Abstract
We have examined the effects on transcription initiation of promoter and enhancer strength and of the curvature of the DNA separating these entities on wild-type and mutated enhancer–promoter regions at the Escherichia coli σ54-dependent promoters glnAp2 and glnHp2 on supercoiled and linear DNA. Our results, together with previously reported observations by other investigators, show that the initiation of transcription on linear DNA requires a single intrinsic or induced bend in the DNA, as well as a promoter with high affinity for σ54-RNA polymerase, but on supercoiled DNA requires either such a bend or a high affinity promoter but not both. The examination of the DNA sequence of all nif gene activator- or nitrogen regulator I-σ54 promoters reveals that those lacking a binding site for the integration host factor have an intrinsic single bend in the DNA separating enhancer from promoter.
The initiation of transcription at nitrogen-responsive, σ54 RNA polymerase-dependent promoters of enteric bacteria is activated by nitrogen regulator I (NRI)-phosphate or by nif gene activator (NifA). The enhancers, the binding sites for these activators, are located 100 bp or more upstream from the transcriptional start sites. In the case of many, but not all, of these systems, a binding site for the abundant integration host factor (IHF) is located between enhancer and promoter (1).
The role of IHF is to introduce a bend in the stretch of DNA separating enhancer from promoter and in this manner to increase the probability of a successful interaction of the activator complex bound to the enhancer with the σ54 RNA polymerase bound to the promoter. This role of IHF is strongly supported by the experimental demonstration that the relative positions of activator, of IHF, and of σ54-RNA polymerase on the face of the DNA determine whether IHF will stimulate or block open complex formation (2).
It also has been shown that the affinity of the promoter for σ54-RNA polymerase determines whether or not IHF is required for open complex formation on supercoiled DNA. The nifHp promoter has much less affinity for RNA polymerase than the glnAp2 promoter and, in contrast to the latter, is associated with a binding site for IHF. It could be shown that the initiation of transcription at nifHp on supercoiled DNA required IHF but that mutations in the promoter that increased its affinity for σ54-RNA polymerase allowed open complex formation to proceed, just as in the case of glnAp2, in the absence of IHF (3). Another promoter, glnHp2, is also associated with a binding site for IHF; open complex formation at this promoter on supercoiled DNA did not require, but was stimulated by, IHF (4). In this case, too, a mutation in the promoter bringing it closer to the consensus for σ54-dependent promoters resulted in maximal initiation of transcription in the absence of IHF (2). On the other hand, not every low affinity promoter is associated with a binding site for IHF. The NRI-phosphate-activated nifLp promoter, which, like the nifHp promoter, has low affinity for σ54-RNA polymerase, is not associated with a binding site for IHF but supports the initiation of transcription on supercoiled DNA (5, 6).
We have investigated the initiation of transcription on linear DNA at glnAp2 and glnHp2. As described in a previous publication, open complex formation at glnHp2 absolutely depended on the presence of IHF, even when this promoter had been replaced by the stronger mutant promoter (2). On the other hand, the initiation of transcription at glnAp2 on linear DNA neither required nor was stimulated by IHF or by another DNA bending protein, HU. We found that the ability of glnAp2 to allow open complex formation on linear DNA depended on the sequence of nucleotides of the DNA separating enhancer from promoter. Replacing this sequence by random DNA did not affect open complex formation on supercoiled DNA but made open complex formation on linear DNA dependent on HU. This result suggested the possibility that the DNA separating the enhancer from the promoter at glnAp2, in contrast to that at glnHp2, was naturally bent and therefore did not require the help of a DNA bending protein on linear DNA (7).
We now present the results of a comparison of the glnAp2 and glnHp2 enhancers and promoters and of the effect of the elimination of the potential natural bend between enhancer and promoter at glnAp2, as well as a computer simulation of the DNA structure at nitrogen-regulated, σ54-dependent promoters to define the roles of DNA curvature and promoter strength in the initiation of transcription on linear and supercoiled DNA.
MATERIALS AND METHODS
Plasmid Construction.
All of the plasmids used in the transcription assays were derived from pTE103 (8). pMC80 was constructed as follows: a SmaI–BamHI fragment containing the sequence (−160-−78) from glnAp2 that included the NRI binding site obtained by PCR was cloned into SmaI–BamHI sites of pFC54 (4). The pMC80 plasmid has the NRI binding sites from glnAp2 located −100 bp from the glnHp2 promoter.
Site-Directed Mutagenesis.
The Altered Site Mutagenesis system from Promega was used for in vitro mutagenesis as described (2). The following oligonucleotides (5′-3′) were used to generate the mutants referred to in the legends to Figs. 3 and 4, respectively: TTGCAAAAGTTGGCACAGATTTCGCTATATGTGAATGTCACG (pMC40) and CGTGGTGCAGATCGCTCGCACGACGATGGTG (pMC70).
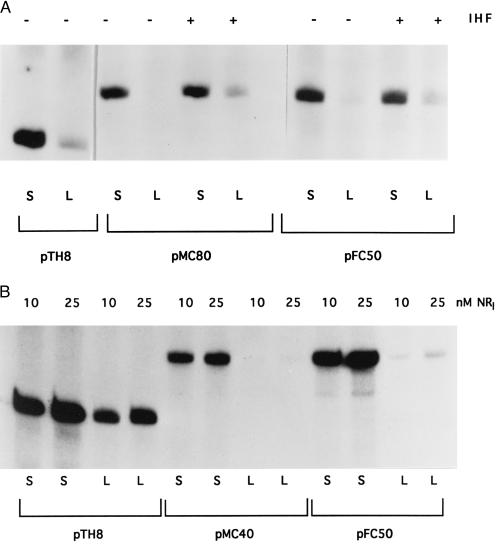
Figure 3.
Transcription initiation at hybrid glnAp2–glnHp2 templates. pTH8, wild-type glnAp2 template; pFC50, wild-type glnHp2 template. In pMC80, the NRI-binding sites (enhancer) from glnAp2 were substituted for the corresponding sites of glnHp2 100 bp from the transcriptional start site. In pMC40, the glnAp2 promoter (−26-−11) has replaced the glnHp2 promoter. S, supercoiled DNA; L, linear DNA. The concentration of NRI in 3A was 10 nM and is indicated in B, Top. The presence (+) or absence (−) of IHF is indicated at A, Top.
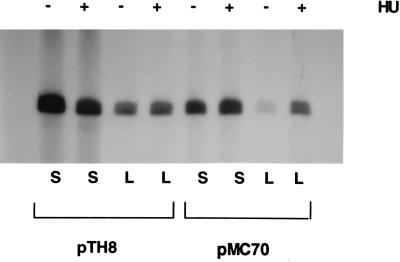
Figure 4.
The effect on transcription of replacing the sequencing CCCTTTT, 73 bp upstream from the transcriptional start site at glnAp2 in plasmid pTH8 by the sequence ATCGCTC in plasmid pMC70. S, supercoiled DNA; L, linear DNA. The concentration of NRI was 10 nM. The presence (+) or absence (−) of HU is indicated at the top.
Proteins.
Purification of core RNA polymerase, σ54, NRI, and NRII has been described (9–11). IHF and HU were kindly supplied by C. Robertson and H. Nash (National Institutes of Health), and by T. Baker (Massachusetts Institute of Technology). The NRI concentration is expressed in terms of the monomer.
In Vitro Transcription Assays.
Computer Simulation.
Cylindrical projections of DNA sequences were generated as described (7, 12).
Footprinting with Dimethyl Sulfate (DMS).
Footprinting with DMS was carried out with supercoiled templates (0.5 μg of DNA) as described (2). Products were analyzed by primer extension using the alkaline denaturing procedure of Sasse–Dwight and Gralla (13) as described (2). Samples were run on 7% (wt/vol) polyacrylamide gels containing 7 M urea.
RESULTS
Comparison of glnAp2 and glnHp2 Enhancers and Promoters.
Strong initiation of transcription at the glnAp2 promoter depends on NRI-phosphate bound to an enhancer consisting of two binding sites for NRI separated by 15 bp and located 100 bp upstream from the transcriptional start site (14). Similarly, effective initiation of transcription at glnHp2 depends on NRI-phosphate bound to an enhancer consisting of two strong binding sites for NRI overlapping by 4 bp and also located 100 bp upstream from the transcriptional start site; in addition, a binding site for the DNA-bending protein IHF is located 34 bp upstream from the transcriptional start site between the glnHp2 promoter and the enhancer (4).
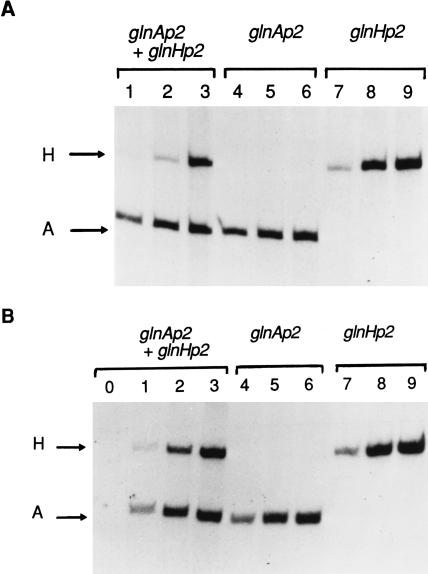
We compared the affinities for NRI-phosphate of the glnAp2 and glnHp2 enhancers by determining the response of the two promoters to increasing concentrations of NRI-phosphate when present separately or together, in the presence or absence of IHF. The results of these experiments, illustrated in Fig. 1, showed that the glnAp2 enhancer had slightly more affinity for NRI than the glnHp2 enhancer. This was particularly apparent at the lowest concentration of NRI, 1.2 nM, when both plasmids were present in the same reaction mixture; in this case, the presence of glnAp2 prevented the initiation of transcription at glnHp2, presumably by competing successfully for NRI-phosphate. Nevertheless, at the next higher concentration of NRI used, 2.5 nM, both promoters were operative.
Figure 1.
Activation of transcription on supercoiled DNA by NRI-phosphate in the absence (A) and presence (B) of IHF. Lanes: 0–4, the transcripts initiated at glnAp2 (A) and initiated at glnHp2 (H) in reactions containing both plasmids pTH8 (glnAp2) and pFC50 (glnHp2); 4–6, reactions containing only pTH8; and 7–9, reactions containing only pFC50. The concentrations of NRI were 0 (lane 0), 1.2 nM (lanes 1, 4, and 7), 2.5 nM (lanes 2, 5, and 8), and 5 nM (lanes 3, 6, and 9).
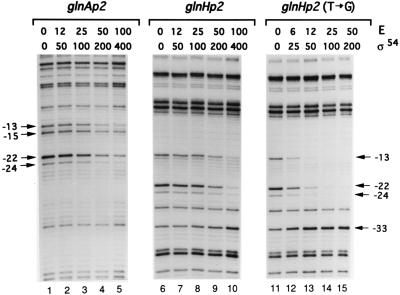
We next compared the affinities of the two promoters for σ54-RNA polymerase using DMS footprinting of the closed promoter-polymerase complexes. As shown in Fig. 2, protection of either promoter became apparent at a 50-nM concentration of RNA polymerase, indicating approximately equal affinities of the two promoters for σ54-RNA polymerase. On the other hand, the mutated glnHp2 promoter in which the T in position −13 had been replaced by the canonical G had much greater affinity; protection could be detected at a 6-nM concentration of the polymerase.
Figure 2.
Determination of promoter affinities for σ54-RNA polymerase by footprinting with DMS. The promoters are glnAp2, glnHp2, and glnHp2 with the canonical G substituted for T in position −13. The plasmids pTH8 (glnAp2), pFC50 (glnHp2), and pFC50-M12 (mutant glnHp2) were incubated with increasing concentrations of core enzyme (E) and σ54 (nM, Top) and treated with DMS. The DNA was cleaved at the methylated guanine residues, and the primer extension products were analyzed in sequencing gels. The numbers on both sides indicate positions on the DNA with respect to the transcriptional start site. The arrows on the left and right hand sites indicate residues protected from methylation by DMS or showing hyperreaction to methylation by DMS.
The fact that both the enhancers and promoters of glnAp2 and glnHp2 did not differ greatly in their abilities to bind, respectively, NRI-phosphate and σ54-RNA polymerase and the additional fact that transcription at the mutant glnHp2 promoter with greatly increased affinity for the RNA polymerase could not be activated on a linear DNA template militate against the idea that promoter or enhancer strength determined whether transcription could be initiated on linear DNA. Nevertheless, we wanted to examine the possibility that some property of enhancers or promoters other than affinity is responsible for transcription initiation on linear DNA. We therefore replaced in one case the glnHp2 enhancer and in the other case the glnHp2 promoter by the corresponding entities of glnAp2. We compared transcription initiation on these plasmids, pMC80 and pMC40, with transcription initiation on the wild-type enhancer promoter plasmids pTH8 carrying glnAp2 and pFC50 carrying glnHp2. The results of these experiments, presented in Fig. 3 A and B, show clearly that neither the substitution of the enhancer nor that of the promoter of glnHp2 has allowed transcription to be initiated on linear DNA in the absence of IHF. These results confirm the conclusion that the DNA region between promoter and enhancer is solely responsible for the transcription initiation at glnAp2 in the absence of a DNA-bending protein.
The Bending Site at glnAp2.
The computer analysis of the DNA region between enhancer and promoter at glnAp2 indicated that the center of the bend is in the sequence CCCTTTT, located 73 bp upstream from the transcriptional start site (see Fig. 5). We therefore attempted to eliminate the bend by replacing this sequence with the sequence ATCGCTC in plasmid pMC70. We then compared the ability of supercoiled and linear plasmids pTH8 and pMC70 to support transcription initiation on DNA in the presence and absence of HU. The results illustrated in Fig. 4 show that the replacement of the CCCTTTT sequence has not affected transcription initiation on supercoiled DNA but has greatly reduced transcription initiation on linear DNA. It can also be seen that HU fails to stimulate transcription on linear DNA with the wild-type promoter region (plasmid pTH8) but stimulates transcription initiation on the linear, altered promoter region (plasmid pMC70). The fact that “straightening out” the bend has made transcription initiation on linear DNA largely dependent on the DNA-bending protein HU confirms that the intrinsic bend in the region between enhancer and glnAp2 promoter is responsible for the ability of this region to support transcription initiation on linear DNA.
Figure 5.
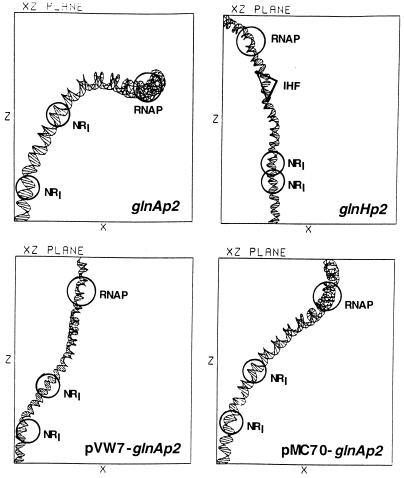
Computer simulation of enhancer–promoter regions using the dna bend program based on the Trifonov algorithm (12). The promoters are: glnAp2, wild-type; glnHp2, wild-type; pVW–glnAp2, region separating enhancer from promoter replaced by a random DNA sequence (7); pMC70–glnAp2, CCCTTTT sequence at −73 replaced by TTCGCTC.
DISCUSSION
Our results show clearly that an intrinsic or induced bend in the DNA region separating enhancer and promoter is essential for open complex formation on linear DNA. A change, specifically elimination of the bend in the case of glnAp2, made open complex formation at this promoter dependent on the DNA-bending protein HU on linear, but not on supercoiled, DNA. Furthermore, there was very little difference in the affinities of the glnAp2 and glnHp2 enhancers for NRI-phosphate and promoters for σ54-RNA polymerase, respectively, yet transcription initiation at glnHp2 required IHF, even when its enhancer or promoter had been replaced by that of glnAp2 or by a mutant promoter with much greater affinity than either one of these promoters. We present in Fig. 5 a computer simulation of the regions upstream of the glnHp2 and glnAp2 promoters, as well as of two altered glnAp2 promoters, pVW7, where the entire region between enhancer and promoter has been replaced by a random DNA sequence (7) and by pMC70, in which the CCCTTTT sequence located in the center of the bend has been replaced by ATCGCTG. The only one of these promoters capable of forming an open complex on linear DNA is glnAp2, which contains a single bend between enhancer and promoter. On the other hand, neither an intrinsic nor an induced bend was required for open complex formation on supercoiled DNA at any one of the promoters shown in this figure.
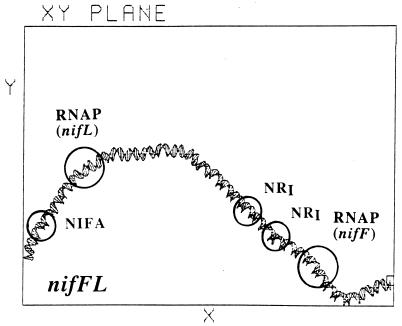
The ability of another NRI-phosphate-activated, σ54-dependent promoter, nifLp, to support open complex formation on supercoiled and linear DNA has been investigated. The enhancer–promoter region of nifLp is overlapped by the divergently transcribed, NifA-activated, σ54-dependent promoter nifFp (15). There is no binding site for IHF associated with these promoters, making nifFp the only NifA-activated promoter without such a site (16). Transcription at the low affinity nifLp promoter can be initiated on supercoiled, but not on linear, DNA; however, in this case, substitution of the higher affinity glnAp2 promoter for nifLp, resulting in a hybrid template with the nifL enhancer and intervening region fused to glnAp2, allowed transcription to be initiated on linear DNA (17, 18). Our computer analysis of the nifF nifL enhancer–promoter region, illustrated in Fig. 6, shows that the DNA is bent in the center of the region, with enhancers and promoters in the same plane and all bound proteins located on the same face of the DNA helix. The inability of the nifLp promoter to support the initiation of transcription on linear DNA is therefore not due to the lack of intrinsic curvature of the DNA separating enhancer and promoter but is due to its low affinity for σ54-RNA polymerase.
Figure 6.
Computer simulation of the nifL nifF enhancer–promoter region.
The expression of nifL and nifF has also been studied in intact cells. The facts that binding of NifA to the nifF enhancer can stimulate expression of nifL and that the presence of NifA does not interfere with the activation of transcription at nifLp by NRI-phosphate are in good accord with the view that the bending of the DNA allows both of these low affinity promoters to be activated despite the lack of a binding site for IHF (15).
The view that the initiation of transcription on linear DNA requires both an intrinsic or induced bend in the DNA and a promoter with high affinity for σ54-RNA polymerase is supported by the results of a study of the promoter for the nac gene of Klebiella aerogenes; transcription at nacP could be initiated on supercoiled, but not on linear, DNA (19). This promoter has low affinity for σ54-RNA polymerase and no binding site for IHF, but our computer analysis revealed the presence of a single intrinsic bend in the DNA separating enhancer and promoter. Furthermore, computer analysis of all known nitrogen-regulated, σ54-dependent promoters of enteric bacteria showed that those without a binding site for IHF, in contrast to those with such a site, have an intrinsic single bend in the DNA separating enhancer and promoter.
Our experimental results, the analysis of the experimental results of other studies, and our computer analysis of DNA structure reveal that transcription initiation on linear DNA requires a bend in the DNA between enhancer and promoter as well as a promoter with high affinity for σ54-RNA polymerase; on the other hand, transcription initiation on supercoiled DNA requires either such a bend, which may be intrinsic or induced by IHF, or a high affinity promoter but not both.
Acknowledgments
We thank H. Nash and T. Baker for the gift of IHF and HU. We thank Hilda Harris-Ransom for the preparation of the manuscript.
ABBREVIATIONS
- NifA
nif gene activator
- NRI
nitrogen regulator I
- IHF
integration host factor
- DMS
dimethyl sulfate
References
- 1.Magasanik B. In: Escherichia coli and Salmonella typhimurium (Cellular and Molecular Biology) Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1344–1356. [Google Scholar]
- 2.Claverie-Martin F, Magasanik B. J Mol Biol. 1992;227:996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- 3.Santero E, Hoover R, North A K, Berger D K, Porter S C, Kustu S. J Mol Biol. 1992;227:602–620. doi: 10.1016/0022-2836(92)90211-2. [DOI] [PubMed] [Google Scholar]
- 4.Claverie-Martin F, Magasanik B. Proc Natl Acad Sci USA. 1991;88:1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin S, Henderson N, Dixon R. Mol Microbiol. 1987;1:92–100. doi: 10.1111/j.1365-2958.1987.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 6.Wong P K, Popham D, Keener J, Kustu S. J Bacteriol. 1987;169:2876–2880. doi: 10.1128/jb.169.6.2876-2880.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmona M, Magasanik B. J Mol Biol. 1996;261:348–356. doi: 10.1006/jmbi.1996.0468. [DOI] [PubMed] [Google Scholar]
- 8.Elliot T, Geiduschek E P. Cell. 1984;36:211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- 9.Reitzer L J, Magasanik B. Proc Natl Acad Sci USA. 1983;80:5554–5558. doi: 10.1073/pnas.80.18.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt T P, Magasanik B. Proc Natl Acad Sci USA. 1985;82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ninfa A J, Ueno-Nishio S, Hunt T P, Robustell B, Magasanik B. J Bacteriol. 1986;168:1002–1004. doi: 10.1128/jb.168.2.1002-1004.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolshoy A, McNamara P, Harrington R E, Trifonov E N. Proc Natl Acad Sci USA. 1991;88:2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasse-Dwight S, Gralla J. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- 14.Ninfa A J, Reitzer L J, Magasanik B. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 15.Minchin S D, Austin S, Dixon R A. Mol Microbiol. 1988;2:433–442. doi: 10.1111/j.1365-2958.1988.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoover T R, Santero E, Porter S, Kustu S. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 17.Whitehall S, Austin S, Dixon R. J Mol Biol. 1992;225:591–607. doi: 10.1016/0022-2836(92)90388-z. [DOI] [PubMed] [Google Scholar]
- 18.Whitehall S, Austin S, Dixon R. Mol Microbiol. 1993;9:1107–1117. doi: 10.1111/j.1365-2958.1993.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 19.Feng J, Goss T J, Bender R A, Ninfa A J. J Bacteriol. 1995;177:5523–5534. doi: 10.1128/jb.177.19.5523-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]