Abstract
The capsid protein of hepatitis B virus, consisting of an “assembly” domain (residues 1–149) and an RNA-binding “protamine” domain (residues 150–183), assembles from dimers into icosahedral capsids of two different sizes. The C terminus of the assembly domain (residues 140–149) functions as a morphogenetic switch, longer C termini favoring a higher proportion of the larger capsids, it also connects the protamine domain to the capsid shell. We now have defined the location of this peptide in capsids assembled in vitro by engineering a mutant assembly domain with a single cysteine at its C terminus (residue 150), labeling it with a gold cluster and visualizing the cluster by cryo-electron microscopy. The labeled protein is unimpaired in its ability to form capsids. Our density map reveals a single undecagold cluster under each fivefold and quasi-sixfold vertex, connected to sites at either end of the undersides of the dimers. Considering the geometry of the vertices, the C termini must be more crowded at the fivefolds. Thus, a bulky C terminus would be expected to favor formation of the larger (T = 4) capsids, which have a greater proportion of quasi-sixfolds. Capsids assembled by expressing the full-length protein in Escherichia coli package bacterial RNAs in amounts equivalent to the viral pregenome. Our density map of these capsids reveals a distinct inner shell of density—the RNA. The RNA is connected to the protein shell via the C-terminal linkers and also makes contact around the dimer axes.
Hepatitis B virus (HBV) causes acute and chronic hepatitis and is an important cause of cirrhosis of the liver and hepatocellular carcinoma (for review see refs. 1 and 2). In its replication cycle, virion assembly begins with polymerization of the capsid protein (Cp) to enclose a complex of the 3.2-kilobase RNA pregenome and reverse transcriptase (3). Upon completion of capsid assembly, the pregenome is retro-transcribed (4, 5). The resulting DNA-containing nucleocapsid then acquires a lipoprotein envelope en route from the cell (6).
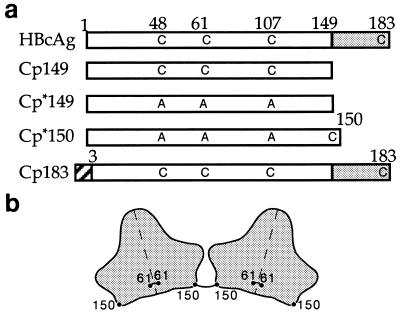
The HBV Cp, also known as core antigen or HBcAg, is a 183-residue protein with two domains: the assembly domain that forms the contiguous shell, and the protamine domain that is responsible for RNA packaging (7, 8) (Fig. 1a). Dimeric Cp (9, 10) spontaneously assembles into icosahedral capsids when expressed in heterologous systems (11) or from purified protein (10). These capsids are mixtures of particles with T = 3 and T = 4 icosahedral symmetry (10, 12, 13). T = 4 capsids predominate in vivo (14). Their structures have been determined in density maps calculated from cryo-electron micrographs (13–15). Both capsids are composed of dimers with the overall shape of an inverted capital T: the stem, 50 Å long, is bisected by the dimer interface, and its outer portion protrudes ≈25 Å outward from the contiguous shell as a spike; the crosspieces are ≈70 Å long and pack together to form the framework of the shell. Much of the substructure of this α-helical protein (10, 16) now has been delineated in cryo-microscopy-derived density maps at resolutions below 10 Å (17, 18).
Figure 1.
Schematics of Cp sequences (a) and disulfide bonding (b). (a) The constructs are referred to as Cp followed by the number of the C-terminal residue, thus, Cp149 is a 149-residue protein. In Cp*149 the cysteines found in the native sequence, residues 47, 61, and 107, are mutated to alanine. Cp*150 has arginine 150 replaced by a C-terminal cysteine. The protamine domain of Cp183, residues 150–183, is shaded. The Cp183 construct has eight N-terminal residues (hatched box) derived from a cloning vector (29). (b) Shown are silhouettes of two Cp150 dimers connected by a disulfide bond between Cys-150 residues. Each molecule also contains an intradimer disulfide bond linking its two Cys-61 residues across the dyad axis (16, 25). The position of the latter bond has still not been fully defined (see refs. 17 and 18). The position of cysteine 150 also marks the beginning of the protamine domain in reconstructions of Cp183.
Using Cp constructs progressively truncated from the C terminus, we found that residues 138–149 play an influential role in morphogenesis in vitro (15). About 95% of the capsids assembled from a 149-residue Cp (Cp149) have the T = 4 morphology; decreasing to ≈20% for Cp140 (see Fig. 1). Further truncating the protein by two residues renders it incapable of assembly (15, 19, 20). When full-length Cp183 is expressed in Escherichia coli, RNA-containing capsids are formed (11, 21), ≈90% T = 4. In our expression system, these capsids contain 25% RNA by weight (10), which corresponds to about 3,400 nucleotides for a T = 4 capsid—the same amount of RNA as the viral pregenome. However, the RNAs in the expressed capsids are bacterial in origin and small in size.
The location of the C terminus is of interest from two perspectives: (i) as an important determinant of capsid assembly; and (ii) for its role in the encapsidation and organization of the pregenome. We have addressed this question experimentally by labeling residue 150 of Cp (Fig. 1a) with an 11-gold atom cluster that has been used successfully in several previous studies (e.g., refs. 22–24). To obtain specific labeling, we used the maleimidyl derivative, which reacts with cysteines. The native assembly domain has three cysteines, at positions 48, 61, and 108. The first two can form disulfide bonds with the corresponding cysteines of the dyad-related monomer (16, 25), but are not required for capsid formation (26). Our experiments were performed with a 150-residue construct in which the three native cysteines were substituted by alanine, and an additional cysteine was appended to the assembly domain. We find that the terminal peptide of the assembly domain is at the ends of the Cp dimers; we also examined its relationship with encapsidated RNA in image reconstructions of RNA-containing capsids.
MATERIALS AND METHODS
Capsid Protein Constructs.
The Cp*149 and Cp*150 proteins (for nomenclature see Fig. 1 legend) were produced in E. coli as previously described (15) using the expression vector pET11a and bacterial strain BL21 (DE3) (27). PCR methods (28) were used to change the cysteine codons to those encoding alanine and to add a cysteine codon at the carboxy terminus (Fig. 1a). The overexpressed protein was purified as described (15), except that for Cp*150, all buffers included 5 mM DTT. Purified Cp*150 dimer was stored in 100 mM sodium bicarbonate, pH 9.5 (storage buffer) with 5 mM DTT at −80°C. RNA-containing capsids of the Cp183 construct (29) were previously characterized (10).
Preparation and Biochemical Studies of Gold-Labeled Cp*150.
Cp*150 capsids were assembled in 100 mM sodium phosphate, pH 6.9, 300 mM sodium chloride, by exchanging buffer in a PD10 column (Pharmacia). Identical results were obtained using 100 mM Hepes, pH 7.5, 300 mM sodium chloride. To prevent sulfhydryl-disulfide exchange on nonreducing SDS/PAGE, samples were alkylated with 20 mM iodoacetamide for 30 min before denaturation in SDS-containing gel-loading buffer. Capsids to be dissociated had their buffer exchanged with 100 mM sodium bicarbonate, pH 9.5, followed by addition of buffered urea to final concentrations of 3.5 M and <1 mg/ml protein. Assembly of some samples was characterized by sedimentation through 5–30% sucrose gradients (15).
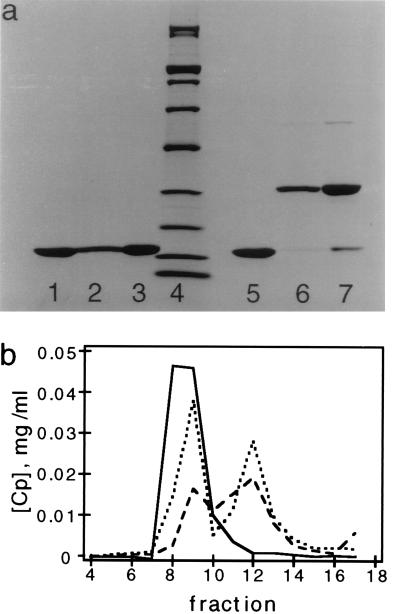
Immediately before labeling, Cp*150 was incubated for 1 hr in fresh 50 mM DTT. The DTT was separated from the protein, and the buffer exchanged for 100 mM sodium phosphate, pH 6.9, using a PD10 column. The concentration of Cp*150 was determined by absorbance, using ɛ280 = 29,500 M−1⋅cm−1. In a typical reaction, 12 nmols of reduced Cp*150 were mixed with 6 nmols of monomaleimidyl-undecagold (Au11) (30) (Nanoprobes, Stony Brook, NY) to a final volume of 300 μl and incubated for 24 hr at 4°C. The reactants were separated on a Sephacryl S-200 HR column (1.4 cm diameter × 6 cm), eluted in 0.5-ml fractions with 100 mM sodium phosphate, pH 6.9. Capsids eluted in the void volume at 3–4 ml, free dimers at 5–6 ml, and free Au11 at about 8 ml (Fig. 2).
Figure 2.
Polymerized Cp*150 forms disulfide bonded dimers. (a) SDS/PAGE of Cp*150 from isolated capsids. Samples of Cp*150 were subjected to electrophoresis under reducing (lanes 1–3) and nonreducing conditions (lanes 5–7). Samples of Cp*150 polymerized at pH 9.5 (lanes 1 and 7) and at pH 7.5 (lanes 2 and 6). Lanes 3 and 5 are Cp*149, which has no cysteines. Molecular mass standards (200, 116, 97, 66, 45, 31, 21.5, 14.4, and 6.5 kDa) are in lane 4. (b) Size exclusion chromatography of capsid protein after exposure to 3.5 M urea. Samples are oxidized Cp*150 (solid line), Cp*150 with 130 mM DTT (dashed line), and Cp*149 (dotted line).
Fractions were assayed for protein and Au11 content by absorbance spectroscopy. Au11 has absorbance maxima at 420 nm and 310 nm with extinction coefficients of 47,000 and 135,000 M−1⋅cm−1, respectively. The extinction coefficient for free Au11 is 178,000 M−1·cm−1 at 280 nm. The protein concentration in the fractions was determined using the same ɛ280 as unlabeled protein after subtracting the calculated Au11 absorbance.
Cryo-Electron Microscopy and Image Reconstruction.
Capsid samples, adjusted to 1.5–2.5 mg/ml protein, were frozen in thin films of vitrified ice on holey carbon films (31, 32). Micrographs were recorded at a magnification of ×38,000 on a Philips CM20-FEG (Philips Electronic Instruments, Mahwah, NJ) fitted with a Gatan 626 cryoholder (Gatan, Pleasanton, CA) and operating at 120 keV. Pairs of micrographs at different defocus values were recorded using low-dose techniques. For gold-labeled capsids (Cp*150-Au11), the focal pair analyzed had first contrast transfer function zeros at spacings of (23.5 Å)−1 and (18.0 Å)−1, respectively; for RNA-filled Cp183 capsids, the zeros were at spacings of (25.5 Å)−1 and (22.0 Å)−1.
Micrographs were digitized on a Perkin–Elmer 1010MG scanner. Images of individual particles were extracted using a semiautomated procedure (33). Focal pairs of images were combined, and three-dimensional density maps were calculated as described (15, 34). Particle orientations were determined with the program empft (35) using a 17-Å resolution density map of an empty T = 4 capsid as a starting model (15). Reconstructions of labeled Cp*150 (Cp*150-Au11) and Cp183 were calculated with data sets of 86 and 151 pairs of capsids, respectively. The resolution of the final maps were determined using the Fourier ring correlation method (33). The Cp147 structure shown for comparison with Cp*150-Au11 and Cp183 capsids was calculated from the 600-particle data set previously used for a 9-Å resolution reconstruction (17) but limited here to 17-Å resolution. The surface shaded maps were contoured to enclose a volume that was 100% of that expected for the protein mass.
RESULTS AND DISCUSSION
Capsids Assembled from Cp*150 and Cp*150-Au11.
Purified Cp*149 and Cp*150 assemble into capsids under the same conditions as other Cp constructs (10, 15), with or without DTT. These capsids were indistinguishable by negative staining electron microscopy and sedimentation on sucrose gradients (data not shown). When reduced CP*150 capsids were stored without DTT for 2 days, >90% of the protein oxidized to form disulfide-bonded dimers (Fig. 2a). These bonds stabilize the quaternary structure of the capsid, as attested by the observation that oxidized Cp*150 capsids—unlike CP*149 capsids or reduced Cp*150 capsids—are resistant to dissociation by 3.5 M urea (Fig. 2b). Knowledge of the location of residue 150 (see below) indicates that this disulfide bond links two dimers (Fig. 1b) and is distinct from the intradimeric disulfide observed in Cp proteins with native cysteines (16, 25).
Generally, when Cp proteins are stored in a low ionic strength, high pH buffer they do not polymerize (10). However, when stored in this buffer without DTT, Cp*150 dimers assemble into capsids, as determined by negative stain electron microscopy and analytical ultracentrifugation. A high proportion of the protein in these capsids is disulfide-bonded (Fig. 2a). These data show that disulfide bond formation by Cp*150 can promote capsid assembly. Without disulfide formation, higher-order structures do not accumulate in storage buffer, i.e., the rate for dissociation is greater than the rate of association. Formation of these disulfide bonds stabilizes complexes against dissociation. Thus, under these conditions, Cp polymerization appears to involve an equilibrium between subunits, assembly intermediates, and capsids (36). We also note that, in capsids, the cysteine 150 residues from adjacent subunits must be close enough to one another to form a covalent bond, a distance of 4.6–7.4 Å between α carbons (37).
Undecagold Labeling of Cp*150.
Assembled capsids of reduced Cp*150 did not react with Au11. Apparently, the C-terminal cysteine is inaccessible (see below). Accordingly, Cp*150 was labeled with Au11 at neutral pH and low ionic strength, where the sample is mainly free dimers (10). After reaction with Au11, >85% of the protein eluted in the void volume of a Sephacryl S-200 column, indicating that the protein was polymerized. Thus it appears that modification of Cp*150 with Au11 promotes polymerization. The Au11 has a single reactive maleimide group and cannot crosslink proteins. Though the molar ratio of Au11 to protein in the labeling reaction was varied from 1:1 to 1:5, a maximum of one label for every 5.3 Cp*150 monomers was observed, corresponding to about 45 Au11 clusters per T = 4 capsid.
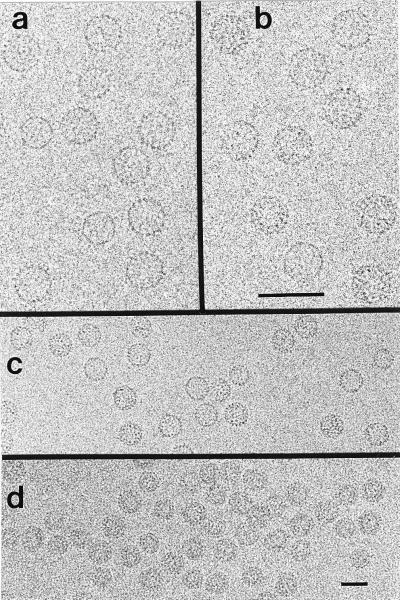
Cryo-electron micrographs of labeled Cp*150 show a mixture of T = 3 and T = 4 capsids, typical for assembled Cp (Fig. 3a). Samples of labeled capsids were very similar to unlabeled ones, except that the proportion of T = 4 capsids was greater (91% vs. 75% T = 4, respectively). In micrographs taken close to focus (i.e. <1 μm defocus)—conditions that suppress the overall contrast of capsids but should enhance the visibility of the undecagold particles—individual clusters appeared to be marginally visible (cf. Fig. 3 a and b).
Figure 3.
Cryo-electron micrographs of unlabeled Cp149 capsids (a), Au11-labeled Cp*150 capsids (b and c), and RNA-filled Cp183 capsids (d). Views of unlabeled (a) and labeled (b) capsids are from closer to focus micrographs to facilitate comparison. Encapsidated RNA is visible as a dark ring within individual capsids in d. The first contrast zero function zeros are at spacings of 18.0, 18.0, 23.5, and 25.5 (Å)−1, respectively. (Bars = 50 nm.)
Localization of the C Terminus by Image Reconstruction.
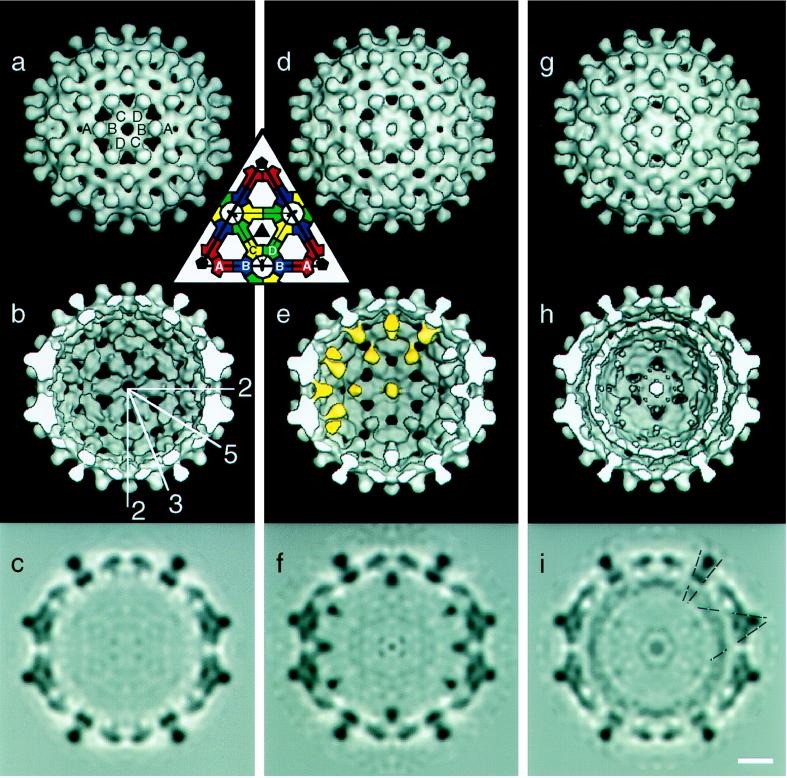
At a resolution of ≈20 Å, the outer surface of the Au11-labeled Cp*150 capsid is indistinguishable from those of unlabeled Cp147 and Cp183 capsids, (cf. Fig. 4 Top). In contrast, the interior view of the labeled Cp*150 capsid shows well defined densities attributable to the Au11 clusters directly beneath all fivefold and quasi-sixfold vertices (Fig. 4 e and f). These masses are ≈20 Å in diameter. Visualized in a central section through the map, the peak label-associated density is at least as strong as that of protein in the contiguous capsid (Fig. 4f): in comparison, only vestigial noise is seen in the corresponding region of the unlabeled capsid (cf. Fig. 4 c and f). Au11 is a ≈20 Å diameter molecule with an 8 Å diameter gold cluster at its center surrounded by an organic shell (30). Moreover, the density of gold is 19 g/cm3 as compared with 1.3 for protein. As visualized, the label is larger and more diffuse than these numbers would suggest. However, the clusters visualized represent averages over the subunits around each fivefold or quasi-sixfold vertex and over all particles included in the reconstruction. Taking into account the measured level of labeling (≈45 clusters per T = 4 capsid) and the fact that the densities seen under the fivefold and quasi-sixfold axes are very similar, it follows that only one subunit per vertex is labeled. The apparent density of the label is broadened and further attenuated by the limited resolution (20 Å) of the reconstruction.
Figure 4.
Image reconstructions of T = 4 HBV capsids: empty Cp147 capsids at 17-Å resolution (a–c), C-terminally labeled Cp*150 capsids at 20 Å (d–f), and Cp183 capsids with RNA at 18.5 Å (g–i). Views are down an icosahedral twofold axis. (Inset) The arrangement of A, B, C, and D quasi-equivalent subunits on one icosahedral facet; the same labeling is used in a. The positions of some icosahedral symmetry axes, in the plane of the page, are shown in b. Some of the gold clusters in e are highlighted. Putative connections between capsid and RNA are identified by dashed lines in i. Surface shaded representations, external (a, d, and g) and cutaway (b, e, and h), are contoured so that 100% of the expected protein volume is enclosed. Gray scale maps (c, f, and i) are shown of the central section of each reconstruction. (Bar = 50 Å.)
As noted above, the 42 Au11 clusters observed in the reconstruction (T = 4 icosahedra have 12 fivefold and 30 quasi-sixfold vertices) agrees well with the 45 Au11 clusters per T = 4 capsid determined spectroscopically. Though less than 20% of the protein is labeled, there is one Au11 per vertex. We infer that labeling of one subunit enhances its propensity for assembly, and its label sterically excludes other labeled subunits from assembling into the ring forming around a vertex. The lack of Au11 reactivity for assembled Cp*150 particles most likely reflects the inaccessibility of the C terminus due to the large diameter of the label. The largest of the holes fenestrating the capsids is 24 Å by 18 Å (15) implying that it would be difficult for the 20 Å diameter Au11 molecule to diffuse through.
The Au11 labels are clearly bound to the ends of the Cp*150 dimers (Fig. 4e). A connection between Cp*150 and Au11 is observed for B subunits. This connection appears to emanate from the small protrusion first identified in reconstructions at 17 Å resolution (15). Our visualization of the arm of connecting density indicates that at least some the C-terminal residues of Cp*150 are icosahedrally ordered (Fig. 4c). The corresponding connections from Au11 to A, C, and D subunits are much weaker. The lack of connectivity to C and D subunits is due to a real preference for Au11-labeled subunits at the B position. This preference reflects the quasi-equivalence around HBV T = 4 quasi-sixfold vertices resulting in distinct differences between the environment of the B subunit and that of the C and D subunits (15). The weak connectivity to C and D subunits is unlike the situation observed for A subunits where the density has been decreased by fivefold averaging (see below).
Why is the resolution of Cp*150-Au11 limited to 20 Å? Part of the answer is that because there is only one Au11 per vertex, labeled capsids are no longer strictly icosahedral. The reconstruction algorithm that we use assumes and imposes icosahedral symmetry (15, 17, 34). For example, only one of the five A subunits actually connects to a given Au11 cluster near a fivefold vertex; after averaging, the connecting density will be 20% of the density of nearby protein. In calculating a reconstruction, the handedness of each particle must be determined (38). For a T = 4 capsid the differences between the left-handed and right-handed enantiomorphs are necessarily subtle (12) and distinguishing between them is further hampered by the asymmetric distribution of Au11 clusters. The likely inclusion of some particles with the wrong hand would limit the resolution of the reconstruction. Nevertheless, our conclusions as to cluster location are unaffected.
Structure of RNA-Containing Capsids.
In micrographs of Cp183 capsids, the encapsidated RNA is evident as a smaller ring within many of the particles (Fig. 3d). The observed double ring implies that encapsidated RNA forms an inner shell. If capsids were packed full of RNA, one would expect to see them as dense disks.
A density map of RNA containing particles was calculated to 18.5-Å resolution. Like the Au11-labeled particles, the external features of the T = 4 Cp183 capsid are as expected for a reconstruction at this resolution (Fig. 4g). In thin sections (e.g., Fig. 4i), an RNA shell is seen to be ≈30 Å thick and its average density is lower than that of the protein shell, implying that the distribution of the RNA is somewhat variable from particle to particle and between symmetry-related sites on a given particle. The highest density of RNA is located under fivefold and quasi-sixfold vertices and beneath the sides of the AB and CD spikes. There are also indications of low-density connections between protein and RNA (Fig. 4i, dashed lines).
CONCLUSIONS AND IMPLICATIONS
The Site Occupied by the C Terminus of the Assembly Domain.
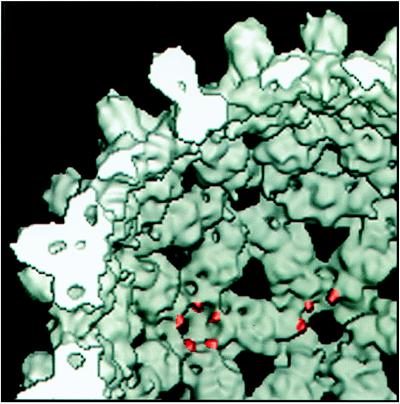
Residue 150 has been localized by image reconstruction of gold-labeled capsids. The appearance of the undecagold cluster in our map (Fig. 4 e and f) is broadened on account of the limited resolution (20 Å). Nevertheless, the position of its center of mass is defined to within a few Å units in three dimensions. For the B subunits, the connecting linker is sufficiently well ordered to be visualized directly (Fig. 4 e and f). We conclude that the labeled residue, cysteine 150, lies 10–15 Å along this linker from the cluster center of mass and, consequently, its location is also defined to within a few Å units (Fig. 5). We infer that the corresponding residues on A, C, and D subunits are similarly positioned. Thus, residue 150 has been localized to small protrusions at either end of the Cp dimer (15). This feature, as defined in our 9-Å resolution map (17), is shown and the C-terminal sites marked (Fig. 5).
Figure 5.
The C terminus of the assembly domain in a 9-Å resolution reconstruction. Some C termini are highlighted. The map is contoured at 100% of expected protein density, a lower contour level than used to trace the protein chain (17).
Recently, image reconstructions of HBV Cp have been determined to unprecedented resolution (17, 18). The analysis of Böttcher et al. (18) tentatively assigned the C terminus to positions near the ends of the dimer and near the outer surface (figure 4 of ref. 18). A similar assignment was made in the model of Bringas (39). Our experimental localization of the C terminus confirms that it resides near the end of the arm, but on its underside.
Control of Assembly.
Our earlier work revealed that residues 140–149 serve as a morphogenetic switch, longer proteins favoring T = 4 and shorter ones, T = 3 geometry (15). This behavior now can be explained in light of the position that the switch occupies. The C-termini of adjacent subunits crowd one another around fivefold and quasi-sixfold vertices. Viewed from the inside, the crowding is more acute around the fivefold vertices, suggesting that the more open geometry of the T = 4 capsid would be preferred for proteins with a bulkier C terminus (see also ref. 18). This argument is consistent with the greater proportion of T = 4 capsids with gold-labeled versus unlabeled capsids (this work).
Other observations imply that the C termini also may influence assembly in more subtle ways. For instance, binding Au11 to Cp*150 induces assembly, though Au11 cannot crosslink subunits nor, because of its organic shell, coordinate C-terminal cysteines. Binding Au11 may cause small changes in the molecular surfaces, near the C termini, that dock together when dimers polymerize and, in this way, stimulate the assembly process. If the binding of the reverse transcriptase-RNA complex to the Cp (5) were to transmit a similar effect, this could enhance the cooperativity of nucleocapsid assembly.
Packaging of RNA and its Interactions with the Capsid.
The protamine domain (residues 150–183) is required for packaging RNA (8, 21) and should greatly influence the organization of encapsidated RNA (cf., ref. 40). This expectation is confirmed in our density map (Fig. 4i), in which RNA-associated density is greatest under fivefold and quasi-sixfold vertices, and presumably represents the binding of RNA to protamine domains, secured to the contiguous shell via the C termini of the assembly domains. Otherwise, the RNA is not closely associated with the capsid inner surface except at sites underlying both AB and CD dimer interfaces (Fig. 4i). This latter contact is consistent with that observed by Kenney et al. (14) in their reconstruction of duck hepatitis virus capsids. This site on the assembly domain with an apparent affinity for nucleic acid may coincide with that previously inferred by biochemical methods (41).
Compared with many other viruses, the packing density of RNA in HBV capsids is decidedly loose (cf., ref. 31), the RNA being confined to an inner shell of density. We make the caveat that the organization of the native RNA pregenome may differ from that of the bacterially derived RNA in the virus-like particles that we have studied. However, we note that HBV capsids isolated from human liver also appear to be hollow in negative stain electron micrographs (42). A similar configuration of encapsidated nucleic acid has been documented in image reconstructions of cauliflower mosaic virus (43), a pararetrovirus, that—like HBV—has a DNA genome with an RNA replicative intermediate (44).
A General Method for Mapping Polypeptide Chains at Moderate Resolution.
As reconstructions calculated from cryo-electron micrographs continue to improve in resolution to 10 Å and beyond (17, 18, 45), the need arises to be able to determine the overall fold of polypeptide chain(s) within them. In general, higher resolution is required for unambiguous chain tracing by the traditional x-ray crystallographic method of recognizing bulky side-chains and correlating this information with the path followed by the backbone. We suggest that the approach followed in this study—of incorporating cysteines at defined sites by site-specific mutagenesis, labeling them with gold clusters, and determining their locations by cryo-electron microscopy—may prove to be useful for this purpose.
Acknowledgments
We acknowledge the superb technical contributions of Mr. Joshua Kaufman and Mr. Ira Palmer in the preparation of genetic constructs and purification of protein. We thank Drs. Frank Booy, Benes Trus, David Belnap, and Jim Hainfeld for helpful discussions and Tim Baker for software. A.Z. was supported by a National Research Council-National Institute of Arthritis and Musculoskeletal and Skin Diseases fellowship.
ABBREVIATIONS
- Au11
monomaleimidyl undecagold
- Cp
capsid protein
- HBV
hepatitis B virus
References
- 1.Hollinger F B. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Philadelphia: Lippincott; 1996. pp. 2738–2808. [Google Scholar]
- 2.Nassal M, Schaller H. Trends Microbiol. 1993;1:221–228. doi: 10.1016/0966-842x(93)90136-f. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Junker-Niepmann M, Schaller H. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Summers J, Mason W S. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Schaller H. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerelsaikhan T, Tavis J E, Bruss V. J Virol. 1996;70:4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petit M A, Pilot J. J Virol. 1985;53:543–551. doi: 10.1128/jvi.53.2.543-551.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nassal M. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou S, Standring D N. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wingfield P T, Stahl S J, Williams R W, Steven A C. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 11.Cohen B J, Richmond J E. Nature (London) 1982;296:677–679. doi: 10.1038/296677a0. [DOI] [PubMed] [Google Scholar]
- 12.Caspar, D. L. D. & Klug, A. (1962) Cold Spring Harbor Symp. Quant. Biol, 1–24. [DOI] [PubMed]
- 13.Crowther R A, Kiselev N A, Böttcher B, Berriman J A, Borisova G P, Ose V, Pumpens P. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 14.Kenney J M, von Bonsdorff C H, Nassal M, Fuller S D. Structure. 1995;3:1009–1019. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 15.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 16.Zheng J, Schodel F, Peterson D L. J Biol Chem. 1992;267:9422–9429. [PubMed] [Google Scholar]
- 17.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Nature (London) 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 18.Böttcher B, Wynne S A, Crowther R A. Nature (London) 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 19.Beames B, Lanford R E. Virology. 1993;194:597–607. doi: 10.1006/viro.1993.1299. [DOI] [PubMed] [Google Scholar]
- 20.Birnbaum F, Nassal M. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasek M, Goto T, Gilbert W, Zink B, Schaller H, MacKay P, Leadbetter G, Murray K. Nature (London) 1979;282:575–579. doi: 10.1038/282575a0. [DOI] [PubMed] [Google Scholar]
- 22.Milligan R A, Whittaker M, Safer D. Nature (London) 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- 23.Crum J, Gruys K J, Frey T G. Biochemistry. 1994;33:13719–13726. doi: 10.1021/bi00250a024. [DOI] [PubMed] [Google Scholar]
- 24.Hainfeld J F. Science. 1987;236:450–453. doi: 10.1126/science.3563522. [DOI] [PubMed] [Google Scholar]
- 25.Nassal M, Rieger A, Steinau O. J Mol Biol. 1992;225:1013–1025. doi: 10.1016/0022-2836(92)90101-o. [DOI] [PubMed] [Google Scholar]
- 26.Nassal M. Virology. 1992;190:499–505. doi: 10.1016/0042-6822(92)91242-m. [DOI] [PubMed] [Google Scholar]
- 27.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahl S, MacKay P, Magazin M, Bruce S A, Murray K. Proc Natl Acad Sci USA. 1982;79:1606–1610. doi: 10.1073/pnas.79.5.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartlett P A, Bauer B, Singer S J. J Am Chem Soc. 1978;100:5085–5092. [Google Scholar]
- 31.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booy F P. In: Viral Fusion Mechanisms. Bentz J, editor. Boca Raton, FL: CRC; 1993. pp. 21–54. [Google Scholar]
- 33.Conway J F, Trus B L, Booy F P, Newcomb W W, Brown J C, Steven A C. J Struct Biol. 1993;111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 34.Crowther R A. Philos Trans R Soc London Ser B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 35.Baker T S, Cheng R H. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 36.Zlotnick A. J Mol Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 37.Thornton J M. J Mol Biol. 1981;151:261–287. doi: 10.1016/0022-2836(81)90515-5. [DOI] [PubMed] [Google Scholar]
- 38.Baker T S, Drak J, Bina M. Biophys J. 1989;55:243–253. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bringas R. J Struct Biol. 1997;118:189–196. doi: 10.1006/jsbi.1997.3846. [DOI] [PubMed] [Google Scholar]
- 40.Cheng R H, Reddy V S, Olson N H, Fisher A, J, Baker T S, Johnson J E. Structure. 1994;2:271–282. doi: 10.1016/s0969-2126(00)00029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuda K, Satoh S, Ohori H. J Virol. 1988;62:3517–3521. doi: 10.1128/jvi.62.9.3517-3521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onodera S, Ohori H, Yamaki M, Ishid N. J Med Virol. 1982;10:147–155. doi: 10.1002/jmv.1890100209. [DOI] [PubMed] [Google Scholar]
- 43.Cheng R H, Olson N H, Baker T S. Virology. 1992;186:655–668. doi: 10.1016/0042-6822(92)90032-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mesnard J M, Carriere C. Virology. 1995;213:1–6. doi: 10.1006/viro.1995.1540. [DOI] [PubMed] [Google Scholar]
- 45.Trus B L, Roden R B S, Greenstone H L, Vrhel M, Schiller J T, Booy F P. Nat Struct Biol. 1997;4:411–418. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]