Abstract
We describe the identification of Neuregulin-3 (NRG3), a novel protein that is structurally related to the neuregulins (NRG1). The NRG1/neuregulins are a diverse family of proteins that arise by alternative splicing from a single gene. These proteins play an important role in controlling the growth and differentiation of glial, epithelial, and muscle cells. The biological effects of NRG1 are mediated by receptor tyrosine kinases ErbB2, ErbB3, and ErbB4. However, genetic studies have suggested that the activity of ErbB4 may also be regulated in the central nervous system by a ligand distinct from NRG1. NRG3 is predicted to contain an extracellular domain with an epidermal growth factor (EGF) motif, a transmembrane domain, and a large cytoplasmic domain. We show that the EGF-like domain of NRG3 binds to the extracellular domain of ErbB4 in vitro. Moreover, NRG3 binds to ErbB4 expressed on cells and stimulates tyrosine phosphorylation of this receptor. The expression of NRG3 is highly restricted to the developing and adult nervous system. These data suggest that NRG3 is a novel, neural-enriched ligand for ErbB4.
The neuregulins (NRG1, also called heregulin, NDF, GGF, and ARIA) belong to a family of membrane-bound or secreted proteins produced by neurons and mesenchymal cells, with multiple effects on a wide range of cell types (1). NRG1 was purified from human breast tumor cells on the basis of its ability to stimulate phosphorylation of ErbB2/Neu (2, 3). NRG1 has also been identified as an acetylcholine receptor-inducing activity (4) and as glial growth factors (5). The NRG1 gene encodes a large number of alternatively spliced transcripts, most of which encode integral membrane proteins containing an extracellular epidermal growth factor (EGF)-like domain. The EGF-like domain is sufficient to bind NRG1 receptors and stimulate cellular responses (2). Although NRG1 was initially proposed to be the ligand for the receptor tyrosine kinase ErbB2, further studies have demonstrated that activation of ErbB2 frequently occurred as a result of NRG1 binding to ErbB3 (6) or ErbB4 (7, 8).
Recent studies have begun to highlight the roles of NRG1, ErbB2, and ErbB4 in the development of the heart. Mice lacking ErbB4, ErbB2, or NRG1 die during mid-embryogenesis [embryonic day (E) 10.5] from the aborted development of myocardial trabeculae in the ventricle (9–11). These results are consistent with the view that NRG1, expressed in the endocardium, is an important ligand required for the activation of ErbB2 and ErbB4 in the myocardium. Interestingly, these same studies suggest that NRG1 and ErbB2 may play a different role than ErbB4 in the development of the hindbrain. NRG1 is expressed in the neuroepithelium and cells arising from rhombomeres 2, 4, and 6, whereas ErbB4 is expressed in rhombomeres 3 and 5 (10). NRG1 and ErbB2 knockout mice exhibit a loss of cells and axons of the cranial sensory ganglia. In contrast, ErbB4-deficient mice do not exhibit a loss of cellularity in the cranial ganglia. Rather, the organization, spacing, and pattern of innervation of these ganglia to and from the central nervous system is disrupted (10). One possible reason for this striking difference in hindbrain phenotypes of NRG1 and ErbB4 knockout mice is that an additional ligand(s) distinct from NRG1 may be recognized by ErbB4 in the central nervous system (10). A newly isolated family of NRG1-like ligands, termed NRG2, has recently been described (12, 13).
In this report, we describe the cloning and characterization of a novel NRG1-related ligand (named NRG3), which displays an expression pattern with enriched signals in neural tissues. We also provide evidence that NRG3 is a ligand for ErbB4.
MATERIALS AND METHODS
Molecular Cloning.
The National Center for Biotechnology Information (NCBI) DNA database was searched for sequences homologous to NRG1 family members using blast. We identified a cDNA clone in the expressed sequence tag database that was predicted to encode a polypeptide bearing 62% amino acid identity to amino acids 232–316 of heregulin-β1. A 50-base single-stranded oligonucleotide probe (5′-TGGTAAAAGCTACAGTCTCAAAGCATCCAGCACAATGGCAAAGTCAGAGA-3′) based on this expressed sequence tag was used to screen 1.5 × 106 plaques from a λgt10 cDNA library prepared from RNA isolated from human fetal brain (HL3003a, CLONTECH). Conditions for plating libraries, hybridizing, and washing filters were as described (14). Inserts from nine positive plaques were characterized and the sequences of both strands of the largest clones determined. A partial cDNA of human NRG3 was obtained. To clone murine NRG3 cDNAs, two degenerate primers were designed to encode regions of human NRG3 proximal to transmembrane domain (NDGECFVI and EFMesEEVY, respectively). A mouse brain cDNA library (ML1042a, CLONTECH) was screened, and a clone (C5a), which contained a partial murine NRG3 cDNA, was obtained. Using a probe derived from the C5a sequence, two additional mouse brain cDNA libraries (ML1034h, CLONTECH and 936309, Stratagene) were screened. Two overlapping clones, SWAJ-3 and ZAP-1, were sequenced in both directions and found to cover the entire ORF. The assembled murine NRG3 cDNA encodes an ORF of 2,139 bp. Additional 5′ sequence of human NRG3 was obtained by anchored PCR using RNA from human hippocampus (CLONTECH).
Northern Blot Analysis.
A multi-tissue RNA blot containing 2 μg each of poly(A)+ RNA from human tissues were purchased from CLONTECH. The hybridization probe was generated by PCR amplification of the region of human NRG3 coding for amino acids 394–536. DNA probes were labeled with [α-32P]dCTP by random priming (Promega). RNA blot was hybridized with 50% formamide, 5 × SSC, 50 mM potassium phosphate (pH 7.0), 5 × Denhardt’s solution, 10% dextran sulfate at 42°C for 20 hr. The blot was washed with 0.1 × SSC/0.1% SDS at 50°C for 30 min and exposed in a phosphorimager.
In Situ Hybridization Analysis.
Formalin-fixed, paraffin-embedded mouse embryos (E13, E14, and E16), and glutaraldehyde-fixed, paraffin-embedded or paraformaldehyde-fixed, frozen adult mouse brain, ovary, jejunum, kidney, adrenal, lung, stomach, spleen, skeletal muscle, liver, and colon were sectioned and processed for in situ hybridization by a modification of the method described (15). [33P]UTP-labeled sense and antisense riboprobes were generated from PCR products of a fragment of cDNA encoding amino acids 292–482 of murine NRG3.
Expression and Purification of NRG3EGF Fusion Protein in Mammalian Cells.
A secreted, epitope-tagged version of the EGF-like domain of murine NRG3284–344 was constructed by linking the coding sequence for the gD signal sequence and epitope tag (16) to the sequences encoding amino acids 284–344 of murine NRG3, which is followed by the coding sequences of the Fc portion of human IgG. The plasmid (containing the N-terminal gD tag NRG3284–344 and the C-terminal Fc in pSAR.SD5 vector) is designated NRG3EGF.Fc. The amino acid sequence of the portion of murine NRG3 used in this construct is identical to that of human NRG3. The NRG3EGF.Fc expression plasmid was transfected using LipofectAMINE (GIBCO/BRL) into DHFR− Chinese hamster ovary cells (CHO/DP12). Selected clones in glycine/hypoxanthine/thymidine minus medium were pooled and expanded. Conditioned media from these cells were collected and the recombinant protein purified by a HiTrap protein A affinity column (Pharmacia). NRG3EGF.H6 fusion protein was produced in the same system and purified through a cobalt affinity column by R. Vandlen (Genentech). Protein concentration was determined by Bio-Rad protein assay.
Generation of K562erbB Cell Lines.
Stable K562 cell lines that expressed human ErbB2, ErbB3, or ErbB4 were generated by standard methods. Receptor expression was confirmed by Western blot analysis. Phorbol ester stimulation was found to significantly enhance receptor expression in both the ErbB3 and ErbB4 transfectants (unpublished results). Therefore, the K562 stable lines were cultured in medium containing 10 ng/ml phorbol 12-myristate 13-acetate overnight prior to use.
Fluorescence-Activated Cell Sorter (FACS) Analysis.
For each binding reaction, 5 × 105 K562 cells were suspended in PBS/2% BSA at 4°C for 30 min followed by incubation with 5 μg of NRG3EGF.Fc (Mr 90 kDa) in a volume of 0.25 ml on ice for 60 min. One microgram of primary antibody (anti-gD or anti-ErbB receptor) and secondary phycoerythrin-conjugated (Caltag, South San Francisco, CA, goat anti-mouse, 1:100 dilution) antibodies were added sequentially with 30–60 min incubation time and extensive washes before each addition. FACS analyses were performed on a Becton Dickinson FACS instrument. Anti-gD (5B6), anti-ErbB2 (4D5), anti-ErbB3 (2F9), and anti-ErbB4 (3B9) mAbs were obtained from the Monoclonal Antibody Group (Genentech).
125I-NRG3EGF.Fc Binding Assay.
Purified NRG3EGF.Fc was iodinated using the lactoperoxidase method as described (6). The average specific activity of the radiolabeled protein was 300 μCi/μg (1 Ci = 37 GBq). The displacement binding assays were performed in Maxisorp C 96-wells (Nunc). Goat anti-human antibody (Boehringer Mannheim) was coated on the plate at a concentration of 0.2 μg per well in 100 μl of 50 mM sodium carbonate buffer (pH 9.6) at 4°C, overnight. The plate was blocked by 1% BSA in TBST buffer (25 mM Tris, pH 7.5/150 mM NaCl/0.02% Tween 20) for 30 min at room temperature. Receptor Fc fusion proteins (e.g., ErbB4.Fc) were added at 15 ng per well in 1% BSA/TBST and incubated for 1.5 hr at room temperature. To prevent radiolabeled protein from interacting with residual goat anti-human antibodies, 1 μM of a humanized mAb (rhuMAB HER2) (17) was added to the plate for 20 min (and was included in the subsequent binding reaction). A competitive binding assay was then initiated by the addition of varying concentration of unlabeled NRG3EGF.Fc or NRG1EGF (Escherichia coli expressed without Fc), along with 80 pM (200,000 cpm) of 125I-NRG3EGF.Fc. The final incubation volume was 100 μl in binding buffer (F-12/DMEM medium/50 mM Hepes, pH 7.5/2% BSA) and the reaction was allowed to proceed at room temperature for 1.5 hr. The unbound material was washed by TBST extensively, and the bound radioactivity was counted on a Beckman IsoData gamma counter. Data were analyzed using a nonlinear regression program. NRG1EGF is the EGF domain of NRG1, corresponding to amino acids 177–244 of the heregulin-β1 isoform (2) (obtained from J. A. Lofgren, Genentech). ErbB4.Fc fusion proteins were also from J. A. Lofgren (Genentech).
Tyrosine Phosphorylation Assay.
K562erbB4 cells or MDA-MB-453 cells were cultured in medium lacking serum for 12 hr and then stimulated with NRG3EGF.Fc, NRG3EGF.H6, or NRG1EGF. Cells were lysed with lysis buffer (20 mM Tris, pH 7.5/100 mM NaCl/30 mM NaF/2 mM EDTA/2 mM EGTA/0.1% SDS/1% Triton X-100/2 mM sodium vanadate/2 mM sodium molybdate/2 mM of phenylmethylsulfonyl fluoride). After clearing the cell lysate by centrifugation, 1 μg of anti-ErbB4 mAb (C-18, Santa Cruz Biotechnology) was added together with 20 μl of protein A-agarose slurry (Sigma). Immunoprecipitation was performed at 4°C overnight, and complexes were collected by centrifugation and washed three times with 1 ml of lysis buffer. Proteins were separated by reducing SDS/PAGE on Novex 4%–12% minigels, transferred to nitrocellulose, and blots were probed with peroxidase conjugated anti-phosphotyrosine antibody (Transduction Laboratories, Lexington, KY). The blot was stripped and reprobed with anti-ErbB4 antibody followed by peroxidase-conjugated goat anti-rabbit IgG antibody (Sigma) to visualize ErbB4 proteins.
RESULTS
NRG3 Gene Has Similar and Distinct Features of Neuregulins (NRG1).
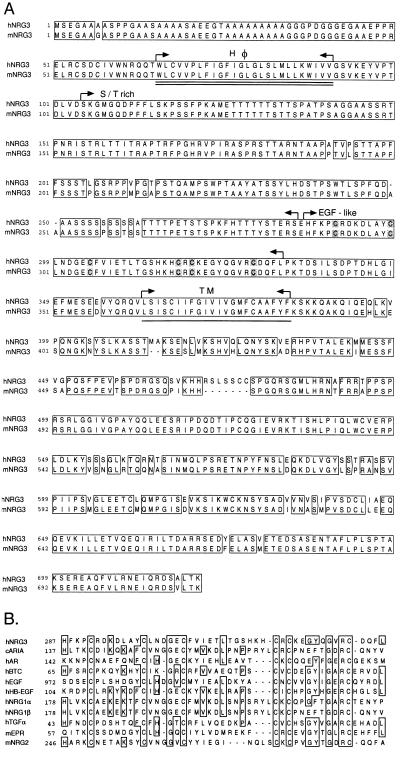
The NCBI database of expressed sequence tags was searched for sequences related to NRG1 family members. A cDNA clone was identified that was predicted to encode a polypeptide bearing 62% amino acid identity to amino acids 232–316 of human heregulin-β1. DNA probes based on these sequences were used to isolate human and then murine NRG3 cDNAs. The cDNAs of human and murine NRG3 contained ORFs encoding proteins of 720 and 713 amino acids, respectively, with predicted Mr of 77,901 Da for human NRG3 and 77,370 Da for murine NRG3 (Fig. 1A). The two species of NRG3 are 93% identical in amino acid sequence.
Figure 1.
cDNA sequence analysis of NRG3. (A) Amino acid sequences of human and murine NRG3 (hNRG3 and mNRG3, respectively). The EGF-like domain, the N-terminal hydrophobic segment (double underline), the serine/threonine-rich portion, and a predicted transmembrane domain (single underline) are highlighted. (B) Sequence alignment analysis for the EGF-like domains of human NRG3 (hNRG3), chicken ARIA (cARIA) (4), human amphiregulin (hAR) (18), human betacellulin (hBTC) (19), human EGF (hEGF) (20), human heparin-binding EGF-like growth factor (hHB-EGF) (21), human neuregulin-1α (hNRG1α), human neuregulin-1β (hNRG1β) (2), mouse NRG2 (mNRG2) (12, 13), human TGF-α (hTGF-α) (43), and mouse epiregulin (mEPR) (22). The sequences were analyzed using Sequence Analysis Programs (Genentech).
Analysis of the amino acid sequence of human NRG3 revealed that it contained homology to NRG1 family members [i.e., 23% and 19% sequence identity to sensory and motor neuron-derived factor (SMDF) (23) and heregulin-β1 (2), respectively]. Phylogenic prediction and fasta/blast analysis also revealed a close relationship between NRG3 and NRG1 family members (not shown). A hydropathy analysis (not shown) indicated two hydrophobic segments: W66–V91 and L362–F383 (numbers according to human NRG3). Similar to the NRG1, the C-terminal hydrophobic segment may serve as the transmembrane domain and the N-terminal region may act as internal signal sequence (24–26). In contrast to many NRG1 family members, the extracellular domain of NRG3 is devoid of Ig-like or kringle domains. Instead, NRG3 contains a unique Ala/Gly rich segment at the N terminus, a mucin-like Ser/Thr rich region containing abundant sites for O-linked glycosylation, and an EGF motif. There are no predicted sites for N-linked glycosylation. The EGF-like domain of NRG3 is distinct from those encoded by the NRG1 (31% identity compared with NRG1-β1 EGF-like domain) and NRG2 (39% identity), suggesting that NRG3 is not an alternatively spliced NRG1 isoform. A diagrammatic comparison of EGF-like domains of EGF family members is shown in Fig. 1B. The putative intracellular domain of NRG3 contains only ≈13% sequence identity to the intracellular domain of NRG1.
Northern Blot and in Situ Hybridization Analyses Reveal a Neural Expression Pattern of NRG3.
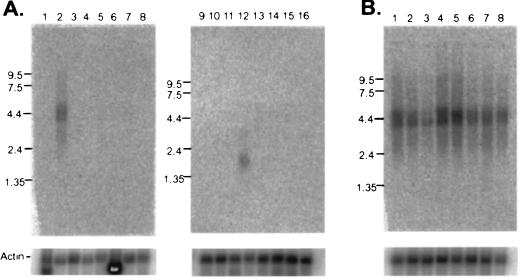
The expression of NRG3 in different human tissues was examined by RNA blot analysis using a probe derived from a portion of the intracellular domain of human NRG3 (encoding amino acids 394–536). A 4.4-kb mRNA transcript was highly expressed in brain. A lower level expression of a 1.9-kb transcript was detected in testis. The 4.4-kb transcript, but not the 1.9-kb transcript, is of sufficient size to encode NRG3. It is conceivable that the smaller transcript encodes an alternatively spliced form of NRG3, but this has not been investigated. The expression of NRG3 in other tissues was below the limit of detection (Fig. 2A). A similar pattern of expression of NRG3 was observed in RNA blots from murine tissues using a probe derived from the region of murine NRG3 that overlaps the EGF-like domain (data not shown). In a Northern blot of various brain tissues, NRG3 expression was detected at high levels in most regions of the brain with the exception of corpus callosum (Fig. 2B).
Figure 2.
Multi-tissue Northern blot analysis of NRG3. (A) Northern blot analysis of mRNA from human tissues hybridized to a human NRG3 specific probe. RNA size markers (in kb) are shown on the left of each blot. Lanes 1–16 represent poly(A)+ mRNA from lanes: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas; 9, spleen; 10, thymus; 11, prostate; 12, testis; 13, ovary; 14, small intestine; 15, colon (mucosal lining); 16, peripheral blood leukocytes. (B) Northern blot analysis of mRNA from human brain tissues using the same probe as in A. Lanes 1–8 represent poly(A)+ mRNA from lanes: 1, amygdala; 2, caudate nucleus; 3, corpus callosum; 4, hippocampus; 5, whole brain; 6, substantia nigra; 7, subthalamic nucleus; 8, thalamus. Northern blot analysis using mRNA from multiple murine tissues and murine NRG3 cDNA probe also revealed a 4.4 kb signal with a similar expression pattern as human NRG3 (not shown). β-actin signal was used for loading control.
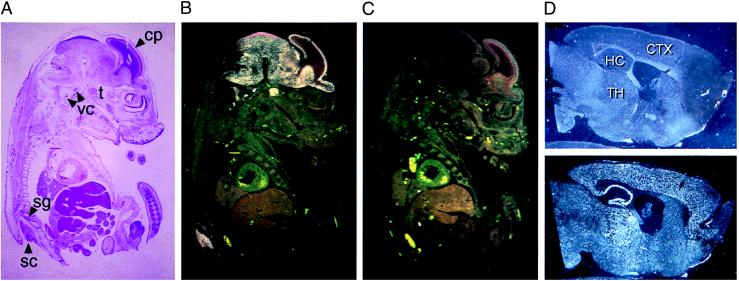
The tissue distribution of NRG3 expression was further characterized by in situ hybridization using tissues of embryonic and adult mice. In E16 mice, a strong signal for NRG3 mRNA was detected in the brain, spinal cord, trigeminal, vestibular-cochlear, and spinal ganglia (Fig. 3 A–C). Regions of the telencephalon containing differentiating cells (e.g., the cortical plate) displayed an intense NRG3 signal, whereas the underlying regions containing proliferating or migrating cells (ventricular and subventricular zones) showed little expression. Thus, NRG3 appeared to be expressed mainly in the nervous system. At E13 (the earliest time point examined), NRG3 mRNA was also enriched to the nervous system (data not shown). In adult animals, NRG3 antisense probes hybridized to mRNA in spinal cord and numerous brain regions including deep cerebellar nuclei, vestibular nuclei, cerebral cortex, piriform cortex, anterior olfactory nucleus, medial habenula, hippocampus, hypothalamus, and thalamus (Fig. 3D).
Figure 3.
In situ hybridization analysis of NRG3 RNA expression. (A) Low-power brightfield view of an E16 embryo section stained with hematoxylin and eosin. Structures labeled by the NRG3 probe are identified: sc, spinal cord; sg, spinal ganglion; t, trigeminal ganglion; vc; vestibular-cochlear ganglion; cp, cortical plate. (B) Darkfield view of the same section demonstrating the pattern of NRG3 hybridization. Intense hybridization signal appears exclusively over neural tissue. In the developing cerebral cortex, NRG3 signal is detected in the cortical plate, but not in underlying regions. (C) Darkfield view of an adjacent section hybridized with the NRG3 sense control probe. (D Upper) A sagittal section of formalin-fixed adult mouse brain hybridized with NRG3 sense probe observed under darkfield illumination. (Lower) An adjacent section hybridized with NRG3 antisense probe and viewed under darkfield illumination. Note the strong signal in cerebral cortex, hippocampus, and thalamus. CTX, cerebral cortex; HC, hippocampus; TH, thalamus.
The EGF-Like Domain of NRG3 Binds to the ErbB4 Receptor Tyrosine Kinase.
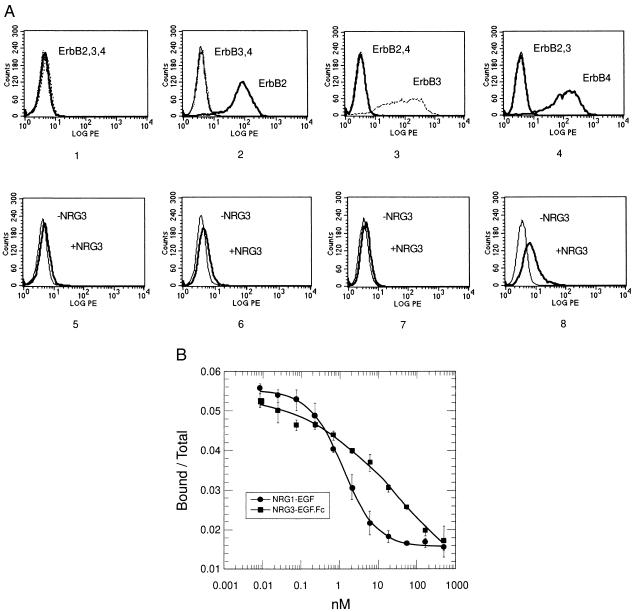
We examined if NRG3 binds to any of the known ErbB family members. Stable cell lines were generated that expressed ErbB2, ErbB3, or ErbB4. As shown in Fig. 4A, the parental cell line K562 does not express detectable levels of ErbB receptors (Panel 1). K562erbB2, K562erbB3, and K562erbB4 cells expressed only the corresponding receptors (Panels 2–4). Because the EGF-like domain determines the binding specificity of NRG1 to their receptors, we expressed and purified a protein containing an epitope-tagged version of the EGF-like domain of NRG3 fused to the Fc portion of human IgG. Using a FACS assay we observed that NRG3EGF.Fc bound to cells expressing ErbB4 (Panel 8). Binding was specific in that NRG3EGF.Fc did not bind to either the parental K562 cells or cells expressing either ErbB2 or ErbB3 (Panels 5–7). Also, a control Fc protein (Rse.Fc), which contains the extracellular domain of the Rse receptor tyrosine kinase (16), did not bind to any of these cell lines (not shown). The binding of NRG3EGF.Fc to K562erbB4 cells was competed in a dose-dependent fashion by NRG1EGF, but not by Rse.Fc (not shown), suggesting that NRG3EGF.Fc interacts directly with ErbB4 receptors on the cell surface. In addition, ErbB4.Fc could be specifically coimmunoprecipitated with NRG3EGF.Fc in vitro (data not shown).
Figure 4.
Binding of NRG3EGF.Fc to ErbB4. (A) Binding of NRG3EGF.Fc to K562erbB cells as analyzed by FACS. (Panels 1-4) Parental K562 cells (1) or K562 cells expressing either ErbB2 (K562erbB2 cells, 2), ErbB3 (K562erbB3 cells, 3), or ErbB4 (K562erbB4 cells, 4) were examined for the expression of corresponding receptors. Cells were incubated with anti-ErbB2, anti-ErbB3, or anti-ErbB4 antibodies as indicated before the phycoerythrin-conjugated secondary antibody was added. The binding of antibodies was analyzed by FACS. LOG PE, relative fluorescent intensity; Counts, cell numbers. (Panels 5-8) NRG3EGF.Fc binds to ErbB4 expressing cells. Parental K562 cells (5), K562erbB2 cells (6), K562erbB3 cells (7), and K562erbB4 cells (8) were incubated with or without NRG3EGF.Fc (containing gD tag) for 1 hr, followed by anti-gD-tag primary antibody and phycoerythrin-conjugated secondary antibody. The binding of antibodies was evaluated by FACS analysis. (B) Competitive inhibition of 125I-NRG3EGF.Fc binding to immobilized ErbB4.Fc by NRG3EGF.Fc or NRG1EGF. Iodination of NRG3EGF.Fc and the binding assay were as described in Materials and Methods. ErbB4.Fc was immobilized on 96-well plates and incubated with various concentrations of unlabeled NRG3EGF.Fc or NRG1EGF and a constant amount of 125I-labeled NRG3EGF.Fc for 1.5 hr at room temperature. Unbound ligand was removed and the plate was extensively washed. The fraction of radioactivity bound over total 125I-NRG3EGF.Fc input is plotted against the concentration of competitor. Data of a representative experiment from four independent assays are shown. Error bars indicate standard deviation of quadruplicate samples of this experiment.
The binding of NRG3EGF.Fc to ErbB4 was analyzed by direct binding assays using 125I-labeled NRG3EGF.Fc. As shown in Fig. 4B, binding of 125I-NRG3EGF.Fc to immobilized ErbB4.Fc can be competed by either NRG3EGF.Fc or NRG1EGF in a concentration-dependent manner. 125I-NRG3EGF.Fc was unable to bind the control, Rse.Fc, in the same experiment (not shown), and Rse.Fc did not compete with 125I-NRG3EGF.Fc for binding to ErbB4.Fc (not shown). The estimated affinity for NRG3EGF.Fc to bind ErbB4.Fc is 9 ± 4 nM (n = 4), and the apparent Ki of NRG1EGF is 1 nM. The shallowness of the displacement curve of NRG3EGF.Fc may be due to the fact that the NRG3EGF.Fc is expressed as a bivalent protein.
NRG3EGF Activates Tyrosine Phosphorylation of ErbB4.
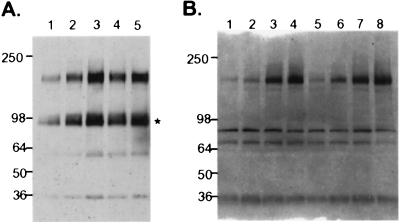
Binding of NRG1 to ErbB4 receptor results in tyrosine phosphorylation and downstream signaling events (7). To elucidate the ability of NRG3EGF to activate ErbB4, K562erbB4 cells were treated with 2.5 nM or 25 nM of NRG3EGF.Fc for 3 min or 8 min. Tyrosine phosphorylation of ErbB4 was detected by immunoprecipitation and Western blot. Indeed, NRG3EGF.Fc stimulated ErbB4 tyrosine phosphorylation at both time points and in a dose-dependent manner (Fig. 5A).
Figure 5.
Tyrosine phosphorylation of transfected and endogenous ErbB4 by NRG3EGF.Fc. (A) K562erbB4 cells were untreated (lane 1), or treated with 2.5 nM (lanes 2 and 4) and 25 nM (lanes 3 and 5) of NRG3EGF.Fc for 3 min (lanes 2 and 3) and 8 min (lanes 4 and 5). The cells were lysed and immunoprecipitated by anti-ErbB4 antibody, and probed with peroxidase-conjugated anti-phosphotyrosine antibody (Transduction Laboratories). *, a truncated ErbB4. The Mr (in kDa) is indicated at left. (B) MDA-MB-453 cells were untreated (lane 1) or treated with 2 nM, 20 nM, or 200 nM of NRG3EGF.Fc (lanes 2, 3, and 4, respectively), with 2 nM, 20 nM, or 200 nM of NRG3EGF.H6 (lanes 5, 6, and 7, respectively) or with 20 nM NRG1EGF (lane 8) for 3 min. The cells were lysed and subjected to immunoprecipitation and immunoblot as described in A. The Mr (in kDa) is indicated at left. For the blots shown in A and B, equal loading was confirmed by reprobing with anti-ErbB4 antibody (not shown).
To confirm the ability of NRG3EGF to activate the ErbB4 tyrosine phosphorylation, we examined the receptor activation in the human breast cancer cell line MDA-MB-453. This cell line expresses high levels of ErbB2 and ErbB3 and low levels of ErbB4 (data not shown). Treatment of MDA-MB-453 cells with NRG3EGF.Fc or with a monomeric form of the EGF domain (NRG3EGF.H6) resulted in a substantial increase of tyrosine phosphorylation of ErbB4 receptor (Fig. 5B).
DISCUSSION
This report describes the identification of NRG3, a novel member of the EGF family of protein ligands that is distinct from, but related to, the NRG1 family. The EGF domain of NRG3 is 31% identical to heregulin β-1. The N-terminal domain of NRG3 is similar to SMDF (23) in that it lacks Ig-like and kringle-like domains that are characteristic of many NRG1 family members. A hydropathy profile of NRG3 reveals that it lacks a hydrophobic N-terminal signal sequence often found in secreted proteins, but does contain a region of nonpolar or uncharged amino acids (W66–V91). A similar region is found in SMDF, and has been proposed to act as an internal, uncleaved signal sequence that mediates translocation across the membrane of the endoplasmic reticulum. The predicted extracellular domain of NRG3 lacks consensus sites for N-linked glycosylation, but contains numerous potential O-linked sites. In the intracellular domain, NRG3 demonstrates limited similarity with the intracellular domain of NRG1. We note that there are seven tyrosines contained within the NRG3 intracellular domain. The chromosomal localization of human NRG3 was mapped to 10q22 by PCR analysis of somatic cell hybrid DNA (D.Z., unpublished data), whereas the NRG1 gene is located at 8p11–22 (27, 28). Thus, NRG3 appears to be a novel member of the EGF-like family of protein ligands.
NRG1 family members and other members in the EGF family display a complex pattern of receptor binding. In most cases, one ligand is able to bind several combinations of receptor homo- and heterodimers (29, 30). For example, NRG1 members bind ErbB2/ErbB3 heterodimers and ErbB4/ErbB4 homodimers with high affinity, but ErbB3/ErbB3 homodimers with low affinity (6, 8, 31–33). Betacellulin binds both EGF receptor (EGFR) and ErbB4 homodimers (34). The EGF-like domains of EGF and NRG1 family members determine the specificity of receptor activation (2, 35). In this report, we have shown that the EGF-like domain of NRG3 binds to ErbB4 and stimulates tyrosine phosphorylation of the receptor. The binding of the EGF-like domain of NRG3 to ErbB4 can be competed by the EGF-like domain of NRG1-β1, suggesting that their binding sites are overlapping. We did not observe binding of NRG3EGF.Fc to K562 cells that express either ErbB2 or ErbB3 (Fig. 4A), or to MDA-MB-486 cells that express high levels of the EGFR (data not shown). We also did not observe an increase in phosphorylation of either the EGFR, ErbB2, or ErbB3 in MDA-MB-453 cells treated with NRG3. However, we noted that the basal level of phosphorylation of these receptors in MDA-MB-453 cells was relatively high. Thus, the possibility that NRG3 might activate these other receptors via its interaction with ErbB4 requires further investigation.
Most variants of NRG1, with the exception of the neural specific form of SMDF, are widely expressed in numerous tissues including brain, heart, skeletal muscle, breast, liver, and lung, among others. Betacellulin, a ligand for both EGFR and ErbB4, also displays broad tissue expression patterns (19, 36). We found that the expression of NRG3 is strikingly enriched to neural tissues as shown by Northern blot analysis and in situ hybridization. Developmentally, NRG3 mRNA can be detected as early as E11 (but not E4) in mouse as judged by Northern blot analysis (data not shown) and E13 by in situ hybridization (the earliest age examined). ErbB4 is predominantly expressed in brain, heart, and skeletal muscle (37). ErbB4 was also shown to be broadly distributed in the brains of chicken embryos (E14 and E17, predominantly in neurons) (38), in rat retina cultures (39), at neuromuscular synapses (44), but not in cultured human and rat Schwann cells (40, 41). Recently, ErbB4 was found to colocalize with GABA+ cells (42). It is conceivable that the same receptor mediates distinct biological functions in different tissues or cell types when interacted with corresponding tissue-specific ligands. For example, NRG1 may serve as a ligand for ErbB4 during heart development, betacellulin may act as a mitogenic ligand for ErbB4 in variety of cell types, whereas a neural specific ligand(s) may function as trophic or guidance molecules on ErbB4+ cells in the central or peripheral nervous systems. The phenotype of NRG3-deficient mice will be informative. In this regard, the properties of NRG3 are consistent with the predicition by Lemke and coworkers for the existence of a novel ligand for ErbB4 expressed in the central nervous system based on analysis of ErbB4-deficient mice (10).
Acknowledgments
We thank Dr. Wiliam I. Wood and Colin Watanabe for their help in searching the expressed sequence tag database, and Dr. Richard Vandlen for purifying the NRG3EGF.H6 protein. We also thank the flow cytometry group of Genentech for technical help.
Footnotes
Abbreviations: EGF, epidermal growth factor; EGFR, EGF receptor; NRG3, Neuregulin-3; NRG1, Neuregulin-1; SMDF, sensory and motor neuron-derived factor; FACS, fluorescence-activated cell sorter; E, embryonic day.
References
- 1.Lemke G. Mol Cell Neurosci. 1996;7:247–262. doi: 10.1006/mcne.1996.0019. [DOI] [PubMed] [Google Scholar]
- 2.Holmes W E, Sliwkowski M X, Akita R W, Henzel W J, Lee J, Park J W, Yansura D, Abadi N, Raab H, Lewis G D, Shepard M, Kuang W-J, Wood W I, Goeddel D V, Vandlen R L. Science. 1992;256:1205–1210. doi: 10.1126/science.256.5060.1205. [DOI] [PubMed] [Google Scholar]
- 3.Wen D, Peles E, Cupples R, Suggs S V, Bacus S S, Luo Y, Trail G, Hu S, Silbiger S M, Levy R B, Koski R A, Lu H S, Yarden Y. Cell. 1992;69:559–572. doi: 10.1016/0092-8674(92)90456-m. [DOI] [PubMed] [Google Scholar]
- 4.Falls D L, Rosen K M, Corfas G, Lane W S, Fischbach G D. Cell. 1993;72:801–815. doi: 10.1016/0092-8674(93)90407-h. [DOI] [PubMed] [Google Scholar]
- 5.Marchionni M A, Goodearl A D, Chen M S, Bermingham-McDonogh O, et al. Nature (London) 1993;362:312–318. doi: 10.1038/362312a0. [DOI] [PubMed] [Google Scholar]
- 6.Sliwkowski M X, Schaefer G, Akita R W, Lofgren J A, Fitzpatrick V D, Nuijens A, Fendly B M, Cerione R A, Vandlen R L, Carraway K L R. J Biol Chem. 1994;269:14661–14665. [PubMed] [Google Scholar]
- 7.Plowman G D, Green J M, Culouscou J M, Carlton G W, Rothwell V M, Buckley S. Nature (London) 1993;366:473–475. doi: 10.1038/366473a0. [DOI] [PubMed] [Google Scholar]
- 8.Carraway K L R, Cantley L C. Cell. 1994;78:5–8. doi: 10.1016/0092-8674(94)90564-9. [DOI] [PubMed] [Google Scholar]
- 9.Meyer D, Birchmeier C. Nature (London) 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 10.Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Nature (London) 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 11.Lee K F, Simon H, Chen H, Bates B, Hung M C, Hauser C. Nature (London) 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 12.Chan H, Riese D J, Gilbert W, Stern D F, McMahan U J. Nature (London) 1997;387:509–511. doi: 10.1038/387509a0. [DOI] [PubMed] [Google Scholar]
- 13.Carraway K L, Weber J L, Unger M J, Ledesma J, Yu N, Gassman M, Lai C. Nature (London) 1997;387:512–516. doi: 10.1038/387512a0. [DOI] [PubMed] [Google Scholar]
- 14.Godowski P J, Leung D W, Meacham L R, Galgani J P, Hellmiss R, Keret R, Rotwein P S, Parks J S, Laron Z, Wood W I. Proc Natl Acad Sci USA. 1989;86:8083–8087. doi: 10.1073/pnas.86.20.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu L H, Gillett N A. Cell Vision. 1994;1:169–176. [Google Scholar]
- 16.Mark M R, Scadden D T, Wang Z, Gu Q, Goddard A, Godowski P J. J Biol Chem. 1994;269:10720–10728. [PubMed] [Google Scholar]
- 17.Carter P, Presta L, Gorman C M, Ridgway J B, Henner D, Wong W L, Rowland A M, Kotts C, Carver M E, Shepard H M. Proc Natl Acad Sci USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plowman G D, Green J M, McDonald V L, Neubauer M G, Disteche C M, Todaro G J, Shoyab M. Mol Cell Biol. 1990;10:1969–1981. doi: 10.1128/mcb.10.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasada R, Ono Y, Taniyama Y, Shing Y, Folkman J, Igarashi K. Biochem Biophys Res Commun. 1993;190:1173–1179. doi: 10.1006/bbrc.1993.1173. [DOI] [PubMed] [Google Scholar]
- 20.Nagai M, Hiramatsu R, Kaneda T, Hayasuke N, Arimura H, Nishida M, Suyama T. Gene. 1985;36:183–188. doi: 10.1016/0378-1119(85)90084-8. [DOI] [PubMed] [Google Scholar]
- 21.Higashiyama S, Abraham J A, Miller J, Fiddes J C, Klagsbrun M. Science. 1991;251:936–939. doi: 10.1126/science.1840698. [DOI] [PubMed] [Google Scholar]
- 22.Toyoda H, Komurasaki T, Ikeda Y, Yoshimoto M, Morimoto S. FEBS Lett. 1995;377:403–407. doi: 10.1016/0014-5793(95)01403-9. [DOI] [PubMed] [Google Scholar]
- 23.Ho W H, Armanini M P, Nuijens A, Phillips H S, Osheroff P L. J Biol Chem. 1995;270:14523–14532. doi: 10.1074/jbc.270.24.14523. [DOI] [PubMed] [Google Scholar]
- 24.Wickner W T, Lodish H F. Science. 1985;230:400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- 25.Sabatini D D, Kreibich G, Morimoto T, Adesnik M. J Cell Biol. 1982;92:1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blobel G. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Wood W I. Genomics. 1993;16:790–791. doi: 10.1006/geno.1993.1272. [DOI] [PubMed] [Google Scholar]
- 28.Orr-Urtreger A, Trakhtenbrot L, Ben-Levy R, Wen D, Rechavi G, Lonai P, Yarden Y. Proc Natl Acad Sci USA. 1993;90:1867–1871. doi: 10.1073/pnas.90.5.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karunagaran D, Tzahar E, Beerli R R, Chen X, Graus-Porta D, Ratzkin B J, Seger R, Hynes N E, Yarden Y. EMBO J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- 30.Beerli R R, Hynes N E. J Biol Chem. 1996;271:6071–6076. doi: 10.1074/jbc.271.11.6071. [DOI] [PubMed] [Google Scholar]
- 31.Tzahar E, Levkowitz G, Karunagaran D, Yi L, Peles E, Lavi S, Chang D, Liu N, Yayon A, Wen D, Yarden Y. J Biol Chem. 1994;269:25226–25233. [PubMed] [Google Scholar]
- 32.Carraway K L R, Sliwkowski M X, Akita R, Platko J V, Guy P M, Nuijens A, Diamonti A J, Vandlen R L, Cantley L C, Cerione R A. J Biol Chem. 1994;269:14303–14306. [PubMed] [Google Scholar]
- 33.Kita Y A, Barff J, Luo Y, Wen D, Brankow D, Hu S, Liu N, Prigent S A, Gullick W J, Nicolson M. FEBS Lett. 1994;349:139–143. doi: 10.1016/0014-5793(94)00644-x. [DOI] [PubMed] [Google Scholar]
- 34.Riese D J N, van Raaij T M, Plowman G D, Andrews G C, Stern D F. Mol Cell Biol. 1995;15:5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbacci E G, Guarino B C, Stroh J G, Singleton D H, Rosnack K J, Moyer J D, Andrews G C. J Biol Chem. 1995;270:9585–9589. doi: 10.1074/jbc.270.16.9585. [DOI] [PubMed] [Google Scholar]
- 36.Shing Y, Christofori G, Hanahan D, Ono Y, Sasada R, Igarashi K, Folkman J. Science. 1993;259:1604–1607. doi: 10.1126/science.8456283. [DOI] [PubMed] [Google Scholar]
- 37.Plowman G D, Culouscou J M, Whitney G S, Green J M, Carlton G W, Foy L, Neubauer M G, Shoyab M. Proc Natl Acad Sci USA. 1993;90:1746–1750. doi: 10.1073/pnas.90.5.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Francoeur J R, Richardson P M, Dunn R J, Carbonetto S. J Neurosci Res. 1995;41:836–845. doi: 10.1002/jnr.490410614. [DOI] [PubMed] [Google Scholar]
- 39.Bermingham-McDonogh O, McCabe K L, Reh T A. Development (Cambridge, UK) 1996;122:1427–1438. doi: 10.1242/dev.122.5.1427. [DOI] [PubMed] [Google Scholar]
- 40.Grinspan J B, Marchionni M A, Reeves M, Coulaloglou M, Scherer S S. J Neurosci. 1996;16:6107–6118. doi: 10.1523/JNEUROSCI.16-19-06107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levi A D, Bunge R P, Lofgren J A, Meima L, Hefti F, Nikolics K, Sliwkowski M X. J Neurosci. 1995;15:1329–1340. doi: 10.1523/JNEUROSCI.15-02-01329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber J, Ledesma J, Battenberg E, Morales M, Bloom F E, Lemke G, Lai C. Soc Neurosci Abstr. 1996;22:1579. [Google Scholar]
- 43.Derynck R, Roberts A B, Winkler M E, Chen E Y, Goeddel D V. Cell. 1984;38:287–297. doi: 10.1016/0092-8674(84)90550-6. [DOI] [PubMed] [Google Scholar]
- 44.Zhu X, Lai C, Thomas S, Burden S J. EMBO J. 1995;14:5842–5848. doi: 10.1002/j.1460-2075.1995.tb00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]