Abstract
The crystal structure of Escherichia coli ornithine transcarbamoylase (OTCase, EC 2.1.3.3) complexed with the bisubstrate analog N-(phosphonacetyl)-l-ornithine (PALO) has been determined at 2.8-Å resolution. This research on the structure of a transcarbamoylase catalytic trimer with a substrate analog bound provides new insights into the linkages between substrate binding, protein–protein interactions, and conformational change. The structure was solved by molecular replacement with the Pseudomonas aeruginosa catabolic OTCase catalytic trimer (Villeret, V., Tricot, C., Stalon, V. & Dideberg, O. (1995) Proc. Natl. Acad. Sci. USA 92, 10762–10766; Protein Data Bank reference pdb 1otc) as the model and refined to a crystallographic R value of 21.3%. Each polypeptide chain folds into two domains, a carbamoyl phosphate binding domain and an l-ornithine binding domain. The bound inhibitor interacts with the side chains and/or backbone atoms of Lys-53, Ser-55, Thr-56, Arg-57, Thr-58, Arg-106, His-133, Asn-167, Asp-231, Met-236, Leu-274, Arg-319 as well as Gln-82 and Lys-86 from an adjacent chain. Comparison with the unligated P. aeruginosa catabolic OTCase structure indicates that binding of the substrate analog results in closure of the two domains of each chain. As in E. coli aspartate transcarbamoylase, the 240s loop undergoes the largest conformational change upon substrate binding. The clinical implications for human OTCase deficiency are discussed.
Keywords: allostery, protein crystallography, urea cycle, ornithine transcarbamoylase deficiency
Anabolic ornithine transcarbamoylase (OTCase) catalyzes the first reaction in the urea cycle, in which l-ornithine is carbamoylated to form citrulline. More than 100 human mutations in OTCase that cause hyperammonemia and subsequent neurological damage or even death have been identified (1–5). Despite extensive modeling (3, 6), functional, and mutagenic studies (7, 8), interpretation of the clinical results has been hampered by the unavailability of an appropriate enzyme/substrate analog structure.
Escherichia coli anabolic OTCase (EC 2.1.3.3) shares ≈49% sequence similarity with its human counterpart and has a similar enzymatic mechanism (9–12). It is also homologous to the catalytic subunit of three transcarbamoylases whose crystal structures have been solved (13–15) [Pseudomonas aeruginosa catabolic OTCase, Bacillus subtilis aspartate transcarbamoylase (ATCase), and E. coli ATCase]. All four enzymes have the same tertiary structural motif, with each polypeptide chain folded into two discrete structural/functional domains, a carbamoyl phosphate (CP) binding domain and an ornithine (OTCase) or aspartate (ATCase) binding domain (6, 13–15). Active sites are shared between subunits of the trimer (16, 17). Chimeric enzymes with the E. coli OTCase CP binding domain exchanged with the CP binding domain of OTCases from other species or the catalytic subunit of E. coli ATCase are active (18, 19). Substrate binding induces conformational changes in these enzymes (20–23).
The quaternary structures of E. coli OTCase, P. aeruginosa catabolic OTCase, E. coli ATCase, and B. subtilis ATCase are quite different. E. coli OTCase and B. subtilis ATCase exist as free catalytic trimers (11, 14), P. aeruginosa catabolic OTCase exists as dodecamers with molecular 23 symmetry (13), whereas E. coli ATCase has pseudo 32 symmetry with two catalytic trimers linked by three regulatory dimers (15). In contrast to B. subtilis ATCase, E. coli ATCase catalytic trimer and E. coli OTCase, which are not cooperative, the more highly aggregated transcarbamoylases are allosterically regulated (ref. 24; reviewed in refs. 25–28).
The allosteric properties of E. coli ATCase have been studied most thoroughly and serve as a model for understanding other allosteric enzymes. Upon substrate binding, E. coli ATCase undergoes a large conformational change, in which the molecule expands ≈12 Å along its three-fold axis, each catalytic trimer and regulatory dimer rotate about the corresponding symmetry axes, the domains of the catalytic chains close about the substrate, and several interchain interfaces are restructured (20, 29–33). Because these changes are concerted and each chain is involved in several protein–protein interactions (33–36), short- and long-range effects are difficult to distinguish. Although the subunits have been studied extensively in solution (16, 20, 37–51), crystal structures of an isolated catalytic trimer of E. coli ATCase in the presence and absence of substrate analogs that would allow conformational changes restricted to the catalytic trimer to be identified have not previously been available.
We report here the crystal structure of E. coli OTCase complexed with its bisubstrate analog N-(phosphonacetyl)-l-ornithine (PALO) determined at 2.8-Å resolution. Active site residues that interact with the bound inhibitor are identified and the conformational change that accompanies substrate binding is described. The functional roles of specific residues are correlated with the kinetic and clinical properties of OTCase mutants and the OTCase/PALO structure is compared with the structures of other members of the transcarbamoylase family. The relevance of this model to understanding the allostery of E. coli ATCase is also discussed.
MATERIALS AND METHODS
Crystallization, Data Collection, and Processing.
The E. coli ArgI gene was engineered into overexpression system pET21a+ (Novagen) and subsequently transformed into E. coli strain BL21(DE3). Overexpressed protein was purified on a DEAE-Sephadex G-50 column with a yield of ≈140 mg of protein/liter of cell culture. Purified OTCase was concentrated to 15 mg/ml with 1 mg/ml PALO and 10 mM Hepes buffer at pH 7.5. The best crystals were obtained with the hanging drop method by mixing 5 μl of the above protein solution with 5 μl of well solution containing 14% PEG 3,350, 90 mM sodium acetate, 70 mM MgCl2, 2% 2-methylpropanediol, and 45 mM Tris⋅HCl (pH 8.5) and equilibrating with the well solution at 18°C for about 2 weeks. X-ray diffraction data were collected with a Siemens area detector. The software package astro (52) was used to design the collecting strategy and 99.4% of the unique reciprocal space from 15 to 2.8 Å was observed. Data frames were indexed, integrated, sorted, and scaled with the software package xengen (53). The space group is P1 with unit cell dimensions of a = 104.03 Å, b = 114.69 Å, c = 93.78 Å, and α = 86.99°, β = 93.11°, and γ=118.81°. The final data set contains 53,778 unique reflections with an weighted Rmerge for the measured intensities of 9.01%.
Structure Determination.
The self rotation function, calculated with the CCP4 program polarref (54) indicated that there were four potential three-fold axes, all at angles of ≈109° with respect to each other. The cross rotation function calculated with the P. aeruginosa catabolic OTCase trimer (Protein Data Bank reference no. 1otc; ref. 13) as the search probe and subsequent Patterson Correlation (PC) refinement implemented in x-plor (55) produced three unique solutions, corresponding to three of the three-fold axes obtained from the self rotation function, and related to each other by the fourth three-fold axis. The trimer corresponding to the highest PC peak was fixed, and a translation function was calculated using this trimer and the trimer with the next highest PC value. This translation function gave a well-defined solution ≈13σ above the average. The position of the third trimer was determined in a similar way. The packing of the trimers is close to that of a 32 screw axis, tilted by 0.84° from the unit cell c axis.
A poly-glycine backbone structure was generated from this molecular replacement (MR) solution (R factor ≈46%), structure factors were calculated to 6-Å resolution and phases were extended to 4-Å resolution by solvent flattening and nine-fold noncrystallographic symmetry (NCS) averaging using the CCP4 program dm. The calculated electron density map was of good quality, enabling a poly-serine/glycine monomer to be built. Cycles of density modification and phase extension were repeated and difference/omit Fourier synthesis was used to facilitate map interpretation, until the entire sequence was built into the model. Positional refinement lowered the R value to 29% under strict NCS constraints. Bound PALO was identified in the difference Fourier map as a region of well defined electron density (see Fig. 5), and PALO was built in with an orientation based upon that of N-(phosphonacetyl)-l-aspartate (PALA) in the ATCase/PALA structure (Protein Data Bank reference no. 8atc) and electrostatic considerations (35, 56). Further cycles of manual adjustments, refinement of NCS matrices, modeling of solvent molecules, minimization by simulated annealing, and grouped and individual B-factor refinement under strict NCS lowered the crystallographic R factor to 21.3% (Rfree = 23.0%), with rmsd of the model from ideal bond lengths and angles of 0.010 Å and 2.664°, respectively. There are only four residues (Ser-79, His-85, Leu-128, and Leu-274) in the generously allowed or disallowed region of the Ramachandran plot. The average temperature factors for backbone, side chain, ligand, and 8 solvent atoms are 30.09, 33.32, 10.24, and 28.70 Å2, respectively.
Figure 5.
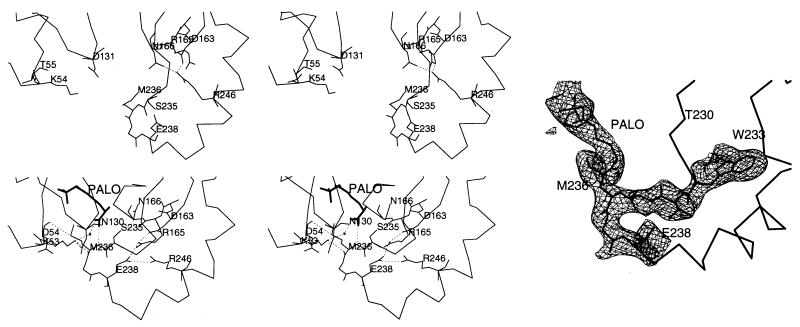
Domain closure. The Cα trace of the loop region and side chains of relevant residues are shown. (Upper Left) Unligated P. aeruginosa catabolic OTCase structure, with the active site open to solvent (13). (Lower Left) E. coli OTCase/PALO structure, with the two domains closed. (Right) The simulated annealing omit Fo − Fc map (contour level 2.0 σ, residues 233–239, and PALO omitted from minimization and phase calculation), confirms the positions of these residues and the ligand bound.
RESULTS AND DISCUSSION
Monomeric and Trimeric Structure.
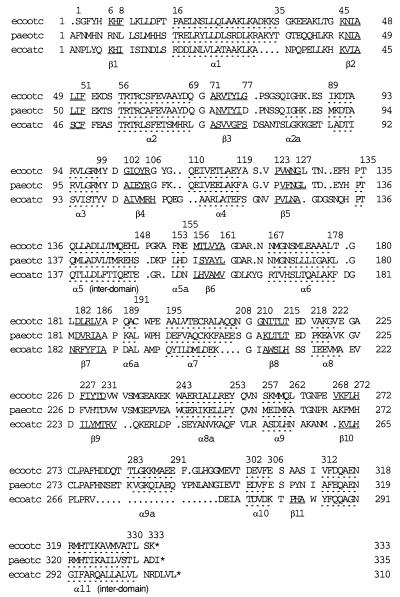
The structure of the E. coli OTCase monomer is similar to that of P. aeruginosa catabolic OTCase (13), B. subtilis ATCase (14), and the catalytic subunit of E. coli ATCase (15) (Fig. 1). In all four, each polypeptide chain folds into two structural domains linked by two interdomain helices. Each domain has a central parallel β-pleated sheet embedded in flanking α-helices and loops with approximate α/β topology. The sequences and secondary structural elements of the three proteins for which high resolution structures are available are compared in Fig. 2. Of the two domains, the CP binding domain (residues 1–135 in E. coli OTCase) has a higher degree of sequence homology (≈33% identity between E. coli OTCase and E. coli ATCase) and structural similarity, reflecting a common functional role. The rmsd of the Cα positions of the α helices and β strands between E. coli OTCase and E. coli ATCase is ≈0.9 Å. The ornithine/aspartate binding domain (residues 148–312) has less homology (≈12% identity) and a higher degree of structural divergence (rmsd of ≈2.1 Å). This domain undergoes a large conformational change upon substrate binding, as discussed below.
Figure 1.
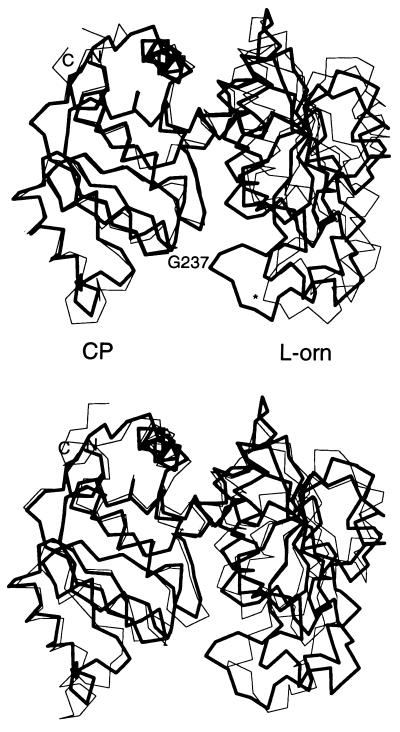
Superposition of E. coli OTCase onto P. aeruginosa catabolic OTCase and E. coli ATCase catalytic subunit, generated with the 78 Cα coordinates of the 5 β-strands and 4 α-helices of the CP binding domain as defined in E. coli ATCase (35). The rmsd between structures are 0.53 Å and 0.91 Å, respectively. CP, CP binding domain; l-orn, l-ornithine binding domain. (Upper) Superposition of E. coli OTCase ligated with PALO (thick line) on unligated P. aeruginosa catabolic OTCase (thin line), with Cα of Gly-237 labeled for comparison. (An ∗ is used in unligated P. aeruginosa catabolic OTCase.) (Lower) Superposition of E. coli OTCase ligated with PALO (thick line) on E. coli ATCase ligated with PALA (thin line).
Figure 2.
Sequence and secondary structure alignment of E. coli OTCase (ecootc), P. aeruginosa catabolic OTCase (paeotc), and E. coli ATCase catalytic subunit (ecoatc). Residues corresponding to α-helices and β-strands are separated by a space from the rest of the sequence and underlined with dashed and solid lines, respectively (13, 35). Residues at the N/C termini and the beginning and end of secondary structural elements in E. coli OTCase are numbered.
Shown in Fig. 3 is the trimeric organization of E. coli OTCase, which is identical to other transcarbamoylase catalytic trimers with the active sites shared between monomers (13–15). In all transcarbamoylases, the trimer appears to be the smallest structural element that is stable, has enzyme activity and can be used to build more complex enzyme assemblies.
Figure 3.
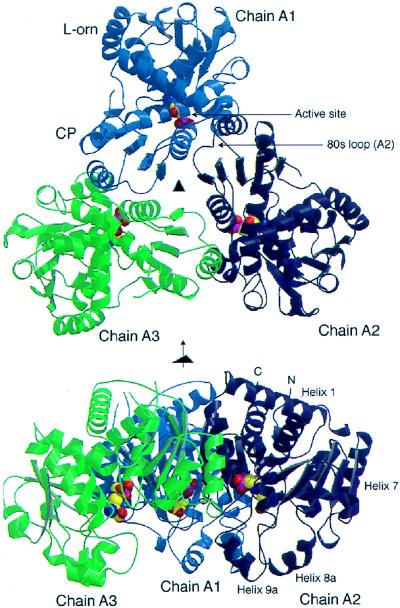
Cartoon drawings of E. coli OTCase catalytic trimer ligated with the bisubstrate analog PALO, shown as a space filling model. These figures were generated by graphic programs molscript (57) and render (58, 59). Chain A1, A2, and A3 are colored light blue, blue, and green respectively. (Upper) Top view, down the molecular three-fold axis. CP and l-ornithine binding domains of chain A1 are labeled. One active site, shared between chain A1 and the 80s loop of chain A2, is also labeled. (Lower) Side view, perpendicular to the three-fold axis. N/C termini and α helices 1, 7, 8a, and 9a of chain A2 are labeled.
E. coli OTCase and P. aeruginosa catabolic OTCase have a helix (α9a), which is absent in the E. coli ATCase catalytic subunit. In the E. coli OTCase/PALO structure, it is on the periphery of the trimer and appears to interact with the 240s loop (residues 232–256, including helix α8a), mainly through hydrophobic bonds. It also interacts with the 80s loop (residues 78–88) from a neighboring polypeptide chain. As discussed below, these two loops are important in catalysis. The position of helix α9a, combined with sequence differences at Lys-10, Gly-109, Gln-110, Glu-114, Glu-118, Glu-131, Thr-199, and Ala-203 would disrupt the potential c1–r1 interface between the catalytic chain and the regulatory chain of ATCase (15, 33–35), thus preventing OTCase from binding to the regulatory chain of E. coli ATCase in vivo.
Active Site Structure.
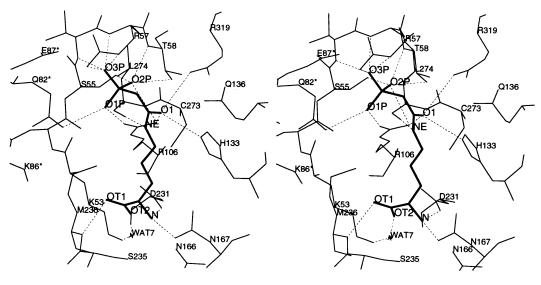
The PALO/OTCase complex is stabilized by an intricate network of interactions (Fig. 4). Ser-55 through Thr-58, Arg-106, Arg-319, and Gln-82 from the neighboring subunit stabilize the negative charge of the phosphonate moiety. One phosphonate oxygen, O1P, forms a salt bridge with the guanidino nitrogen NH2 of Arg-106 and is hydrogen bonded to the amide nitrogen NE2 of Gln-82 from the neighboring chain. Another phosphonate oxygen, O2P, may form hydrogen bonds with the hydroxyl oxygens of Ser-55 and Thr-58, the backbone amide nitrogens of Thr-58 and guanidino nitrogens NH1 and NH2 of Arg-106. The last phosphonate oxygen, O3P, may be hydrogen bonded to guanidino nitrogens NE and NH1 of Arg-57 and the backbone amide nitrogens of Thr-56 and Arg-57. The dipole moment of helix α2 also appears to stabilize the phosphonate group, as in the ATCase/PALA structure (33).
Figure 4.
Stereoview showing the interaction of the bisubstrate analog PALO with active site residues and interactions of Arg-57 and PALO with Gln-82, Lys-86, and Glu-87 of the 80s loop from an adjacent polypeptide chain. The interactions between NZ of Lys-86 with OE1 of Gln-82 and OT1 of PALO appear to be bridged by disordered water molecules.
The carbonyl oxygen O1 from the acetyl group of PALO is polarized by forming hydrogen bonds with NE2 of His-133 and the guanidino nitrogens of two arginines, Arg-106 and Arg-319. The ɛ nitrogen of the l-ornithine moiety is hydrogen bonded to the main chain carbonyl oxygen of Leu-274, which is located at a conserved loop (His-272, Cys-273, Leu-274, and Pro-275) between strand β10 and helix α9a. These residues are likely to act as general acids/bases in the catalytic mechanism.
In contrast to PALA, which is negatively charged, PALO has a positive charge on the α-amino group. This charged nitrogen forms a salt bridge with the carboxyl oxygen OD2 of Asp-231, and is hydrogen bonded to the carbonyl oxygen OD1 of Asn-167. One carboxyl oxygen of l-ornithine, OT1, is hydrogen bonded to the backbone nitrogen of Met-236, while the other carboxyl oxygen, OT2, may interact with the amide nitrogen ND2 of Asn-167 and the positively charged NZ of Lys-53 through a water molecule. The inhibitor/protein interactions and possible bonding parameters are summarized in Table 1.
Table 1.
Hydrogen bonds and salt bridges between the protein and the bisubstrate analog PALO
| PALO atoms | Protein atoms (distance, Å; angles,* °) |
|---|---|
| O1P | Arg-106 NH2† (2.68, 144°), Gln-82‡ NE2§ (3.01, 123°) |
| O2P | Ser-55 OG§ (2.72, 121°), Thr-58 N§ (2.99, 108°), Thr-58 OG1§ (2.71, 98°), Arg-106 NH1§ (3.11, 91°), Arg-106 NH2§ (3.01, 87°) |
| O3P | Thr-56 N§ (3.17, 104°), Arg-57 N§ (2.69, 108°), Arg-57 NE§ (2.78, 125°), Arg-57 NH2§ (3.03, 87°) |
| O1 | Arg-106 NH1§ (3.15, 108°), His-133 NE2§ (2.81, 105°), Arg-319 NH1§ (3.60, 114°) |
| NE§ | Leu-274 O (2.99, 128°) |
| N†§ | Asn-167 OD1 (3.03, 132°), Asp-231 OD2 (2.82, 100°) |
| OT1 | Met-236 N§ (2.92, 121°) |
| OT2 | Asn-167 ND2§ (3.23, 110°), via Wat 7 O1§ (2.78, 139°) to Lys-53 NE§ (2.71, 129°) |
The angles are defined by X-hydrogen donor–hydrogen acceptor. For backbone amide nitrogens, X = CA; for histidine NE2, X = CE1; for arginine NE, X = CD; for PALO NE, X = CD.
The cation of the salt bridge.
Gln-82 from an adjacent polypeptide chain in the catalytic trimer.
Hydrogen bond donor.
In E. coli ATCase, Ser-80 and Lys-84 in the 80s loop of an adjacent chain directly interact with the bound substrate analog PALA (33–35). In the E. coli OTCase/PALO structure, Gln-82, the counterpart of Ser-80 in E. coli ATCase, forms a hydrogen bond with the phosphonate oxygen O1P of PALO (Fig. 4). Lys-86, which corresponds to Lys-84 in E. coli ATCase, is adjacent to both the carbonyl oxygen OE1 of Gln-82 and the carboxyl oxygen OT1 of PALO, but not close enough to form direct bonds (Fig. 4). It is possible that they are bridged by solvent molecules, though no clear density was observed in the current map. These interactions appear to be functionally significant since mutating Lys-86 to Gln lowers Vmax by two orders of magnitude (6). Residues in the 80s loop of OTCases are not as well conserved as others at the active site, with the exception of Glu-87 (Glu-86 in E. coli ATCase), which is hydrogen bonded to Arg-57 from the adjacent chain, helps orient the side chain of Arg-57, and maintains the conformation of the 80s loop and its proximity to the adjacent chain.
The interactions between PALO and the protein described above correlate well with sequence alignments and the results of mutagenic and clinical studies (3, 7, 8). Residues conserved in the 26 OTCases that have been sequenced include Lys-53, Ser-55, Arg-57, Thr-58, Glu-87, Arg-106, His-133, Gln-136, Asn-167, Asp-231, Ser-235, Met-236, Leu-274, and Arg-319. These residues interact directly or indirectly with the bound substrate analog. Substitutions at positions 53 (human mutation K88N), 57 (R92Q), 58 (T93A), 106 (R141Q, R141P), 133 (H168Q), 231 (D263N), 236 (M268T), 237 (G269E), 272 (H302Q, H302L), 273 (C303Y), 274 (L304F), and 319 (R330G), all of which are located at or near the active site, affect catalysis to varying degrees, producing clinical symptoms (1–5, 7, 8). The model also accounts for the functional effects of mutations remote from the active site; for example, substitutions at Asp-163 (D196V) or Arg-246 (R277W, R277Q) (3, 7) would be expected to alter the conformation of the 240s loop and the movement of this loop that accompanies substrate binding, as discussed below.
Domain Closure.
Fig. 1 compares the back bone traces of E. coli OTCase, P. aeruginosa catabolic OTCase, and E. coli ATCase. As shown, the position of the 240s loop (residues 232–256) is different in the E. coli OTCase structure than it is in the other two structures. The rmsd of the Cα positions of residues 232–256 between the ligated and unligated OTCases is 6.5 Å and the positions of the Cα of Gly-237, which has one of the largest displacements, differ by 9.5 Å. Because there is 68% identity in the 240s loops of E. coli and P. aeruginosa OTCases, the loop conformation is not affected by crystal contacts in either structure, and this movement brings the CP and l-ornithine binding domains together (as well as rotating one relative to the other by ≈9°), this conformational difference is most reasonably attributed to the binding of PALO. A similar conformational change occurs when PALA binds to the catalytic subunit of E. coli ATCase; the 240s loop swings toward the CP binding domain and the aspartate binding domain rotates ≈6° relative to the CP binding domain, triggering the T to R quaternary structural transition (33–35).
Movement of the 240s loop alters many noncovalent bonds, particularly in and near the 240s loop. In the absence of bound substrate (P. aeruginosa catabolic OTCase) (13), Arg-246 forms a salt bridge with Asp-163 and interacts with Asn-166, whereas the side chains of Arg-165 and Glu-238 extend into the solvent. No residue in the 240s loop interacts with those in the CP binding domain, and the active site is open to the solvent (Fig. 5). When PALO binds to E. coli OTCase, Ser-235 and Met-236 are drawn into the active site to interact with the l-ornithine moiety of the inhibitor. These movements lead to a change in the position of the side chain of Glu-238, which moves to interact with Arg-246. Substrate binding also leads to a slight rotation of Arg-165 and Asn-166, enabling Arg-165 to form a salt bridge with Glu-238 and to interact with Asp-163, while breaking the interaction of Asn-166 with Arg-246. Several interdomain interactions also form. The sulfur atom of Met-236 interacts with the carbonyl oxygen of Asp-54. The amino nitrogen NZ of Lys-53 may interact with the backbone carbonyl oxygen of Met-236 and form a hydrogen bond with the side chain amide oxygen OD1 of Asn-130 (Fig. 5). The importance of these interactions and domain closure in the catalytic mechanisms of all OTCases are reflected in the conservation of Lys-53, Asp-163, Asn-166, Ser-235, Met-236 and Arg- (or Lys) 246 in the 26 OTCase sequences examined (3).
Although the closed conformation has only been observed in transcarbamoylase structures with both substrates/analogs bound (13–15, 33–36), transcarbamoylases may differ in the ease with which they undergo the transition between the closed and open states. Binding of the first substrate CP alone to E. coli OTCase induces ≈80% of the ultraviolet absorbance change that results from binding of the bisubstrate analog PALO (23). In contrast, most of the ultraviolet absorbance change in the E. coli ATCase holoenzyme or catalytic trimer does not occur until the second substrate, aspartate, binds (40, 43). It will be of interest to determine the conformational change that E. coli OTCase undergoes upon binding CP.
Protein–Protein Interactions and Conformational Change in the Transcarbamoylase Family.
Because the quaternary structure change that accompanies binding of substrate analogs such as PALA to E. coli ATCase has served as a model for conformational change in many other allosteric enzymes (25–28), there continues to be great interest in understanding its mechanism in detail. The T → R transition in E. coli ATCase is extremely complex, involving changes in domain interactions in both the catalytic and regulatory chains, elimination of most of the interactions between the 240s loops of the two catalytic subunits and the c1–r4 interaction between catalytic and regulatory chains, and the formation of new interactions at the c1–r1 interface between catalytic and regulatory chains (28). For many years, it has been recognized that detailed knowledge of the structure of the isolated catalytic trimer in the presence and absence of substrate analogs would be extremely useful because it lacks much of the structural and functional complexity of the holoenzyme. However, only one unligated crystal structure of a free catalytic subunit (from B. subtilis) is currently solved (14, 60). The OTCase crystal structures that are becoming available (61) provide a new opportunity to examine the properties of trimeric transcarbamoylases, the structural and functional effects of assembly into larger aggregates and the relationship between sequence and function.
Although domain closure is a major feature of the quaternary structure change that E. coli ATCase undergoes when substrates bind (33–35), the magnitude of the conformational change that accompanies substrate binding to isolated catalytic trimers has not been well defined experimentally. Methods such as ultraviolet difference spectroscopy (40) and hydrogen exchange kinetics (45) have provided evidence for a conformational change; however, the change in sedimentation coefficient is small (≈1%) (20, 41). The results reported here indicate that domain closure accompanies binding of substrate analogs to isolated catalytic trimers much as it does in the holoenzyme. However, wild-type ATCase catalytic trimers are noncooperative. Hence, domain closure is necessary but not sufficient for cooperativity.
Cooperativity can be introduced into the E. coli ATCase catalytic trimer, B. subtilis ATCase, and E. coli OTCase by mutating Arg-105 to Ala, Arg-99 to Ala, and Arg-106 to Gly, respectively (62–64). Binding of l-ornithine to E. coli OTCase also becomes cooperative when Zn(II) binds to Cys-273 (65–67). Apparently the signal transduction pathways that exist between the active sites and interchain interfaces of the holoenzyme of E. coli ATCase exist in the trimeric catalytic subunits of E. coli ATCase, B. subtilis ATCase and E. coli OTCase, but are latent in the wild type. The fact that they can be activated by mutation indicates that there is further scope for using OTCases as a tool for understanding cooperativity in ATCase.
Acknowledgments
We thank Drs. L. Banaszak and D. Ohlendorf for facilitating our use of the diffraction equipment in the Kahlert Center for Structure Biology at the University of Minnesota, and for their critical reading of the manuscript. Some intensive calculations were carried out on a Cray X-MP EA at the Minnesota Supercomputer Center. We thank Drs. G. Barany and D. Venugopal for synthesizing PALO and Dr. N. Glansdorff for providing the plasmid with the ArgI gene. We thank Dr. B. Rajagopal for verifying the sequence of the engineered ArgI gene and Dr. H. Morizono for helpful discussions. This work was supported by National Institutes of Health Grant DK-47870 (to M.T.).
ABBREVIATIONS
- OTCase
ornithine transcarbamoylase
- ATCase
aspartate transcarbamoylase
- CP
carbamoyl phosphate
- PALO
N-(phosphonacetyl)-l-ornithine
- PALA
N-(phosphonacetyl)-l-aspartate
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 [reference 2otc; embargo period: 1 year (till June 15, 1998).
References
- 1.Tuchman M, Morizono H, Rajagopal B S, Plante R J, Allewell N M. J Inherited Metab Dis. 1997;20:525–527. doi: 10.1023/a:1005301513465. [DOI] [PubMed] [Google Scholar]
- 2.Tuchman M, Plante R J, Garcia-Perez M A, Rubio V. Hum Genet. 1996;97:274–276. doi: 10.1007/BF02185751. [DOI] [PubMed] [Google Scholar]
- 3.Tuchman M, Morizono H, Reish O, Yuan X, Allewell N M. J Med Genet. 1995;32:680–688. doi: 10.1136/jmg.32.9.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuchman M, Plante R J. Hum Mutat. 1995;5:293–295. doi: 10.1002/humu.1380050404. [DOI] [PubMed] [Google Scholar]
- 5.Tuchman M. Hum Mutat. 1993;2:174–178. doi: 10.1002/humu.1380020304. [DOI] [PubMed] [Google Scholar]
- 6.Murata L B, Schachman H K. Protein Sci. 1996;5:709–718. doi: 10.1002/pro.5560050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morizono H, Tuchman M, Rajagopal B S, McCann M T, Listrom C D, Yuan X, Venugopal D, Barany G, Allewell N M. Biochem J. 1997;322:625–631. doi: 10.1042/bj3220625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morizono H, Listrom C D, Rajagopal B S, Aoyagi M, McCann M T, Allewell N M, Tuchman M. Hum Mol Genet. 1997;6:963–968. doi: 10.1093/hmg/6.6.963. [DOI] [PubMed] [Google Scholar]
- 9.Kalousek F, Francois B, Rosenberg L E. J Biol Chem. 1978;253:3939–3944. [PubMed] [Google Scholar]
- 10.Horwich A L, Fenton W A, Williams K R. Science. 1984;224:1068–1074. doi: 10.1126/science.6372096. [DOI] [PubMed] [Google Scholar]
- 11.Legrain C, Stalon V. Eur J Biochem. 1976;63:289–301. doi: 10.1111/j.1432-1033.1976.tb10230.x. [DOI] [PubMed] [Google Scholar]
- 12.Piette J, Cunin R, Van Vliet F, Charlier D, Crabeel M, Ota Y, Glansdorff N. EMBO J. 1982;1:853–857. doi: 10.1002/j.1460-2075.1982.tb01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villeret V, Tricot C, Stalon V, Dideberg O. Proc Natl Acad Sci USA. 1995;92:10762–10766. doi: 10.1073/pnas.92.23.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens R C, Reinisch K M, Lipscomb W N. Proc Natl Acad Sci USA. 1991;88:6087–6091. doi: 10.1073/pnas.88.14.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honzatko R B, Crawford J L, Monaco H L, Ladner J E, Edwards B F P, Evans D R, Warren S G, Wiley D C, Ladner R C, Lipscomb W N. J Mol Biol. 1982;160:219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- 16.Gerhart J C, Schachman H K. Biochemistry. 1965;4:1054–1062. doi: 10.1021/bi00882a012. [DOI] [PubMed] [Google Scholar]
- 17.Robey E A, Schachman H K. Proc Natl Acad Sci USA. 1985;82:361–365. doi: 10.1073/pnas.82.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Major J G, Wales M E, Houghton J E, Maley J A, Davidson J N, Wild J R. J Mol Evol. 1989;28:442–450. doi: 10.1007/BF02603079. [DOI] [PubMed] [Google Scholar]
- 19.Houghton J E, Bencini D A, O’Donovan G A, Wild J R. Nature (London) 1989;338:172–174. doi: 10.1038/338172a0. [DOI] [PubMed] [Google Scholar]
- 20.Gerhart J C, Schachman H K. Biochemistry. 1968;7:538–552. doi: 10.1021/bi00842a600. [DOI] [PubMed] [Google Scholar]
- 21.Collins K D, Stark G R. J Biol Chem. 1971;246:6599–6605. [PubMed] [Google Scholar]
- 22.Ladner J E, Kitchell J P, Honzatko R B, Ke H M, Volz K W, Kalb (Gilboa) A J, Ladner R C, Lipscomb W N. Proc Natl Acad Sci USA. 1982;79:3125–3128. doi: 10.1073/pnas.79.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller A W, Kuo L C. J Biol Chem. 1990;265:15023–15027. [PubMed] [Google Scholar]
- 24.Baur H, Tricot C, Stalon V, Hass D. J Biol Chem. 1990;265:14728–14731. [PubMed] [Google Scholar]
- 25.Schachman H K. Biochem Soc Trans. 1987;15:772–775. doi: 10.1042/bst0150772. [DOI] [PubMed] [Google Scholar]
- 26.Allewell N M. Annu Rev Biophys Biophys Chem. 1989;18:71–92. doi: 10.1146/annurev.bb.18.060189.000443. [DOI] [PubMed] [Google Scholar]
- 27.Kantrowitz E R, Lipscomb W N. Trends Biochem Sci. 1990;15:53–59. doi: 10.1016/0968-0004(90)90176-c. [DOI] [PubMed] [Google Scholar]
- 28.Lipscomb W N. Adv Enzymol Related Areas Mol Biol. 1994;68:67–151. doi: 10.1002/9780470123140.ch3. [DOI] [PubMed] [Google Scholar]
- 29.Howlett G J, Schachman H K. Biochemistry. 1977;16:5077–5083. doi: 10.1021/bi00642a021. [DOI] [PubMed] [Google Scholar]
- 30.Dubin S B, Cannell D S. Biochemistry. 1975;14:192–195. doi: 10.1021/bi00672a032. [DOI] [PubMed] [Google Scholar]
- 31.Hervé G, Moody M F, Tauc P, Vachette P, Jones P T. J Mol Biol. 1985;185:189–200. doi: 10.1016/0022-2836(85)90190-1. [DOI] [PubMed] [Google Scholar]
- 32.Moody M F, Vachette P, Foote A M. J Mol Biol. 1979;133:517–532. doi: 10.1016/0022-2836(79)90405-4. [DOI] [PubMed] [Google Scholar]
- 33.Krause K L, Volz K W, Lipscomb W N. Proc Natl Acad Sci USA. 1985;82:1643–1647. doi: 10.1073/pnas.82.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krause K L, Volz K W, Lipscomb W N. J Mol Biol. 1987;193:527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- 35.Ke H, Lipscomb W N, Cho Y, Honzatko R B. J Mol Biol. 1988;204:725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- 36.Gouaux J E, Stevens R C, Ke H, Lipscomb W N. Proc Natl Acad Sci USA. 1989;86:8212–8216. doi: 10.1073/pnas.86.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerhart J C, Pardee A B. J Biol Chem. 1962;237:891–896. [PubMed] [Google Scholar]
- 38.Porter R W, Modebe M O, Stark G R. J Biol Chem. 1969;244:1846–1859. [PubMed] [Google Scholar]
- 39.Schmidt P G, Stark G R, Baldeschwieler J D. J Biol Chem. 1969;244:1860–1868. [PubMed] [Google Scholar]
- 40.Collins K D, Stark G R. J Biol Chem. 1969;244:1869–1877. [PubMed] [Google Scholar]
- 41.Kirschner M W, Schachman H K. Biochemistry. 1971;10:1919–1926. doi: 10.1021/bi00786a028. [DOI] [PubMed] [Google Scholar]
- 42.Bothwell M A, Schachman H K. Proc Natl Acad Sci USA. 1974;71:3221–3225. doi: 10.1073/pnas.71.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knier B L, Allewell N M. Biochemistry. 1978;17:784–790. doi: 10.1021/bi00598a005. [DOI] [PubMed] [Google Scholar]
- 44.Vickers L P, Donovan J W, Schachman H K. J Biol Chem. 1978;253:8493–8498. [PubMed] [Google Scholar]
- 45.Lennick M, Allewell N M. Proc Natl Acad Sci USA. 1981;78:6759–6763. doi: 10.1073/pnas.78.11.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns D L, Schachman H K. J Biol Chem. 1982;257:8648–8654. [PubMed] [Google Scholar]
- 47.Mallikarachchi D, Burz D S, Allewell N M. Biochemistry. 1989;28:5386–5391. doi: 10.1021/bi00439a011. [DOI] [PubMed] [Google Scholar]
- 48.Markby D W, Zhou B-B, Schachman H K. Proc Natl Acad Sci USA. 1991;88:10568–10572. doi: 10.1073/pnas.88.23.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou B-B, Schachman H K. Protein Sci. 1993;2:103–112. doi: 10.1002/pro.5560020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterson C B, Zhou B B, Hsieh D, Creager A N, Schachman H K. Protein Sci. 1994;3:960–966. doi: 10.1002/pro.5560030611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou B-B, Waldrop G L, Lum L, Schachman H K. Protein Sci. 1994;3:967–974. doi: 10.1002/pro.5560030612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siemens Energy & Automation, Inc. (1995) astro Manual (Siemens Energy & Automation, Inc., Madison, WI).
- 53.Siemens Energy & Automation, Inc. (1990) xengen Manual (Siemens Energy & Automation, Inc., Madison, WI).
- 54.Collaborative Computational Project, Number 4. The CCP 4 Suite, Computer Programs for Protein Crystallography: Overview and Manual. Warrington, U.K.: Daresbury Lab; 1995. [Google Scholar]
- 55.Brünger, A. T. (1992) x-plor Manual (Yale Univ., New Haven, CT), Version 3.0.
- 56.Oberoi H, Trikha J, Yuan X, Allewell N M. Proteins. 1996;25:300–314. doi: 10.1002/(SICI)1097-0134(199607)25:3<300::AID-PROT3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 57.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 58.Bacon D J, Anderson W F. J Mol Graph. 1988;6:219–220. [Google Scholar]
- 59.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 60.Foote A M, Winkler F K, Moody M F. J Mol Biol. 1981;146:389–391. doi: 10.1016/0022-2836(81)90395-8. [DOI] [PubMed] [Google Scholar]
- 61.Kuo L C, Seaton B A. J Biol Chem. 1989;264:16246–16248. [PubMed] [Google Scholar]
- 62.Stebbins J W, Xu W, Kantrowitz E R. Biochemistry. 1989;28:2592–2600. doi: 10.1021/bi00432a037. [DOI] [PubMed] [Google Scholar]
- 63.Stebbins J W, Kantrowitz E R. Biochemistry. 1992;31:2328–2332. doi: 10.1021/bi00123a017. [DOI] [PubMed] [Google Scholar]
- 64.Kuo L C, Zambidis I, Caron C. Science. 1989;245:522–524. doi: 10.1126/science.2667139. [DOI] [PubMed] [Google Scholar]
- 65.Kuo L C, Lipscomb W N, Kantrowitz E R. Proc Natl Acad Sci USA. 1982;79:2250–2254. doi: 10.1073/pnas.79.7.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuo L C, Caron C M, Lee S, Herzberg W. J Mol Biol. 1990;211:271–280. doi: 10.1016/0022-2836(90)90026-I. [DOI] [PubMed] [Google Scholar]
- 67.Lee S, Shen W-H, Miller A W, Herzberg W, Kuo L C. J Mol Biol. 1990;211:255–269. doi: 10.1016/0022-2836(90)90025-H. [DOI] [PubMed] [Google Scholar]