Abstract
Sigma 54 is a required factor for bacterial RNA polymerase to respond to enhancers and directs a mechanism that is a hybrid between bacterial and eukaryotic transcription. Three pathways were found that bypass the enhancer requirement in vitro. These rely on either deletion of the sigma 54 N terminus or destruction of the DNA consensus −12 promoter recognition element or altering solution conditions to favor transient DNA melting. Each of these allows unstable heparin-sensitive pre-initiation complexes to form that can be driven to transcribe in the absence of both enhancer protein and ATP β–γ hydrolysis. These disparate pathways are proposed to have a common basis in that multiple N-terminal contacts may mediate the interactions between the polymerase and the DNA region where melting originates. The results raise possibilities for common features of open complex formation by different RNA polymerases.
Keywords: sigma factor, transcription, DNA melting
Bacteria contain two types of sigma factors and each directs a distinct transcription mechanism (1–4). The sigma 70 family of factors directs transcription from most bacterial promoters. These promoters can be controlled by repression or activation with their activation sites adjacent to the basal promoter elements near −10 and −35 (5). Sigma 54 is used at promoters that are rarely subject to repression and have their activation sites at some distance. These sites are typically within 200 bp of the basal elements at −12 and −24; they can however be hundreds of base pairs distant, and studies suggest that they can act analogously to eukaryotic enhancer elements (3, 4, 6). Because the two sigmas associate with the same simple core RNA polymerase, it is thought that sigma 54 converts the bacterial RNA polymerase into a form that uses an alternative mechanism that is responsive to enhancers (7).
The mechanism used by the RNA polymerase holoenzyme containing sigma 54 has hybrid properties. Like eukaryotic RNA polymerase II, it responds to enhancers and requires ATP hydrolysis for DNA melting and transcription (8, 9). However, other aspects of the transcription initiation mechanism are very like the sigma 70 holoenzyme, including the need for no other factors to bring polymerase to the promoter and the simplicity of the initiation process. The key determinant of the difference between the two types of holoenzymes is thought to reside within the small N-terminal domain of sigma 54, which has an unusual amino acid composition of 40% leucines and glutamines. Point mutations within a patch of four leucines between amino acids 25 and 31 have the drastic effect of allowing transcription in vitro to occur in the absence of enhancer protein and ATP (7, 10). These “bypass” forms of sigma 54 are relatively normal for activated transcription in that they still respond to enhancers, but they direct basal transcription more like sigma 70 since ATP and activator are not required (7, 10, 11).
Study of these mutants has recently led to a two-step model for enhancer activation of transcription (11). In step 1, the enhancer protein overcomes the inhibitory action of the leucine patch to allow formation of an unstable heparin-sensitive pre-initiation complex. The second step is the conversion to the stable heparin-resistant open complex. In its most extreme form, this model implies that the small N-terminal domain could have at least three separate functions. First, it has the inhibitory activity associated with the leucine patch (7, 10). Second, it may contain determinants that allow the enhancer protein to overcome this inhibitory activity. Third, it is known to be required for optimal recognition of the −12 promoter element on DNA (12–15). Thus, the properties of the N-terminal region are complex and are intimately related to the unique ability of sigma 54 to program RNA polymerase to respond to enhancers.
This model assumes that enhancer proteins trigger conformational changes, involving the N-terminal region, which lead to the two-step activation. We reasoned that it might be possible to mimic some of these changes by altering solution conditions, thus allowing separate study of the remaining steps in the process. Below we optimize conditions that allow wild-type sigma 54 to transcribe in the absence of enhancer activation (16). These conditions allow for new aspects of the mechanism to be uncovered, leading to an explanation for how the three or four seemingly disparate properties of the N terminus might work together to mediate the enhancer response.
MATERIALS AND METHODS
Plasmids.
Plasmid pTH8, bearing the glnAp2 promoter, and plasmids pFC50 and pFC50-M12, bearing the glnHp2 and Hp2-M12 promoters, respectively, were from B. Magasanik (16, 17). Supercoiled plasmids were prepared using Plasmid Maxi Kit (Qiagen, Chatsworth, CA) following the manufacturer’s instructions. All transcription reactions used supercoiled DNA, except as shown in Fig. 1, lane 7.
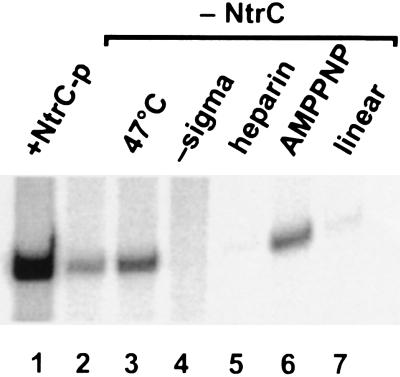
Figure 1.
Characteristics of bypass glnAp2 transcription using wild-type sigma 54. Unless otherwise indicated, reactions were at standard conditions (37°C, supercoiled DNA, 10 mM added KCl; ATP, CTP, and GTP added before heparin and UTP; no activator) yielding the RNA shown in lane 2. Lanes: 1, activated transcription with phosphorylated NtrC and only ATP added prior to heparin; 3, temperature increased to 47°C; 4, sigma 54 omitted; 5, heparin (100 μg/ml) added prior to nucleotides; 6, ATP substituted with nonhydrolyzable β–γ bond analog AMP–PNP; 7, linear template.
N-Terminal Deletion Mutants.
The Sal58 deletion mutant has been described (14). The coding region of Sal58 was recloned into expression plasmid pJF5401 (the sigma 54 gene inserted into expression vector pJLA503) (18, 19) and overexpressed in a sigma 54 minus strain YMC18 (thi, endA, hsr, ΔLacU169, rpoN::Tn10) and purified from inclusion bodies as described below. The ΔN mutant was constructed by first introducing a NdeI site at codon 40 to complement an existing site at the first codon. This facilitated removal of amino acids 2–40 by NdeI digestion. The resulting mutant ΔN was also overexpressed in YMC18 and purified from inclusion bodies as described below.
Protein.
Standard purifications were used for wild-type sigma 54 (20) and NtrC (21). Core polymerase was obtained commercially (Epicentre Technologies, Madison, WI). Both deletion mutant proteins (Sal58 and ΔN) were found in inclusion bodies and, therefore, were purified by methods similar to those described previously (11). Briefly, 1 liter of Luria–Bertani medium with 100 μg/ml ampicillin and 12.5 μg/ml tetracycline was inoculated with YMC18 cells transformed with expression vector that carries the appropriate sigma 54 mutant. The cell culture was grown to 1 OD at 30°C with very vigorous aeration. The culture was shifted to 43°C and grown for another 3–4 hr to induce expression (19). Cells were collected and suspended in 50 ml of buffer S (10 mM Tris⋅HCl, pH 8.0/200 mM KCl/0.1 mM EDTA/1 mM DTT/5% glycerol) and disrupted in a French press. After centrifugation of the cell lysate, the pellet was dissolved in 100 ml of buffer S plus 4 M guanidine⋅HCl and 0.1% Nonidet P-40 (nonionic detergent). Sonication was used to assist in dissolving the pellet. The dissolved material was dialyzed against 1 liter of buffer S with 1 M guanidine⋅HCl overnight. The dialysate was centrifuged and the undissolved material was discarded. The crude lysate was further dialyzed in 2 liters of buffer S alone. Then the crude material was loaded onto a Q-Sepharose ion exchange column (1.5 cm × 15 cm; Sigma). After washing the column with 10 column volumes of buffer S, the protein was eluted with a linear gradient of 0.2–0.8 M KCl in buffer S. Fractions of 5 ml were collected at 1 ml/min for a total of 200 ml elution volume. The protein elutes near fractions 13–17. The peak fractions were checked by SDS/7% PAGE. Fractions with the highest purity of protein were pooled and precipitated with 70% saturated ammonium sulfate (0.436 g/ml at 4°C). The protein pellet was dissolved in 1–2 ml of buffer S and dialyzed against buffer S + 10 mM MgCl2 and 40% glycerol. The concentration of the sigma 54 protein was determined by A280 absorption (1 A280 = 1.26 mg/ml) and also checked by Coomassie blue-stained SDS/PAGE against known protein markers. The mutant sigma 54 proteins from inclusion bodies are found to be >90% pure with this single chromatography step purification method. Because wild-type sigma 54 did not appear in inclusion bodies, its properties were tested after the purified protein (20) was denatured and renatured as described above. The renatured protein lost a small degree of activity but exhibited normal activator-dependent transcription under conditions where appropriate sigma mutants exhibited the bypass phenotype (not shown).
In Vitro Transcription.
Standard in vitro transcription reactions are as follows: 20 μl contains a final concentration of 50 mM Hepes (pH 7.9), 10 mM KCl (unless stated otherwise), 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 50 μg/ml BSA, and 3.5% (wt/vol) polyethylene glycol, 5 nM DNA, 100 nM sigma, 36 nM core polymerase (1 unit; Epicentre Technologies), 10 mM carbamyl phosphate (see ref. 22; Sigma). NtrC is at 100 nM when present; ATP was also added at 4.5 mM final (unless substituted with AMPPNP, which is at 0.5 mM when present). When NtrC is present, no other nucleotides are added with it except ATP. When NtrC is absent, nucleotides GTP and CTP are added to 0.5 mM each; ATP is already present in all reactions (except Fig. 1, lane 6). In addition, dinucleotide GpU is present at 0.1 mM when the DNA template is pFC50 (glnHp2) or M12. The reactions are assembled on ice, transferred to 37°C, and incubated for 20 min. Then heparin and the missing elongation nucleotides are added followed by a 10 min incubation. The elongation mixture includes 4 μCi (1 Ci = 37 GBq) of [α-32P]-labeled UTP (50 μM). Analysis was via 6% polyacrylamide–urea gels followed by PhosphorImaging (Molecular Dynamics). The transcripts generated from plasmid pTH8 (glnAp2) are 300 nucleotides long and those from pFC50 (glnHp2) and M12 are 418 nucleotides long.
RESULTS
We explored changes in solution conditions that might allow transcription by sigma 54 holoenzyme in the absence of activator. The protocol involves addition of holoenzyme to supercoiled DNA along with a nucleotide combination that allows a short transcript to form. Such “initiated complexes” (18) form efficiently only when activator is present (6, 11). Once formed, the initiated complexes produce full-length transcript upon the addition of the missing nucleotide. One goal of these experiments is to identify conditions that allow such complexes to form efficiently in the absence of activator, which normally does not occur (6).
Efficient formation of initiated complexes without activator does occur with bypass mutants of sigma 54 (7, 10, 11). However, formation of these initiated complexes is blocked by heparin unless activator is present (11). Thus, the formation of heparin-resistant pre-initiation complexes (without initiating nucleotides) is a marker of the ability of sigma 54 holoenzyme to respond to activator. A second goal was to learn what mediates this response. This required a heparin-challenge protocol (heparin and nucleotides added together) to learn if heparin blocked formation of initiated complexes.
Fig. 1, lane 2, shows the glnAp2 transcript made by wild-type sigma 54 polymerase without activator in low ionic strength solution. This low level bypass transcription represents 5–10% of the activated level (compare with lane 1). Leaky transcription under normal conditions is highly dependent on DNA supercoiling (16) and bypass transcription at low ionic strength is severely reduced on linear DNA (lane 7). It is also severely diminished in a heparin-challenge protocol (lane 5). As discussed previously, this property is associated with the instability of the open transcription complex (11). The bypass transcription does not require ATP hydrolysis, since substitution of ATP with non-β–γ-hydrolyzable AMP–PNP does not inhibit RNA synthesis (lane 6).
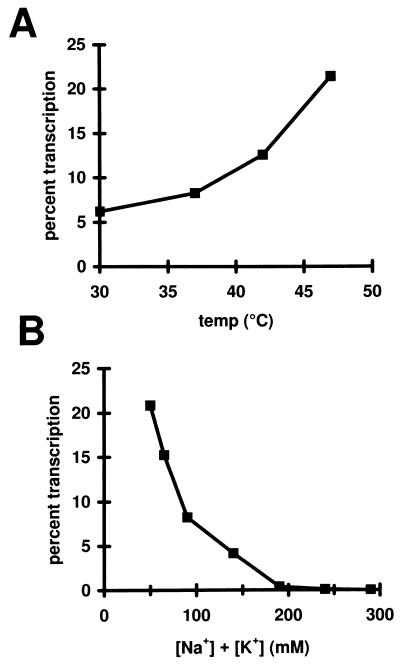
The level of bypass transcription can be increased by raising the temperature. The highest temperature assayed was 47°C (Fig. 1, lane 3). Higher temperatures appeared to partially inactivate protein components (data not shown), but the trend shown in Fig. 2A indicates that the enhancing effect of temperature is not saturated by 47°C. Even at 47°C, the use of higher ionic strength conditions suppresses bypass transcription (Fig. 2B).
Figure 2.
Effects of salt and temperature on bypass transcription. (A) Transcription reactions were done as in Fig. 1, lane 2, but at different temperatures. The amount of RNA produced at each temperature was normalized to activated transcription as in Fig. 1, lane 1. (B) Transcription reactions were done as in Fig. 1, lane 3. To increase the ionic strength of the reaction, KCl was added to supplement the approximately 40 mM NaCl contributed by various components of the reaction. Transcription is normalized to activated wild type and is plotted against total K+ and Na+ ion.
These data show that four factors work together to allow wild-type sigma 54 to direct transcription in the absence of activator: low ionic strength, high temperature, DNA supercoiling, and nucleotides that facilitate formation of initiated complexes. The first three of these have in common an ability to stimulate transient DNA melting and the fourth can drive the open DNA into stable elongation complexes. At the highest temperature tested (47°C) bypass transcription is approximately one-fifth as strong as activated transcription. Fig. 2A suggests that it might be even stronger if higher temperatures did not lead to protein inactivation.
Weak bypass transcription has been observed using premelted heteroduplex templates that retain promoter recognition elements (23). Strong bypass transcription has been observed using N-terminal leucine patch mutants of sigma 54 (7). Previous experiments showed that the N terminus is required for optimal recognition of the −12 promoter element (14). The strong bypass mutations are to varying degrees deficient in this recognition (11–13). These observations raise the possibility that defects in −12 recognition might be related to the strong bypass phenomenon. In an extreme model one might predict that a promoter with a defective −12 element could exhibit stronger transcription under bypass conditions.
To investigate the role of the consensus 12 promoter sequence we use two templates whose promoter sequences are shown in Fig. 3. The glnHp2 promoter has a nonconsensus −12 element due to the inclusion of a top strand T at position −14. The M12 mutation converts this to a consensus G (17). The two templates are otherwise identical. The M12 consensus change facilitates closed and open complex formation in vitro as shown by dimethyl sulfate footprinting and transcription (17). Using heparin-challenge one-round transcription conditions (see also ref. 17), which requires formation of heparin-resistant pre-initiation complexes to produce RNA, we find that the Hp2 and M12 supercoiled templates make roughly equal numbers of transcripts in the presence of activator and ATP (Fig. 3B, lanes 1 and 2).
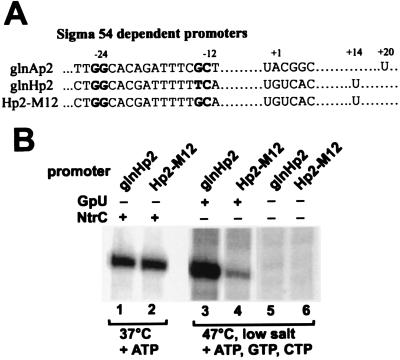
Figure 3.
Comparison of promoters that differ in the −12 consensus sequence. (A) The sequences of the various promoters used in this study are shown. (B) Standard activated transcription (heparin added with NTPs), using phosphorylated NtrC and ATP, was done using supercoiled templates glnHp2 (lane 1) and Hp2-M12 (lane 2). Unactivated transcription under bypass conditions (lower ionic strength, 47°C, and nucleotides; also see Fig. 1, lane 3) was done for supercoiled templates glnHp2 (lane 3) and Hp2-M12 (lane 4). Analysis of several experiments showed that the lane 1 and 2 levels were similar, whereas lane 3 gave 4- to 5-fold more RNA than lane 4. Lanes 5 and 6 are without the GpU dinucleotide in the preincubation, causing inactivation by heparin.
Fig. 3 also shows a comparison of the two templates in the absence of activator but under bypass assay conditions. The conditions include low salt concentrations, elevated temperature, supercoiled DNA, and pre-incubation in the presence of GpU, ATP, CTP, and GTP (except comparison lanes 5 and 6, which lack GpU) to trap an initiated complex prior to addition of heparin and radioactive UTP. The result shows that the unusual prediction is met; the nonconsensus glnHp2 promoter is transcribed much better than the consensus M12 promoter under bypass conditions (lane 3 is 4- to 5-fold stronger than lane 4). Without the initiating dinucleotide GpU required to form initiated complexes, the added heparin abolishes the bypass transcription (lanes 5 and 6). Thus, weakening the consensus −12 element can lead to an enhanced ability to transcribe in the absence of activator. This bypass transcription arises, however, from heparin sensitive pre-initiation complexes.
If, as the results suggest, −12 recognition is intimately related to establishing the requirement for activator, then mutation of sigma 54 to weaken −12 recognition might have the same effect as mutation of the −12 region on the DNA. The N-terminal region sigma 54 mutant reported to be the most defective in −12 recognition is the Sal58 deletion (14, 24). This deletion removes amino acids 18–31 (14), which includes the entire leucine patch and also elements known to be important for −12 recognition (12). Sal58 is nonfunctional in vivo (12, 14).
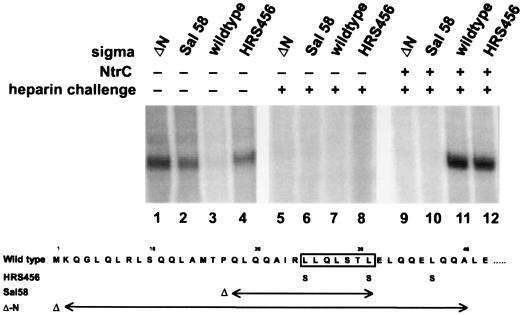
Nonetheless, experiments showed that Sal58 exhibits a strong bypass phenotype in vitro (Fig. 4, lane 2). In this experiment solution conditions are adjusted (by adding 50 mM KCl and lowering the temperature to 37°C) to minimize bypass transcription by wild-type holoenzyme, which no longer shows significant transcription in the absence of NtrC (lane 3, without NtrC, compared with lane 11, with NtrC). However, Sal58 polymerase transcribes in the absence of NtrC, even under these stringent conditions (lane 2), indicating that it is a strong bypass mutation similar to the previously described bypass mutant HRS456 (lane 4 and ref 7). Sal58 transcription does not occur if heparin is added prior to initiating nucleotides (lane 6 and ref. 24), as has been reported previously for bypass mutants (HRS456) and as shown for bypass transcription of the −12 region-deficient glnHp2 promoter (Fig. 3, lanes 5 and 6). Thus, Sal58 polymerase forms pre-initiation complexes that are transcriptionally active but are destroyed if heparin is added before initiating nucleotides. This is similar to mutant HRS456 in that it bypasses the need for NtrC and ATP in step 1 of the transcription mechanism (11), which is the formation of such unstable open complexes.
Figure 4.
Properties of N-terminal deletion mutants. The four indicated forms of sigma 54 were transcribed using supercoiled glnAp2 template, 95 mM total Na+ and K+ at 37°C, and limited elongation substrates, with deviations as indicated. NtrC (lanes 9–12) refers to the addition of phosphorylated activator and ATP. Heparin challenge (lanes 5–12) refers to the addition of heparin prior to adding nucleotides. The amino acid changes associated with HRS456, Sal58, and ΔN are shown below the figure. The leucine patch is boxed.
The data also demonstrate one important difference between the two bypass mutants Sal58 and HRS456. Sal58 polymerase has lost the ability to accomplish step 2 in the mechanism (11), the formation of stable heparin-resistant pre-initiation complexes (without initiating nucleotides) in response to NtrC and ATP. As shown previously, adding NtrC and ATP to HRS456 holoenzyme allows it to form a heparin-resistant pre-initiation complex (see Fig. 4, lane 12 and ref. 11) at levels comparable to wild type (lane 11). By contrast, Sal58 holoenzyme has lost the step 2 response to NtrC and ATP. Even with these components, no RNA is made when heparin is added prior to initiating nucleotides (lane 10). In this regard, Sal58 differs from all prior bypass mutants that still respond to NtrC (7, 10, 11). This is the first direct demonstration that the determinants that allow activator to trigger stable pre-initiation complex formation rely on the N terminus of sigma 54, an important extension of the association of the N terminus with the leucine patch inhibition determinants.
Because the N-terminal region has been associated with so many functions we wondered what would be the properties of a mutant sigma missing the entire N terminus. Amino acids 2–40 were deleted to form mutant ΔN. The purified ΔN protein was found to have properties like those of Sal58. ΔN polymerase can form large amounts of RNA without activator (Fig. 4, lane 1), but the pre-initiation complex formed with DNA is unstable in that it is completely inactivated by heparin (Fig. 4, lane 5, similar to Sal58 in lane 6 and HRS456 in lane 8). NtrC and ATP cannot trigger formation of heparin-resistant pre-initiation complexes involving ΔN polymerase (lane 9), like Sal58 but unlike HRS456 (lane 12).
Thus, removal of the N-terminal domain of sigma 54 does not destroy the catalytic function of the holoenzyme in vitro, as illustrated by its ability to form RNA. RNA synthesis is, however, fully deregulated because it neither depends on, nor responds to, activator NtrC. RNA synthesis also proceeds via unusual heparin-sensitive pre-initiation complexes. By contrast, in vivo assays show that both ΔN and Sal58 are nonfunctional. Both differ from wild type in that they fail to support growth on nitrogen-limiting media, and Sal58 mRNA was not detected in vivo in preliminary experiments (see ref. 12 and A.S. and J.D.G., unpublished data). This difference between in vivo and in vitro RNA production highlights the likely functional importance of forming a proper heparin-resistant pre-initiation complex. In both in vivo and in vitro contexts, however, N-terminal deletions apparently severely diminish the ability to respond to enhancer protein.
DISCUSSION
Sigma 54 directs RNA polymerase to promoters but inhibits DNA melting until enhancer protein is activated (6, 25, 26). There are several surprising experimental results shown here that relate to how sigma 54 directs this control of activation by melting. First, it was found that altering solution conditions in ways that favor DNA melting can lead to detectable transcription in the absence of enhancer activation. Second, this enhancer-independent transcription can be made even stronger by destroying the DNA sequence corresponding to the consensus −12 promoter recognition element. Third, this strengthening can also be achieved by deletion of the N terminus of sigma 54.
These three perturbations are shown to lead to common consequences; RNA synthesis proceeds in vitro without enhancer protein but occurs via a heparin-sensitive unstable pre-initiation complex. This contrasts with the heparin-resistant pre-initiation transcription that occurs when enhancer protein activates the wild-type sigma 54 holoenzyme. That type of transcription occurs only in the presence of enhancer protein even with bypass mutant forms of sigma 54 (11). It also correlates best with transcription in vivo for both sigma 54 and sigma 70 forms of polymerase. In the following discussion we will attempt to integrate these disparate observations.
Our data show that a variety of conditions that favor DNA melting, such as DNA supercoiling, elevated temperature, and lower ionic strength, have a common ability to promote bypass transcription by wild-type sigma 54 holoenzyme. Nonetheless, bypass transcription in vitro is associated with quite low levels of DNA melting (7, 11). This suggests that even low level transient melting must be inhibited during normal regulation (see also ref. 11). That inhibitory role appears to be primarily associated with the leucine patch sequence in the sigma 54 N terminus (7).
These data show other functions of the N terminus. First, it is required for the response to enhancer protein NtrC. Deletion of the N terminus destroys this response, but point mutants in the leucine patch (enhancer bypass mutants; refs. 7 and 10) do not destroy it. This confirms that at least some sequence determinants are different for the inhibition of unregulated bypass transcription via transient melting and the enhancer response activities. Both determinants, however, rely on sequences within the N terminus. Second, N-terminal deletions destroy the ability to form stable heparin-resistant pre-initiation complexes, suggesting that N-terminal sequences play an active role in stabilizing such complexes.
As discussed above, the N terminus of sigma 54 is also required for optimal recognition of the promoter −12 element (12–14), although the recognition is probably not direct. The above data show that destroying the consensus −12 element or making protein mutants with defective −12 recognition both stimulate bypass transcription. An intact −12 element in the context of an artificially melted −10 to +4 region still gives low levels of unregulated transcription (23). Thus, interactions with the consensus −12 element appear to be associated with the inhibition of unregulated bypass transcription. We are currently exploring which nucleotides within the −12 recognition element are important for this inhibition.
These observations may be merged with the other properties of the N terminus by proposing that it is situated between the core polymerase and the −12 region of the promoter and helps keeps them apart. DNA melting is believed to initiate just downstream from the −12 element (24). If the N terminus is held in place by interactions relying on both the polymerase and the DNA, then destroying either set will promote bypass transcription by allowing polymerase enhanced access to the site of melting initiation. Thus, either N-terminal protein mutations or −12 DNA mutations could lead to bypass transcription, as observed.
As discussed in the Introduction, sigma 54 directs a hybrid mechanism with aspects resembling both prokaryotic sigma 70 and eukaryotic RNA polymerase II transcription. With regard to sigma 70, the proposed dual role of the sigma 54 N terminus in promoter recognition and melting has some similarities to the proposed role of helix 14 in region 2 of sigma 70 (27). The two segments appear to be very different in that they have no sequence similarity, recognize different DNA sequences, and helix 14 plays no apparent inhibitory role. However, helix 14, like the above proposal for the sigma 54 N terminus, is postulated to have a joint role in DNA recognition and melting (28). Both sigma regions are required for optimal recognition of the DNA region between −9 and −13 (12, 14, 29, 30). In both cases this is the promoter region where DNA melting nucleates (24, 31). Both protein regions are the locus of mutations that alter DNA melting (7, 31), although the effects of mutation on melting are the opposite. Both regions appear to interact with a distinct strand preference, although the strands may also be opposite (32, 33).
Thus, both of these sigma segments may play critical roles in the nucleation of melting within the double helix and in the stabilization of the melted state by single-strand binding. Mutations in helix 14 region 2.3 cause sigma 70 to behave somewhat analogously to sigma 54; holoenzyme binds the DNA without melting it at low temperature (31). We also note that helix 14 appears to be situated between RNA polymerase and the region where melting nucleates (27), as we suggest for the sigma 54 N terminus. In the case of sigma 54, two steps in activation of melting have been proposed (11) and the two-step model is strongly supported by the results of this study. Step 1 involves formation of a heparin-sensitive nucleated open complex and step 2 leads to a stable open complex. Although some intermediates along the sigma 70 open complex pathway have been detected (34), these have not been tested for heparin sensitivity. Nor has the heparin sensitivity of pre-initiation complexes involving the melting-deficient sigma 70 mutants been tested. Comparative studies of the two systems should lead to the knowledge of which steps are likely to be preserved and which subject to regulation in the fundamental process of prokaryotic promoter DNA melting.
The sigma 54 mechanism may also be compared with mammalian transcription. Recent evidence suggests that the mammalian RNA polymerase II open complex (9) also forms in two steps (35). We have determined that the first open complex is less stable than the second (M. Yan and J.D.G., unpublished data) as observed in these studies of sigma 54 transcription. Because the sigma 54 and mammalian systems also have in common a responsiveness to enhancers and a need for ATP hydrolysis, it is worth considering the applicability of the details of the two-step sigma 54 mechanism to the mammalian case.
Acknowledgments
We thank Y. Song for technical assistance and Boris Magasanik for gifts of plasmids. This research was supported by U.S. Public Health Service Grant GM35754.
References
- 1.Gross C A, Lonetto M, Losick R. In: Bacterial Sigma Factors. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 129–176. [Google Scholar]
- 2.Helmann J D, Chamberlin M J. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 3.Kustu S, North A K, Weiss D S. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 4.Magasanik B. J Cell Biochem. 1993;51:34–40. doi: 10.1002/jcb.240510108. [DOI] [PubMed] [Google Scholar]
- 5.Gralla J D, Collado-Vides J. In: Organization and Function of Transcription Regulatory Elements. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1232–1245. [Google Scholar]
- 6.Ninfa A J, Reitzer L J, Magasanik B. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 7.Wang J T, Syed A, Hsieh M L, Gralla J D. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 8.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Carey M, Gralla J D. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 10.Syed A, Gralla J D. Mol Microbiol. 1997;23:987–996. doi: 10.1046/j.1365-2958.1997.2851651.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang J T, Gralla J D. J Biol Chem. 1996;271:32707–32713. doi: 10.1074/jbc.271.51.32707. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh M, Gralla J D. J Mol Biol. 1994;239:15–24. doi: 10.1006/jmbi.1994.1347. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh M, Tintut Y, Gralla J D. J Biol Chem. 1994;269:373–378. [PubMed] [Google Scholar]
- 14.Sasse-Dwight S, Gralla J D. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 15.Cannon W, Claverie-Martin F, Austin S, Buck M. Mol Microbiol. 1994;11:227–236. doi: 10.1111/j.1365-2958.1994.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 16.Hunt T P, Magasanik B. Proc Natl Acad Sci USA. 1985;82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claverie-Martin F, Magasanik B. J Mol Biol. 1992;227:996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- 18.Feng J, Goss T J, Bender R A, Ninfa A J. J Bacteriol. 1995;177:5523–5534. doi: 10.1128/jb.177.19.5523-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schauder B, Blocker H, Frank R, McCarthy J E. Gene. 1987;52:279–283. doi: 10.1016/0378-1119(87)90054-0. [DOI] [PubMed] [Google Scholar]
- 20.Popham D, Keener J, Kustu S. J Biol Chem. 1991;266:19510–19518. [PubMed] [Google Scholar]
- 21.Moore J B, Shiau S P, Reitzer L J. J Bacteriol. 1993;175:2692–2701. doi: 10.1128/jb.175.9.2692-2701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng J, Atkinson M R, McCleary W, Stock J B, Wanner B L, Ninfa A J. J Bacteriol. 1992;174:6061–6070. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wedel A, Kustu S. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 24.Morris L, Cannon W, Claverie-Martin F, Austin S, Buck M. J Biol Chem. 1994;269:11563–11571. [PubMed] [Google Scholar]
- 25.Popham D L, Szeto D, Keener J, Kustu S. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 26.Sasse-Dwight S, Gralla J D. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malhotra A, Severinova E, Darst S A. Cell. 1996;87:127–137. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 28.Severinova E, Severinov K, Fenyo D, Marr M, Brody E N, Roberts J W, Chait B T, Darst S A. J Mol Biol. 1996;263:637–667. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- 29.Waldburger C, Gardella T, Wong R, Susskind M M. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- 30.Siegele D A, Hu J C, Walter W A, Gross C A. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 31.Juang Y L, Helmann J D. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 32.Tintut Y, Wang J T, Gralla J D. Genes Dev. 1995;9:2305–2313. doi: 10.1101/gad.9.18.2305. [DOI] [PubMed] [Google Scholar]
- 33.Roberts C W, Roberts J W. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 34.Suh W C, Ross W, Record M T J. Science. 1993;259:359–361. doi: 10.1126/science.8420002. [DOI] [PubMed] [Google Scholar]
- 35.Holstege F C, van der Vliet P C, Timmers H T. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]