Activation.
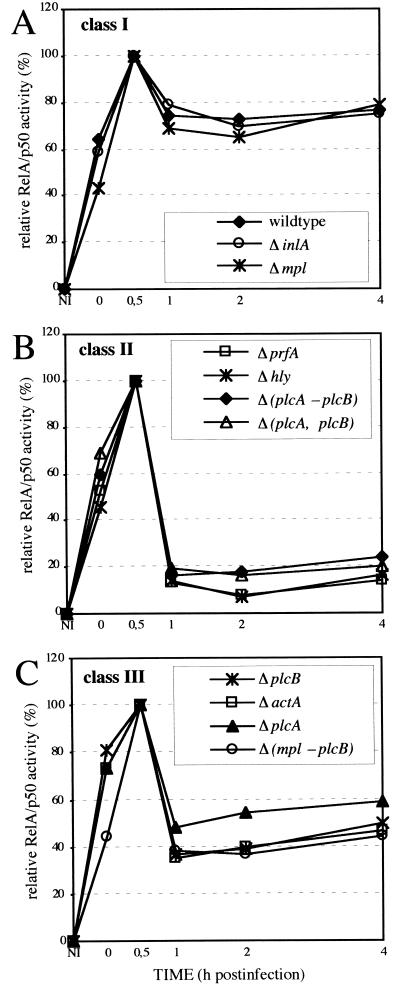
The persistent NF-κB activity in L. monocytogenes-infected macrophages probably relies either on a signal induced during escape from the phagosome or by intracellularly replicating bacteria. This hypothesis was tested by infecting macrophages with a set of in-frame deletion mutants affecting the known virulence genes of L. monocytogenes, most of which are located in the so-called virulence gene cluster. The kinetics of the NF-κB activity induced by these strains fall in three classes (Fig. 4). The strains of class I (Fig. 4A), ΔinlA and Δmpl, behave like the wild-type strain inducing a persistent activation and show that neither internalin nor the bacterial metalloprotease are required for persistent NF-κB activation. Class II (Fig. 4B) is represented by three nonhemolytic strains, Δhly, ΔprfA, and Δ(plcA,hly,mpl,actA,plcB) which cannot escape from the phagocytic vacuole and do not grow intracellularly and the hemolytic double mutant Δ(plcA,plcB). These strains induce a transient NF-κB activation that is similar to the kinetics found after L. innocua infection (Fig. 1B). Class III (Fig. 4C) comprises the four strains ΔplcA, ΔplcB, ΔactA, and Δ(mpl,actA,plcB) of which all four induce comparable, intermediate NF-κB activation. The NF-κB activity after infection with these strains is between 1 and 4 h pi, clearly higher than that of the class II mutants but also significantly lower as compared with the class I strains. Because the Δmpl strain behaves like the wild type, the effect seen with the class three mutants is obviously due to the absence of functional plcB, plcA, or actA gene products either alone or in combination with each other. There is no additive effect when the actA gene and the plcB gene are deleted, but the deletion of plcA and plcB results in an additive effect on the persistence of NF-κB activation. The pore-forming bacterial protein LLO is critical for escape of the bacteria from the phagocytic vacuole (8, 9), but does not induce the persistent NF-κB activation. This is shown by infection of P388D1 macrophages with a recombinant, hemolytic L. innocua strain that grows intracellularly (data not shown) but does not induce a persistent NF-κB DNA-binding activity (Fig. 5).
Figure 4.
Kinetics of NF-κB DNA-binding activity after infection with L. monocytogenes and different in-frame deletion mutants of L. monocytogenes. Nuclear extracts were prepared at the indicated time points, NF-κB activity was determined with EMSAs, and the RelA/p50 containing complexes C1 were analyzed densitometrically. The mean of at least three independent experiments is shown. (A) Class I strains. (B) Class II strains. (C) Class III strains.
Figure 5.
Kinetics of NF-κB DNA-binding activity in macrophages infected with the LLO producing L. innocua strain INN-LLO. Nuclear extracts were prepared at the indicated time points, and NF-κB activity was determined with EMSA. NI, noninfected control.
IκBα and IκBβ Are the Regulators of the Biphasic NF-κB Activation.
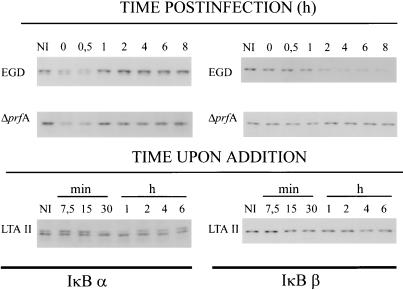
Transient and persistent NF-κB activation has recently been shown to be mediated by either IκBα or IκBβ degradation (7). We estimated the amount of IκBα and IκBβ proteins in cytoplasmic extracts of L. monocytogenes infected macrophages at different time points pi with Western blots. The results, outlined in Fig. 6, clearly indicate that infection with hemolytic and nonhemolytic strains resulted in a transient decrease of IκBα, in parallel to the transient phase of NF-κB activation (Fig. 1). In contrast, the IκBβ protein level in the cytoplasm was not altered after infection with the strain L. monocytogenes ΔprfA, which does not express the virulence gene cluster genes, but dropped continuously after infection with L. monocytogenes EGD beginning at 2 h pi and lasting for several hours (Fig. 6), corresponding to the long-lasting NF-κB activation observed after infection with this strain (Fig. 1). As expected, listerial LTA II treatment of the macrophages resulted in transient IκBα degradation, but had no effect on the amount of IκBβ in the cytoplasm throughout the duration of the experiment (Fig. 6).
Figure 6.
Immunoblot analysis of the kinetics of IκBα and IκBβ degradation after infection with L. monocytogenes and L. monocytogenes ΔprfA, and after L. monocytogenes-derived LTA II treatment as described in the legends of Figs. 3 and 4. Cytoplasmic proteins were isolated as described, and IκBα and IκBβ proteins were detected with specific antisera. NI, noninfected control.
DISCUSSION
Transcription factor NF-κB (RelA/p50) DNA-binding activity was recently shown to be rapidly induced by the infection of mouse P388D1 macrophages with L. monocytogenes (26). Comparing the kinetics of the activation of NF-κB DNA-binding activity of L. monocytogenes- and L. innocua-infected macrophages we demonstrate here that the nonhemolytic and nonvirulent species L. innocua is also able to induce a rapid NF-κB activation upon infection. However, this L. innocua-induced activation is mainly transient in contrast to the persistent activation of NF-κB after L. monocytogenes infection. These differences imply that two independent mechanisms are underlying the first transient and the second persistent phase of NF-κB activation of which only the strong and persistent one is specific for L. monocytogenes.
We have recently shown that inhibition of phagocytosis by cytochalasin B does not interfere with NF-κB activation upon infection (26). It is therefore obvious that binding alone of the bacteria to the target cell triggers rapid NF-κB nuclear localization. In search for ligands present on the surface of virulent and nonvirulent Listeria strains, purified LTA preparations were tested for their ability to induce NF-κB DNA-binding activity. The LTAs consist of alternating glycerol-phosphate units of variable chain length with sugar (galactose, glucose, and glucosamine), amino acid (d-alanine), and fatty acid substitutions. The two fractions obtained after octyl-Sepharose chromatography differ in their fatty acid substitutions, with fraction I having two fatty acids and fraction II four fatty acids linked to one end of the glycerol-phosphate backbone, probably in form of a glycolipid moiety (33). Fraction II (LTA II) indeed showed NF-κB-inducing activity, but only at relatively high concentrations compared with other inducers like lipopolysaccharide or lipoarabinomannan. Fraction I was totally ineffective. The same is true for LTAs derived from L. innocua, where again only fraction II was able to activate NF-κB. LTAs are inserted into the bacterial cytoplasmic membrane via their fatty acids and the glycerol-phosphate chain passing through the murein sacculus, probably resulting in a highly ordered complex. The loss of this ordered structure, when adding the LTA in solution to the macrophages, might be one explanation for the high concentrations of LTA needed for NF-κB activation. Two fractions of LTA with similar fatty acid substitutions were isolated recently from Enterococcus hirae, and in this case again only fraction II showed biological activity resulting in cytokine induction in eukaryotic cells (36).
The NF-κB activity induced by LTA II was largely transient and followed roughly the kinetics of NF-κB activation observed after infection with L. innocua. This result strongly supports the view that the transient phase of NF-κB activation induced equally by virulent and nonvirulent listeriae as well as the persistent low level NF-κB activation in L. innocua-infected macrophages is mainly mediated by listerial LTA II. LTA from different bacterial species including Listeria has been shown to bind to the murine scavenger receptor (SR) (37). The murine scavenger receptors I and II are trimeric proteins that differ in their C-terminal end where 110 amino acids present in SR I are absent in SR II (38). In P388D1 macrophages SR II is the dominant form (39). It is possible that in our system the soluble LTA as well as the bacteria bind to membrane-bound SR II, resulting in NF-κB activation. However, besides some reports of protein kinase C activity and Ca2+ release being linked to SR-mediated gene expression (40–42), nothing is known on signal transduction pathways originating from scavenger receptors and up to now no link of SRs with NF-κB activation has been reported. However, SR knockout mice show increased susceptibility to L. monocytogenes infection (43), pointing to a role for the SR in controlling L. monocytogenes infections in vivo. Because the transient phase of NF-κB activation correlates with IκBα decrease in the cytoplasm, we propose that SR signaling after LTA binding results in IκBα phosphorylation and subsequent degradation followed by the translocation of NF-κB to the nucleus. Whether newly synthesized IκBα also accumulates in the nucleus and thereby removes NF-κB from the DNA, as it has been detected upon tumor necrosis factor α stimulation of U937 macrophages (44), was not investigated in this study.
The high level persistent NF-κB activity observed after infection with L. monocytogenes is dependent on the escape from the phagosome. Strains that do not express functional LLO, either by in-frame deletion in the hly gene (Δhly), deletion of the positive regulating factor necessary for virulence gene expression (ΔprfA), or by deletion of the whole hly gene together with others [Δ(plcA,hly,mpl,actA,plcB)], are obviously unable to induce high level persistent NF-κB activation and are essentially behaving like L. innocua, where the whole virulence gene cluster is naturally absent (45). All nonhemolytic strains tested are unable to lyse the phagocytic vacuole, which is a prerequisite for intracellular growth. On the other hand, escape from the vacuole and intracellular growth is clearly not sufficient for high level persistent induction of NF-κB. This is shown either by infection with strains lacking only functional ActA, PC-PLC, or PI-PLC [ΔactA, ΔplcB, ΔplcA, Δ(mpl,actA,plcB)] which are, despite extensive intracellular growth, clearly diminished in persistent NF-κB activation. In addition, an L. innocua strain engineered for high LLO expression grows intracellularly but behaves like the parental nonhemolytic L. innocua strain concerning NF-κB activation. The mechanism leading to high level persistent NF-κB activation is clearly ActA, PC-PLC, and PI-PLC dependent. The listerial enzyme PC-PLC is a broad spectrum phospholipase with activity also on sphingomyelin, and PI-PLC is a phosphatidylinositol-specific phospholipase C. Both enzymes are expressed while L. monocytogenes resides inside the phagocytic vacuole because they both contribute to the efficient lysis of the phagosome (at least in some cell types) (46, 47). During the lysis of the vacuole, PC-PLC could generate diacylglycerol from phosphoglycerides and ceramide from sphingomyelin (48). Enzymatic activity of the PI-PLC would also release diacylglycerol from phosphatidylinositol. Activation of the acidic sphingomyelinase (found mainly inside lysosomes) by diacylglycerol would also generate ceramide from sphingomyelin (49). The activities of the two bacterial enzymes PI-PLC and PC-PLC during the escape of L. monocytogenes into the host cell cytoplasm therefore most likely result in enhanced intracellular levels of ceramide by independent mechanisms. A PC-PLC-dependent increase of intracellular ceramide, and to a lesser extent, of diacylglycerol, has been reported previously in L. monocytogenes infected J774 macrophages (47). Diacylglycerol and ceramide are discussed as potential second messengers in signaling pathways leading to NF-κB activation, either by stimulating IκB kinases or by other yet unknown mechanisms (49–51). It was also shown that a PC-PLC purified from Bacillus cereus resulted in phosphatidylcholine hydrolysis, diacylglycerol accumulation, and NF-κB activation upon addition to human monocytic cell lines (52). In contrast to the L. monocytogenes PC-PLC, the B. cereus-derived enzyme, however, cannot use sphingomyelin as a substrate and therefore will not directly liberate ceramide from sphingomyelin, as it was shown for the L. monocytogenes PC-PLC. Furthermore, in our system the listerial PI-PLC obviously also contributes to NF-κB activation in contrast to the study mentioned above where the addition of PI-PLC to the cells resulted in diacylglycerol production but not in NF-κB activation (51). These differences may be due to the different localization of the enzymes (and their products) if they are either added externally or synthesized by intracellular bacteria.
The metalloprotease, encoded by the mpl gene, is necessary for maturation of the listerial PC-PLC in the supernatant when the bacteria are grown in broth (53, 54). However, inside the host cell, Mpl is not absolutely necessary for PC-PLC maturation because the proteolytic processing can also be performed by a host cell protease (55). This assumption is in line with the reported observation that a mpl deletion mutant behaved like the wild-type strain concerning the activation of NF-κB upon infection of macrophages. The possible role of ActA in L. monocytogenes-induced NF-κB activation is less obvious. The fact that a deletion in actA and plcB does not have a greater effect (not additive) than a deletion in either gene alone suggests that ActA and PC-PLC might act in a consequential fashion. However, no mechanism has yet been described in which ActA influences the expression, maturation or activity of PC-PLC.
IκBα protein levels are indistinguishable after infection with L. monocytogenes EGD or L. monocytogenes ΔprfA. IκBβ, in contrast, only decreases after infection with wild-type L. monocytogenes strains expressing the PrfA-dependent virulence proteins. This pattern suggests that (i) IκBα is not affected by intracellular growing Listeria, (ii) or by intracellular ceramide levels, and (iii) that the postulated second messenger ceramide specifically leads to degradation of IκBβ resulting in a high level persistent NF-κB activation upon L. monocytogenes infection.
In conclusion, the data presented here show that the biphasic L. monocytogenes-induced NF-κB activation is triggered by several independent mechanisms, the first of which resulting in transient NF-κB activation most likely starts with a classical ligand-receptor interaction. The persistent NF-κB activation, however, seems to be mediated by direct modulation of signal transduction pathway(s) in the host cell through bacterial virulence gene products expressed within the mammalian cell.
Acknowledgments
We thank J. Daniels and A. Demuth for critical reading of the manuscript, E. Serfling for stimulating discussions, L. Greiffenberg for editorial help, T. Chakraborty for plasmid pERL3-503, and R. Böckmann, S. H. E. Kaufmann, B. Middendorf, C. Ochs, and D. A. Portnoy for providing L. monocytogenes strains. This work was supported by a fellowship from the Graduiertenkolleg Infektiologie of the University of Würzburg to N.H. and by the Deutsche Forschungsgemeinschaft through Grant SFB 165-B4.
ABBREVIATIONS
- EMSA
electrophoretic mobility shift assay
- LTA
lipoteichoic acid
- pi
postinfection
- PC-PLC
phosphatidylcholine-specific phospholipase C
- PI-PLC
phosphatidylinositol-specific phospholipase C
- LLO
listeriolysin O
References
- 1.Siebenlist U, Franzoso G, Brown K. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 2.Thanos D, Maniatis T. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 4.Alkalay I, Yaron A, Hatzubai A, Orians A, Ciechanover A, Ben-Neriah Y. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherer D C, Brockmann J A, Chen Z, Maniatis T, Ballard D W. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finco T S, Baldwin A S. Immunity. 1995;3:263–272. doi: 10.1016/1074-7613(95)90112-4. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J E, Phillips R J, Erdjument-Bromage H, Tempst P, Ghosh S. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn M, Goebel W. Genet Eng. 1995;17:31–51. [PubMed] [Google Scholar]
- 9.Sheehan B, Kocks C, Dramsi S, Gouin E, Klarsfeld A D, Mengaud J, Cossart P. Curr Top Microbiol Immunol. 1994;192:187–216. doi: 10.1007/978-3-642-78624-2_9. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn M, Kathariou S, Goebel W. Infect Immun. 1988;56:79–82. doi: 10.1128/iai.56.1.79-82.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Portnoy D A, Jacks P S, Hinrichs D J. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mengaud J, Vicente M-F, Chenevert J, Pereira J M, Geoffroy C, Giquel-Sanzey C, Baquero F, Perez-Diaz J-C, Cossart P. Infect Immun. 1988;56:766–772. doi: 10.1128/iai.56.4.766-772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leimeister-Wächter M, Domann E, Chakraborty T. Mol Microbiol. 1991;5:361–366. doi: 10.1111/j.1365-2958.1991.tb02117.x. [DOI] [PubMed] [Google Scholar]
- 14.Mengaud J, Braun-Breton C, Cossart P. Mol Microbiol. 1991;5:367–372. doi: 10.1111/j.1365-2958.1991.tb02118.x. [DOI] [PubMed] [Google Scholar]
- 15.Mengaud J, Geoffroy C, Cossart P. Infect Immun. 1991;59:1043–1049. doi: 10.1128/iai.59.3.1043-1049.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domann E, Leimeister-Wächter M, Goebel W, Chakraborty T. Infect Immun. 1991;59:65–72. doi: 10.1128/iai.59.1.65-72.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domann E, Wehland J, Rohde M, Pistor S, Hartl M, Goebel W, Leimeister-Wächter M, Wuenscher M, Chakraborty T. EMBO J. 1992;11:1981–1990. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kocks C, Gouin E, Tabouret M, Berch P, Ohayon H, Cossart P. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez-Boland J, Kocks C, Dramsi S, Ohayon H, Geoffroy C, Mengaud J, Cossart P. Infect Immun. 1992;60:219–230. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaillard J L, Berche P, Frehel C, Gouin E, Cossart P. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 21.Engelbrecht F, Chun S-K, Ochs C, Lottspeich F, Hess J, Goebel W, Sokolovic Z. Mol Microbiol. 1996;21:823–837. doi: 10.1046/j.1365-2958.1996.541414.x. [DOI] [PubMed] [Google Scholar]
- 22.Kreft J, Bohne J, Gross R, Kestler H, Sokolovic Z, Goebel W. In: Signal Transduction and Bacterial Virulence. Rappuoli R, Scarlato V, Arico B, editors. Austin, TX: Landes; 1995. pp. 129–142. [Google Scholar]
- 23.Busam K, Gieringer C, Freudenberg M, Hohmann H P. Infect Immun. 1992;60:2008–2015. doi: 10.1128/iai.60.5.2008-2015.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dyer R B, Collaco C R, Niesel D W, Herzog N K. Infect Immun. 1993;61:4427–4433. doi: 10.1128/iai.61.10.4427-4433.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spellerberg B, Rosenow C, Sha W, Tuomanen E I. Microbiol Pathol. 1996;20:309–317. doi: 10.1006/mpat.1996.0029. [DOI] [PubMed] [Google Scholar]
- 26.Hauf N, Goebel W, Serfling E, Kuhn M. Infect Immun. 1994;62:2740–2747. doi: 10.1128/iai.62.7.2740-2747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber E, Matthias P, Müller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreck R, Rieber P, Baeuerle P A. EMBO J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuenscher M, Koehler S, Goebel W, Chakraborty T. Mol Gen Genet. 1991;228:177–182. doi: 10.1007/BF00282463. [DOI] [PubMed] [Google Scholar]
- 30.Camilli A, Tilney L G, Portnoy D A. Mol Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darji A, Chakraborty T, Niebuhr K, Tsonis N, Wehland J, Weiss S. J Biotechnol. 1995;43:205–212. doi: 10.1016/0168-1656(95)00138-7. [DOI] [PubMed] [Google Scholar]
- 32.Fischer W, Koch H U, Haas R. Eur J Biochem. 1983;133:523–530. doi: 10.1111/j.1432-1033.1983.tb07495.x. [DOI] [PubMed] [Google Scholar]
- 33.Ruhland G J, Fiedler F. System Appl Microbiol. 1987;9:40–46. [Google Scholar]
- 34.Brown M C, Taffet S M. Infect Immun. 1995;63:1960–1968. doi: 10.1128/iai.63.5.1960-1968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiedler F, Ruhland G J. Bull Inst Pasteur. 1987;85:287–300. [Google Scholar]
- 36.Takada H, Kawabata Y, Arakaki R, Kusumoto S, Fukase K, Suda Y, Yoshimura T, Kokegushi S, Kato K, Komuro T, Tanaka N, Saito M, Yoshida T, Sato M, Kotani S. Infect Immun. 1995;63:57–65. doi: 10.1128/iai.63.1.57-65.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunne D W, Resnick D, Greenberg J, Krieger M, Joiner K A. Proc Natl Acad Sci USA. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieger M, Acton S, Ashkenas J, Pearson A, Penman M, Resnick D. J Biol Chem. 1993;268:4569–4572. [PubMed] [Google Scholar]
- 39.Aftring R P, Freeman M W. J Lipid Res. 1995;36:1305–1314. [PubMed] [Google Scholar]
- 40.Schaefer H I, Hold K M, Egas-Kenniphaas J M, van der Laarse A. Cell Calcium. 1993;14:507–516. doi: 10.1016/0143-4160(93)90009-u. [DOI] [PubMed] [Google Scholar]
- 41.Palkama T, Majuri M L, Mattila P, Hurme M, Renkonen R. Clin Exp Immunol. 1993;92:353–360. doi: 10.1111/j.1365-2249.1993.tb03404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falcone D J, McCaffrey T A, Vergilio J A. J Biol Chem. 1991;266:22726–22732. [PubMed] [Google Scholar]
- 43.Suzuki H, Karihara Y, Takeya M, Kamada M, et al. Nature (London) 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 44.Arenzana-Seisdedos F, Thompson J, Rodriguez M S, Bachelerie F, Thomas D, Hay R T. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gouin E, Mengaud J, Cossart P. Infect Immun. 1994;62:3550–3553. doi: 10.1128/iai.62.8.3550-3553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marquis H, Doshi V, Portnoy D A. Infect Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith G A, Marquis H, Jones S, Johnston N C, Portnoy D A, Goldfine H. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geoffroy C, Raveneau J, Beretti J L, Lecroisey A, Vazquez-Boland J A, Alouf J E, Berche P. Infect Immun. 1991;59:2382–2388. doi: 10.1128/iai.59.7.2382-2388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schütze S, Wiegmann K, Machleidt T, Krönke M. Immunobiology. 1995;193:1193–203. doi: 10.1016/s0171-2985(11)80543-7. [DOI] [PubMed] [Google Scholar]
- 50.Schütze M, Potthoff K, Machleidt T, Berkovic D, Wiegmann K, Krönke M. Cell. 1992;71:765–776. doi: 10.1016/0092-8674(92)90553-o. [DOI] [PubMed] [Google Scholar]
- 51.Kolesnick R, Golde D W. Cell. 1994;77:325–328. doi: 10.1016/0092-8674(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 52.Arenzana-Seisdedos F, Fernandez B, Dominguez I, Jacqué J M, Thomas D, Diaz-Meco M T, Moscat J, Virelizier J L. J Virol. 1993;67:6596–6604. doi: 10.1128/jvi.67.11.6596-6604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raveneau J, Geoffroy C, Beretti J L, Gaillard J L, Alouf J E, Berche P. Infect Immun. 1992;60:916–921. doi: 10.1128/iai.60.3.916-921.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poyart C, Abachin E, Razafimanantsoa I, Berche P. Infect Immun. 1993;61:1576–1580. doi: 10.1128/iai.61.4.1576-1580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marquis H, Goldfine H, Portnoy D A. J Cell Biol. 1997;137:1381–1392. doi: 10.1083/jcb.137.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones S, Portnoy D A. Infect Immun. 1994;62:5608–5613. doi: 10.1128/iai.62.12.5608-5613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Böckmann R, Dickneite C, Middendorf B, Goebel W, Sokolovic Z. Mol Microbiol. 1996;22:643–653. doi: 10.1046/j.1365-2958.1996.d01-1722.x. [DOI] [PubMed] [Google Scholar]
- 58.Böckmann, R. (1994) Diploma thesis (Univ. Würzburg, Würzburg, Germany).