Abstract
The m7GpppN cap structure of eukaryotic mRNA is formed cotranscriptionally by the sequential action of three enzymes: RNA triphosphatase, RNA guanylyltransferase, and RNA (guanine-7)-methyltransferase. A multifunctional polypeptide containing all three active sites is encoded by vaccinia virus. In contrast, fungi and Chlorella virus encode monofunctional guanylyltransferase polypeptides that lack triphosphatase and methyltransferase activities. Transguanylylation is a two-stage reaction involving a covalent enzyme-GMP intermediate. The active site is composed of six protein motifs that are conserved in order and spacing among yeast and DNA virus capping enzymes. We performed a structure–function analysis of the six motifs by targeted mutagenesis of Ceg1, the Saccharomyces cerevisiae guanylyltransferase. Essential acidic, basic, and aromatic functional groups were identified. The structural basis for covalent catalysis was illuminated by comparing the mutational results with the crystal structure of the Chlorella virus capping enzyme. The results also allowed us to identify the capping enzyme of Caenorhabditis elegans. The 573-amino acid nematode protein consists of a C-terminal guanylyltransferase domain, which is homologous to Ceg1 and is strictly conserved with respect to all 16 amino acids that are essential for Ceg1 function, and an N-terminal phosphatase domain that bears no resemblance to the vaccinia triphosphatase domain but, instead, has strong similarity to the superfamily of protein phosphatases that act via a covalent phosphocysteine intermediate.
mRNA capping occurs by a series of three enzymatic reactions in which the 5′-triphosphate terminus of a primary transcript is first cleaved to a diphosphate by RNA triphosphatase, then capped with GMP by RNA guanylyltransferase, and methylated at the N7 position of guanine by RNA (guanine-7)-methyltransferase (1). To date, only the guanylyltransferase reaction mechanism has been dissected in detail. Transfer of GMP from GTP to the 5′-diphosphate terminus of RNA occurs in a two-stage reaction involving a covalent enzyme–GMP intermediate (2). The GMP is linked to the enzyme through a phosphoamide (P–N) bond to the ɛ-amino group of a lysine residue. This structure is analogous to the enzyme (-Lys)-AMP intermediate formed when DNA ligase reacts with ATP.
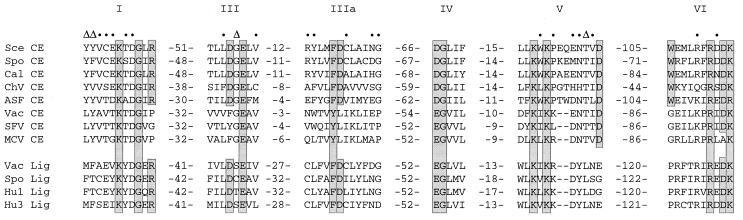
The GTP-dependent capping enzymes and ATP-dependent ligases make up a superfamily of covalent nucleotidyltransferases (3). The guanylate or adenylate moiety is covalently bound to an invariant lysine residue within a conserved KxDG element (motif I in Fig. 1). Five other sequence motifs are conserved in the same order and with similar spacing in the capping enzymes and ligases (motifs III, IIIa, IV, V, and VI in Fig. 1). The amino acid sequence similarity between the capping enzymes and polynucleotide ligases is limited to these segments, suggesting a common core structure in which the six motifs are brought together at the enzyme’s active site. This prediction has been borne out by the recent reports of the crystal structures of bacteriophage T7 DNA ligase with bound ATP and Chlorella virus capping enzyme with bound GTP (4, 5).
Figure 1.
Conserved sequence elements define a superfamily of covalent nucleotidyltransferases. Six collinear sequence elements, designated motifs I, III, IIIa, IV, V, and VI, are conserved in guanylyltransferases and ATP-dependent DNA ligases as shown. The amino acid sequences are aligned for capping enzymes (CE) encoded by S. cerevisiae (Sce), Sc. pombe (Spo), C. albicans (Cal), Chlorella virus PBCV-1 (ChV), African swine fever virus (ASF), vaccinia virus (Vac), Shope fibroma virus (SFV), and molluscum contagiosum virus (MCV). Grouped below the capping enzymes are aligned sequences for the DNA ligases (Lig) of vaccinia, Sc. pombe, human ligase I (Hu1), and human ligase 3 (Hu3). The numbers of amino acid residues separating the motifs are indicated. Residues in the yeast capping enzyme Ceg1 that were found by mutational analysis to be essential for function are shown in shaded boxes. Where these residues are conserved in other family members, they are also shaded. Ceg1 residues judged to be nonessential (i.e., tolerant of alanine substitution) are denoted by dots. Ceg1 positions at which alanine replacement caused a temperature-sensitive growth defect are denoted by Δ.
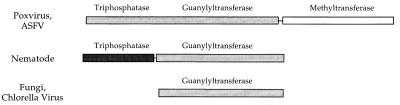
The guanylyltransferases of Saccharomyces cerevisiae (Ceg1; 459 amino acids), Schizosaccharomyces pombe (Pce1; 402 amino acids), Candida albicans (Cgt1; 449 amino acids), and Chlorella virus PBCV-1 (330 amino acids) are monofunctional polypeptides that catalyze GMP transfer to RNA but do not catalyze cap methylation or γ-phosphate cleavage (6–9). In budding yeast, the triphosphatase and methyltransferase functions are encoded separately (6). In contrast, the vaccinia virus capping enzyme is a multifunctional protein that catalyzes all three capping reactions (10). The triphosphatase, guanylyltransferase, and methyltransferase catalytic domains are arrayed in a modular fashion within a single 844-amino acid polypeptide (Fig. 2) (11–16). This trifunctional domain structure also applies to the capping enzyme of African swine fever virus (17).
Figure 2.
Phylogeny of RNA Guanylyltransferases. The organization of functional domains within RNA guanylyltransferases is illustrated in cartoon form. ASFV, African swine fever virus.
We are interested in the evolution of the RNA capping machinery and are focusing on four questions. What structural features are essential for the guanylyltransferase, methyltransferase, and triphosphatase activities? To what extent are these features conserved? How do the essential structural elements illuminate the reaction mechanisms? How have the physical and functional organizations of the component activities diverged from viral to cellular systems? This analysis requires that the essential structural features be defined by mutagenesis and that genes encoding the capping proteins be identified from a wide variety of sources. In practice, the results of mutational analyses can be helpful in genomics-based identification of novel gene family members—i.e., the predictive value of initial protein sequence searches can be enhanced by visual screening for specific essential residues. For example, mutagenesis of the vaccinia capping enzyme helped us to identify the S. cerevisiae ABD1 gene encoding the cellular cap methyltransferase (18). Abd1 is a 436-amino acid monomeric protein that is similar in size and sequence to the C-terminal methyltransferase domain of the vaccinia enzyme. In turn, mutagenesis of Abd1 led to the identification of the cap methyltransferase of Caenorhabditis elegans (19).
In this study, we have performed an extensive structure–function analysis of Ceg1, the RNA guanylyltransferase of S. cerevisiae. The guanylyltransferase activity of Ceg1 is essential for cell viability (20–22). Hence, mutational effects on Ceg1 function in vivo can be evaluated by simple exchange of mutant CEG1 alleles for the wild-type gene. After locating essential amino acids by alanine scanning, we determined by conservative replacements the essential features of individual amino acid side chains. Consideration of the results in light of the crystal structure of the Chlorella virus capping enzyme (5) elucidates the mechanism of catalysis and the basis for nucleotide binding. Moreover, insights gained from the mutational analysis permitted the identification of the cDNA encoding the capping enzyme of C. elegans. The nematode polypeptide is remarkable in that it includes a phosphatase domain that bears no resemblance to the vaccinia triphosphatase domain but, instead, has strong similarity to the superfamily of protein phosphatases that act via a covalent phosphocysteine intermediate.
METHODS AND MATERIALS
Site-Directed Mutagenesis.
Missense mutations in the CEG1 gene were programmed by synthetic oligonucleotides using the two-stage PCR-based overlap extension strategy. An NdeI–BamHI restriction fragment of each PCR-amplified CEG1 gene was inserted into pET16b. The presence of the desired mutation was confirmed in every case by dideoxy sequencing. Restriction fragments of each CEG1 mutant were exchanged with the corresponding segment in the yeast plasmid pGYCE-358 (CEN TRP1 CEG1). Expression of CEG1 in this context is driven by its natural promoter. We sequenced the entire CEG1 insert in each pGYCE plasmid to exclude the occurrence of PCR-generated mutations outside the targeted region.
Test of CEG1 Function by Plasmid Shuffle.
Strain YBS2 (MATa ura3 trp1 lys2 leu2 ceg1::hisG pGYCE-360), which is deleted at the chromosomal CEG1 locus, is viable when it maintains an extrachromosomal copy of CEG1 on a CEN URA3 plasmid (pGYCE-360) (20). YBS2 was transformed with pGYCE-358 plasmids bearing mutant alleles of CEG1. Trp+ transformants were selected on medium lacking tryptophan. Individual colonies were patched on medium lacking tryptophan. Cells from each patch were then streaked on medium containing 0.75 mg/ml fluoroorotic acid (FOA). The plates were incubated at 25°C and 30°C. Mutations scored as lethal were those that did not support colony formation after 7 days. Individual colonies of the viable CEG1 alleles were picked from the FOA plate and patched to yeast extract/peptone/dextrose (YPD). Two isolates of each mutant were tested for growth on YPD agar at 25°C and 37°C.
RESULTS AND DISCUSSION
Alanine-Substitution Mutations Define Essential Residues.
Amino acid residues essential for Ceg1 function were identified by alanine-scanning mutagenesis of the six conserved motifs that define the covalent nucleotidyltransferase superfamily. We reported previously that alanine substitutions at Lys-70 and Gly-73 (motif I), Asp-130 and Glu-132 (motif III), Asp-225 and Gly-226 (motif IV), and Lys-249 and Asp-257 (motif V) were lethal (7, 20). Here, we introduced alanine mutations at 22 new positions within motifs I, IIIa, V, and VI. The CEG1-Ala alleles were tested for in vivo function using the plasmid shuffle procedure (Table 1). Eight of the mutations were lethal: these were at positions Arg-75 (motif I), Phe-151 and Asp-152 (motif IIIa), Lys-247 (motif V), Trp-363, Arg-369, Asp-371, and Lys-372 (motif VI). Thirteen of the mutations had no apparent effect on cell growth. One mutation, Y65A, conferred a temperature-sensitive (ts) growth phenotype.
Table 1.
Effect of alanine-substitution mutations on CEG1 function in vivo
| Motif | Mutation | Growth |
|---|---|---|
| I | Y65A | ts |
| V67A | ++ | |
| C68A | ++ | |
| E69A | ++ | |
| R75A | Lethal | |
| IIIa | R147A | ++ |
| Y148A | ++ | |
| F151A | Lethal | |
| D152A | Lethal | |
| C153A | ++ | |
| N157A | ++ | |
| G158A | ++ | |
| V | K247A | Lethal |
| W248A | ++ | |
| P250A | ++ | |
| V256A | ++ | |
| VI | W363A | Lethal |
| R367A | ++ | |
| R369A | Lethal | |
| D370A | ++ | |
| D371A | Lethal | |
| K372A | Lethal |
YBS2 was transformed with CEN TRP1 plasmids containing the indicated mutant alleles. Trp+ transformants were selected and then streaked on medium containing fluoroorotic acid (FOA) (0.75 mg/ml). The plates were incubated at 25°C and 30°C. Lethal mutations were those that formed no colonies after 7 days. All other alleles supported colony formation in 3 days. Individual colonies were picked from the FOA plate and patched to yeast extract/peptone/dextrose (YPD). Two isolates of each mutant were tested for growth on YPD agar at 37°C. Mutants that formed colonies at both temperatures were scored as two-plus (++). Temperature-sensitive (ts) mutants were those that did not form colonies after 7 days at 37°C.
The results of alanine scanning at 39 residues of the conserved motifs are summarized in Fig. 1. Nineteen nonessential amino acids are denoted by dots above the yeast sequence. Four positions at which alanine replacement resulted in ts growth are denoted by Δ. The 16 essential amino acids are shown in shaded boxes.
Structure–Function Relationships at Essential Residues.
Alanine substitution eliminates the side chain beyond the β-carbon. This mutational approach provides an indication of the essentiality of the side chain for protein function but does not reveal the properties of the missing side chain that are important. This issue can be addressed by introducing conservative substitutions for the essential residues. For example, replacement of Lys-70 by arginine, histidine, or threonine is lethal, implying a strict requirement for lysine as the active site nucleophile in GMP transfer from GTP to RNA (20–22). In the present study, we tested 20 conservative substitutions at 13 essential positions. These included 6 acidic, 5 basic, and 2 aromatic amino acids. (The two essential glycines were not analyzed further.) Instructive results were obtained for each position, as shown in Table 2.
Table 2.
Effect of conservative substitutions on CEG1 function in vivo
| Motif | Mutation | Growth |
|---|---|---|
| I | R75K | Lethal |
| III | D130E | ++ |
| D130N | Lethal | |
| E132D | ++ | |
| E132Q | Lethal | |
| IIIa | F151Y | ++ |
| F151L | Lethal | |
| D152E | ++ | |
| D152N | ts | |
| IV | D225N | Lethal |
| D225E | Lethal | |
| V | K247R | Lethal |
| K249R | Lethal | |
| D257E | ++ | |
| D257N | Lethal | |
| VI | W363F | ++ |
| R369K | Lethal | |
| D371E | ++ | |
| D371N | Lethal | |
| K372R | ++ |
See Table 1 for definitions.
Among the acidic residues, we found that Asp-130 in motif III, Asp-257 in motif V, and Asp-371 in motif VI could be replaced by glutamate, but not by asparagine. Similarly, Glu-132 could be replaced by aspartate, but not by glutamine. We surmise that at each of these positions an acidic side chain is essential for Ceg1 function. Asp-225 of motif IV was strictly essential. Substitution by either asparagine or glutamate was lethal. This is noteworthy, because the equivalent position is a Glu in the vaccinia capping enzyme and the DNA ligases (Fig. 1). Context-dependent steric constraints may account for the failure of the bulkier Glu residue to replace Asp-225. At Asp-152 of motif IIIa, a glutamate substitution was viable, whereas replacement by asparagine resulted in a ts growth phenotype. The fact that D152N cells grew normally at 25°C indicates that an acidic side chain is not essential. Asp-152 may engage in hydrogen bonding, a capacity shared with Glu and Asn, but not with Ala.
Four of the essential basic residues were intolerant of conservative substitutions. Lysine mutations were lethal at Arg-75 in motif I and Arg-369 in motif VI. Similarly, Lys-247 and Lys-249 of motif V could not be substituted by arginine. Only Lys-372 in motif VI was tolerant of replacement by arginine. A requirement for an aromatic residue was evident at Phe-151 of motif IIIa. A F151Y mutant was viable, whereas replacement by leucine was lethal. Trp-363 of motif VI could be replaced by phenylalanine.
Mechanistic Implications.
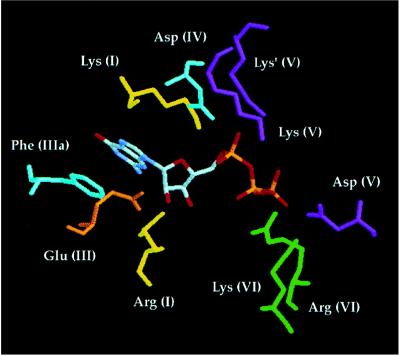
Insights into substrate binding and catalysis emerge when the Ceg1 mutational findings are interpreted in light of the crystal structure of PBCV-1 capping enzyme, which has been solved with GTP bound at the active site and with GMP bound covalently (5).
The ɛ-amino group of the active site Lys in motif I is positioned near the α-phosphate of GTP in the crystal structure, as one might expect (Fig. 3). It is likely that formation of the enzyme–GMP intermediate proceeds through a pentacoordinate phosphorane transition state in which the active site lysine and the β-phosphate are positioned apically. The PBCV-1 enzyme structure reveals a large conformational change in the GTP-bound enzyme, from an “open” to a “closed” state, that reorients the phosphates for in-line attack by the lysine (5). Adoption of the closed conformation brings motif VI into direct contact with the β- and γ-phosphates of GTP and also moves the essential Asp at the end of motif V (Asp-257 in Ceg1) close to the β-phosphate. Motif VI makes several key contacts (Fig. 3). The motif VI Arg residue (Arg-369 in Ceg1) interacts with the β-phosphate and also hydrogen bonds to the essential Asp side chain situated nearby in motif VI (Asp-371 in Ceg1). A requirement for bidentate hydrogen bonding by Arg-369 would explain why lysine substitution at this position is lethal. The essential Lys of motif VI (Lys-372 in Ceg1) contacts the γ-phosphate of GTP. As noted above, Arg can functionally substitute for Lys at this position.
Figure 3.
Interactions between essential amino acid side chains and GTP at the capping enzyme active site. The figure shows the interactions of essential amino acids with GTP in the context of the closed conformation of the PBCV-1 capping enzyme–GTP cocrystal (5). The amino acids are indicated in three-letter code and by the motif in which they reside—e.g., the active site lysine nucleophile is Lys (I).
Contacts with the α-phosphate of GTP are made by the two essential basic residues in motif V. The first lysine of the KxK sequence (Lys-247 in Ceg1) is hydrogen-bonded to the α-phosphate in the open conformation; this contact is attenuated during the conformational change. In the closed form of the enzyme, the distal Lys of motif V (Lys-249 in Ceg1) hydrogen bonds with the α-phosphate (this residue is denoted as Lys′ in Fig. 3). In the covalent enzyme–GMP intermediate, the α-phosphate oxygens interact with both positively charged side chains and a divalent cation (5). We hypothesize that the two lysines and the divalent cation enhance catalysis by stabilizing the equatorial phosphate oxygens in the transition state.
The essential Asp residue of motif IV (Asp-225 in Ceg1) hydrogen bonds to the active site lysine residue (Fig. 3). We speculate that this side chain acts as a general base to withdraw a proton from the -NH2 of lysine during formation of the P–N bond to GMP. The phosphoamide bond should, in principle, be stabilized when the amide nitrogen is unprotonated. The Asp chain would donate a proton back to the Lys, leaving the group as the 5′ diphosphate of RNA attacks the enzyme–GMP intermediate to form the cap structure.
Three of the side chains we found to be essential in Ceg1 (the analogs of Arg-87, Glu-131, and Phe-146) make direct contact with the nucleoside moiety of GTP in the PBCV-1 enzyme structure (5). The Arg residue of motif I hydrogen bonds with the ribose 3′ OH (Fig. 3). In addition, this Arg side chain hydrogen bonds with the essential Asp of motif III. (The Asp itself does not interact with GTP.) A requirement for bidentate contacts by Arg-87 would explain why lysine substitution is lethal. It would also explain why the Arg of motif I is coordinately conserved with the Asp of motif III in the capping enzymes of yeast, African swine fever virus, and PBCV-1 and in the DNA ligases. This Arg is conspicuously not conserved in motif I of the poxvirus-encoded capping enzymes (where it is a Pro or Gly) and neither is the Asp in motif III (Fig. 1). The absence of Arg implies either that the vaccinia capping enzyme does not require the 3′ OH sugar interactions made by other guanylyltransferases and by DNA ligase (4) or that it achieves these contacts via divergent structural elements. The essential Glu side chain of motif III hydrogen bonds with the ribose 2′ OH of GTP, whereas the essential Phe of motif IIIa is stacked on the guanine base. The mutational effects confirm the functional importance of these contacts.
Identification of a Capping Enzyme from C. elegans.
Guanylyltransferase activities have been isolated from several higher eukaryotes (23); however, no genes encoding these enzymes have been identified to date. We found that the structure–function relationships revealed by the present mutational analysis of Ceg1 conferred predictive power to a genomics-based search for candidate capping enzymes in higher eukaryotes. By imposing complete conservation of residues essential for Ceg1 function, we identified the capping enzyme from C. elegans. The C. elegans C03D6.3 gene product is a 573-amino acid (66-kDa) polypeptide derived by conceptual translation after computer-modeled joining of 11 exons distributed over 2.4 kb of genomic DNA (GenBank accession no. Z75525). This is in good agreement with the sizes of the other higher eukaryotic guanylyltransferases [human (68 kDa), rat liver (69 kDa), calf thymus (65 kDa), brine shrimp (73 kDa), and wheat germ (77 kDa)], which were determined by SDS/PAGE analysis of the covalent enzyme–GMP catalytic intermediate (23–28).
To confirm that this polypeptide is truly the product of a single gene, and that the gene is expressed in vivo, we generated cDNA clones by reverse transcription–PCR amplification of C. elegans total RNA using oligonucleotide primers flanking the predicted translation initiation and stop codons. These primers were also used to amplify specific cDNAs from a C. elegans cDNA library. Both approaches led to the isolation of 1.7-kbp cDNAs. We determined by sequencing several cDNA clones that the exons were spliced as predicted by the nematode genome project and that the ORF was continuous. Analysis of the protein sequence shows that the nematode capping enzyme consists of an N-terminal phosphatase domain fused to a C-terminal guanylyltransferase domain (Fig. 2).
Alignment of the sequence of the C terminus of the C. elegans protein with the guanylyltransferases encoded by S. cerevisiae, Sc. pombe, C. albicans, and Chlorella virus PBCV-1 reveals conservation at 75/301 positions (Fig. 4). The nematode protein contains all six defining motifs of the covalent nucleotidyltransferase superfamily (shaded boxes in Fig. 4). All 16 residues that we have identified as essential for Ceg1 function are strictly conserved in the C. elegans protein. Moreover, the amino acids of the Chlorella virus capping enzyme that contact GTP in the cocrystal (arrowheads in Fig. 4) are conserved in the nematode protein; these include several amino acids outside of the six motifs.
Figure 4.
A putative capping enzyme from C. elegans. The amino acid sequence of the C. elegans C03D6.2 gene product from residues 273 to 573 is aligned with the sequences of the guanylyltransferases of Sc. pombe (spo), S. cerevisiae (sce), C. albicans (cal), and Chlorella virus PBCV-1 (chv). Gaps in the sequence are indicated by dashes (-). Amino acids conserved in all five proteins are denoted by asterisks. The six nucleotidyltransferase motifs are shown in shaded boxes. Residues in proximity to the GTP moiety in the PBCV-1 cocrystal are indicated by arrowheads.
A Putative RNA Triphosphatase Domain of the C. elegans Capping Enzyme.
The N-terminal portion of the C. elegans capping enzyme contains the (I/V)HCxAGxGR(S/T)G signature motif of the dual-specificity protein phosphatase/protein tyrosine phosphatase enzyme family (Fig. 5). These proteins catalyze phosphoryl transfer from a protein phosphomonoester substrate to the thiol of a cysteine on the enzyme to form a covalent phosphocysteine intermediate (29). The intermediate is then attacked by water to liberate phosphate. The cysteine within the signature motif is the active site of phosphoryl transfer and is thus essential for reaction chemistry. The conserved Arg side chain makes bidentate contacts with the phosphate oxygens and is also critical for phosphatase activity (29).
Figure 5.
A phosphatase domain in the C. elegans capping enzyme. The amino acid sequence of the C. elegans capping enzyme (Cel CE) from residues 59 to 171 is aligned with the 167-amino acid baculovirus-encoded protein phosphatase (Bac PP; GenBank accession no. M96763) and the sequences of two phosphatase-like C. elegans gene products: T23G7.5 (GenBank accession no. Z68319) and F54C8.4 (GenBank accession no. Z22178). Gaps in the sequences are indicated by dashes (-). Amino acids conserved in all four proteins are denoted by asterisks. The protein phosphatase signature motif is highlighted in the shaded box. The active site cysteine is in boldface type.
An alignment of the amino segment of the C. elegans capping enzyme with the baculovirus-encoded protein phosphatase (34) is shown in Fig. 5. (Two other predicted C. elegans gene products with homology to the capping enzyme N terminus are included in the alignment.) The similarity extends well beyond the signature motif and includes a conserved aspartate (Asp-64) located upstream of the putative active site cysteine (Cys-124). Biochemical and structural studies of several prototypal protein phosphatases have shown that this aspartate acts as a general acid during formation of the cysteinyl phosphate intermediate (29, 30). The Asp is situated within a WxD motif in numerous protein tyrosine phosphatases, and this is also the case in the nematode protein (Fig. 5). This extent of conservation makes it likely that the C. elegans enzyme is a phosphatase. More specifically, we suggest that the N-terminal domain of the protein is an RNA triphosphatase that removes the γ-phosphate of triphosphate-terminated RNA.
Available biochemical data support the idea that higher eukaryotes encode a bifunctional capping enzyme with triphosphatase and guanylyltransferase activities. The guanylyltransferases from rat liver and brine shrimp copurify with an RNA triphosphatase activity (28, 31). In the case of the brine shrimp protein, Yagi et al. (28) showed that both catalytic activities reside within a single 73-kDa polypeptide that was converted by partial proteolysis into catalytically active domains: a 20-kDa triphosphatase module could be separated from a 44-kDa fragment guanylyltransferase domain. The sizes of these two active fragments are consistent with those of the two putative domains of the nematode capping enzyme.
It is noteworthy that the RNA triphosphatase activity of the rat liver capping enzyme is optimal in the absence of a divalent cation and that EDTA has no effect on γ-phosphate cleavage (31, 33). A distinctive characteristic of the protein phosphatases to which the C. elegans protein is related is that they do not require metal cofactors for catalysis (29).
Phylogeny of the Guanylyltransferases.
The results of this study underscore the conserved structural basis for covalent nucleotidyl transfer by cellular and DNA virus-encoded guanylyltransferases. The guanylyltransferases can now be subgrouped according to two criteria: (i) conservation of specific residues within the six motifs and (ii) sequence similarities outside the motifs. Based on intra-motif conservation, we delineate two subgroups: the first consists of the enzymes from fungi, C. elegans, Chlorella virus, and African swine fever virus, and the second consists of the poxvirus enzymes (of vaccinia, Shope fibroma virus, and molluscum contagiosum virus). The first group contains four essential amino acids (Arg in motif I, Asp in motif III, Asp in motif IIIa, and Trp in motif VI) that are replaced by unrelated functional groups in the poxvirus proteins. The essentiality of these four amino acids for achieving RNA capping is apparently context dependent. (Three of the four residues—Arg in motif I, Asp in motif III, and Asp in motif IIIa—are also conserved in the DNA ligases.)
On the basis of sequence conservation outside the motifs, we have placed the enzymes from S. cerevisiae, Sc. pombe, C. albicans, C. elegans, and Chlorella virus into a discrete subfamily. A sequence alignment highlights two motifs that are unique to these capping enzymes, which we have designated motif P and motif Vc (Fig. 4). Motif P is a proline-containing segment [FPGx(Q/N)PVS(L/F/I)] located 17–18 amino acids upstream of the active site lysine. Motif Vc [(K/R)I(I/V)EC] is situated between motifs V and VI. The alignment in Fig. 4 provides a blueprint for further structure–function analysis of conserved functional groups in the new motifs and at conserved positions outside the motifs.
Evolution of the Capping Apparatus.
The physical organizations of the component activities of the capping apparatus have diverged in viral and cellular systems. The poxviruses and African swine fever virus have collected all three active sites within a single multidomain polypeptide. Fungi and higher eukaryotes have segregated the guanylyltransferase and methyltransferase functions to distinct gene products. Yet, it is clear from the few genes available that the guanylyltransferase and methyltransferase proteins of fungi and C. elegans are conserved with respect to the corresponding vaccinia domains. In general, the cellular proteins are more similar to each other than to the vaccinia protein, suggesting that the poxviruses diverged earlier from ancestral nucleotidyl transferase and methyltransferase domains.
Lower and higher eukaryotes differ clearly with respect to the physical linkage of the guanylyltransferase and triphosphatase functions. Yeasts encode a monofunctional guanylyltransferase, whereas C. elegans encodes a bifunctional phosphatase-guanylyltransferase. As discussed above, biochemical evidence suggests that bifunctional triphosphatase-guanylyltransferase enzymes are present in many higher eukaryotes. The linear arrangement of N-terminal phosphatase and C-terminal guanylyltransferase domains in the C. elegans protein is similar to that of the vaccinia capping enzyme (11, 13, 35). Yet, the sequences of the vaccinia and C. elegans N-terminal domains are entirely dissimilar. Moreover, the biochemical properties of the vaccinia triphosphatase differ from those of the triphosphatase from higher eukaryotes in one key respect. The rat liver and brine shrimp triphosphatases require no divalent cation for activity (in fact, they are inhibited by divalent cations), whereas the triphosphatase activity of the vaccinia capping enzyme depends absolutely on a divalent cation cofactor (13, 31–33). The sequence of the C. elegans protein implies that γ-phosphate cleavage occurs through a phosphoenzyme intermediate. Strenuous efforts to detect a phosphoenzyme intermediate for the vaccinia triphosphatase have been unsuccessful, which suggests that covalent catalysis does not apply in this case (S.S., unpublished data). Mutational analysis of the vaccinia triphosphatase provides additional evidence for a distinct mechanism. We have pinpointed four acidic side chains that are essential for catalysis by vaccinia triphosphatase and are conserved among the poxvirus and African swine fever virus enzymes (ref. 13; A. Martins, Y. Yu, and S.S., unpublished data). These acidic residues are likely to bind the essential metal ion(s). An RNA triphosphatase has been isolated from S. cerevisiae, which also depends completely on a divalent cation cofactor (33). We surmise that higher eukaryotes have diverged from vaccinia and yeast with respect to mechanism and structure of the triphosphatase component of the capping machinery.
Note. After this paper was submitted, Takagi et al. (36) reported that an N-terminal fragment of the C. elegans capping enzyme polypeptide, from residues 1 to 236, possesses RNA triphosphatase activity.
Acknowledgments
We thank Dale Wigley, Kjell Hakansson, and Aidan Doherty for helpful discussions and for providing the coordinates of the PBCV-1 capping enzyme-GTP cocrystal. This work was supported by National Institutes of Health Grant GM52470.
References
- 1.Shuman S. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 2.Shuman S, Hurwitz J. Proc Natl Acad Sci USA. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuman S, Schwer B. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 4.Subramanya H S, Doherty A J, Ashford S R, Wigley D B. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 5.Hakansson K, Doherty A J, Shuman S, Wigley D B. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 6.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 7.Shuman S, Liu Y, Schwer B. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada-Okabe T, Shimmi O, Doi R, Mizumoto K, Arisawa M, Yamada-Okabe H. Microbiology. 1996;142:2515–2523. doi: 10.1099/00221287-142-9-2515. [DOI] [PubMed] [Google Scholar]
- 9.Ho C K, Van Etten J L, Shuman S. J Virol. 1996;70:6658–6664. doi: 10.1128/jvi.70.10.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkatesan S, Gershowitz A, Moss B. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 11.Cong P, Shuman S. Mol Cell Biol. 1995;15:6222–6231. doi: 10.1128/mcb.15.11.6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myette J R, Niles E G. J Biol Chem. 1996;271:11936–11944. doi: 10.1074/jbc.271.20.11936. [DOI] [PubMed] [Google Scholar]
- 13.Yu L, Shuman S. J Virol. 1996;70:6162–6168. doi: 10.1128/jvi.70.9.6162-6168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higman M A, Christen L A, Niles E G. J Biol Chem. 1994;269:14974–14981. [PubMed] [Google Scholar]
- 15.Mao X, Shuman S. J Biol Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 16.Mao X, Shuman S. Biochemistry. 1996;35:6900–6910. doi: 10.1021/bi960221a. [DOI] [PubMed] [Google Scholar]
- 17.Pena L, Yanez R, Revilla Y, Vinuela E, Salas M L. Virology. 1992;193:319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- 18.Mao X, Schwer B, Shuman S. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S P, Shuman S. J Biol Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- 20.Schwer B, Shuman S. Proc Natl Acad Sci USA. 1994;91:4328–4332. doi: 10.1073/pnas.91.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fresco L D, Buratowski S. Proc Natl Acad Sci USA. 1994;91:6624–6628. doi: 10.1073/pnas.91.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibagaki Y, Gotoh H, Kato M, Mizumoto K. J Biochem (Tokyo) 1995;118:1303–1309. doi: 10.1093/oxfordjournals.jbchem.a125023. [DOI] [PubMed] [Google Scholar]
- 23.Mizumoto K, Kaziro Y. Prog Nucleic Acid Res Mol Biol. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- 24.Venkatesan S, Moss B. Proc Natl Acad Sci USA. 1982;79:340–344. doi: 10.1073/pnas.79.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuman S. J Biol Chem. 1982;257:7237–7245. [PubMed] [Google Scholar]
- 26.Wang D, Furuichi Y, Shatkin A. Mol Cell Biol. 1982;2:993–1001. doi: 10.1128/mcb.2.8.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishikawa Y, Chambon P. EMBO J. 1982;1:485–492. doi: 10.1002/j.1460-2075.1982.tb01195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagi Y, Mizumoto K, Kaziro Y. J Biol Chem. 1984;259:4695–4698. [PubMed] [Google Scholar]
- 29.Denu J M, Stuckey J A, Saper M A, Dixon J E. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 30.Denu J M, Lohse D L, Vijayalakshmi J, Saper M A, Dixon J E. Proc Natl Acad Sci USA. 1996;93:2493–2498. doi: 10.1073/pnas.93.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yagi Y, Mizumoto K, Kaziro Y. EMBO J. 1983;2:611–615. doi: 10.1002/j.1460-2075.1983.tb01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shuman S, Surks M, Furneaux H, Hurwitz J. J Biol Chem. 1980;255:11588–11598. [PubMed] [Google Scholar]
- 33.Itoh N, Mizumoto K, Kaziro Y. J Biol Chem. 1984;259:13930–13936. [PubMed] [Google Scholar]
- 34.Hakes D J, Martell K J, Zhoa W, Massung R F, Esposito J J, Dixon J E. Proc Natl Acad Sci USA. 1993;90:4017–4021. doi: 10.1073/pnas.90.9.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myette J R, Niles E G. J Biol Chem. 1996;271:11945–11952. doi: 10.1074/jbc.271.20.11945. [DOI] [PubMed] [Google Scholar]
- 36.Takagi T, Moore C R, Diehn F, Buratowski S. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]