Abstract
Aflatoxin B1 (AFB1) is a potent human carcinogen implicated in the etiology of hepatocellular carcinoma. Upon metabolic activation to the reactive epoxide, AFB1 forms DNA adducts primarily at the N7 position of guanines. To elucidate more fully the molecular mechanism of AFB1-induced mutagenesis, an intercalation inhibitor was designed to probe the effects of intercalation by AFB1 epoxide on its reaction with DNA. DNA duplexes were prepared consisting of a target strand containing multiple potentially reactive guanines and a nontarget strand containing a cis-syn thymidine-benzofuran photoproduct. Because the covalently linked benzofuran moiety physically occupies an intercalation site, we reasoned that such a site would be rendered inaccessible to AFB1 epoxide. By strategic positioning of this intercalation inhibitor in the intercalation site 5′ to a specific guanine, the adduct yield at that site was greatly diminished, indicating that intercalation by AFB1 epoxide contributes favorably to adduct formation. Using this approach it has been possible to simplify the production of site-specifically modified oligonucleotides containing AFB1 adducts in the sequence context of a p53 mutational hotspot. Moreover, we report herein isolation of site-specifically AFB1-modified oligonucleotides in sequences containing multiple guanines. Use of intercalation inhibitors will facilitate both investigation of the ability of other carcinogens to intercalate into DNA and the synthesis of specific carcinogen-DNA adducts.
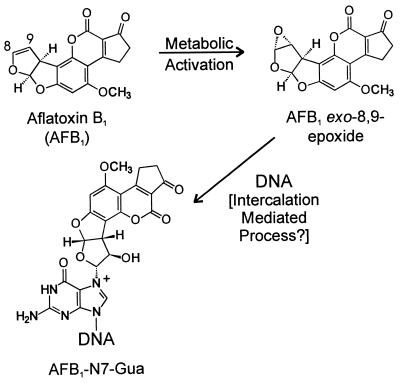
Exposure to aflatoxin B1 (AFB1) and infection by hepatitis B virus are known risk factors for hepatocellular carcinoma (1). The molecular mechanism of human hepatocellular carcinogenesis is poorly defined, but AFB1 is known to be a potent mutagen and hence may induce the mutations that appear in end-stage tumors. Upon metabolic activation to the exo-8,9-epoxide, AFB1 reacts almost exclusively with the N7 of guanine to form guanine DNA adducts (AFB1-N7-Gua) (Fig. 1) (2). Analysis of bacterial mutational spectra and phage genomes containing a specific AFB1-N7-Gua adduct reveals G→T substitutions as the predominant mutagenic event (3, 4). Interestingly, approximately 50% of hepatocellular carcinomas in portions of eastern Asia and sub-Saharan Africa, where exposure to AFB1 through contaminated food is a frequent event, possess G:C→T:A transversions in the third position of codon 249 of the p53 tumor suppressor gene (5–7).
Figure 1.
Metabolic activation and DNA binding of AFB1. AFB1 is oxidized by cytochrome P450 predominantly to the exo-8,9-epoxide, which reacts almost exclusively with guanine residues in DNA. The reaction is believed to be preceded by intercalation of the epoxide specifically on the 5′ face of a target guanine residue.
A recent advance in chemical synthesis has facilitated the production of AFB1 epoxide, which has enabled investigations of the mechanism of the reaction of AFB1 epoxide with DNA (8). Current evidence suggests that formation of AFB1-N7-Gua DNA adducts proceeds through a transition state in which the AFB1 epoxide is intercalated on the 5′ side of the target guanine. This hypothesis is supported by the observation that the reactivity of AFB1 epoxide with double-stranded B-form DNA is greatly enhanced as compared with single-stranded DNA or alternative duplex structures (9, 10). From NMR structural studies on the adduct, it is speculated that 5′ intercalation facilitates adduct formation by positioning the epoxide for in-line nucleophilic attack by the guanine N7 (11, 12). Additional evidence for a 5′ intercalation event arises from stoichiometric studies with the two self-complementary hexamers ATCGAT and ATGCAT. Reaction with excess AFB1 epoxide yields only a 1:1 ratio of AFB1 to the duplex oligonucleotide (ATCGAT)2, where the two guanines share the same 5′ intercalation site; in contrast, in the case where distinct 5′ intercalation sites exist in each strand, a 2:1 AFB1/(ATGCAT)2 ratio is observed (13). Furthermore, modifications of the ring systems in AFB1 that decrease planarity and therefore intercalation ability, a situation that exists with aflatoxin G1, decrease affinity for DNA and result in lower reactivity (14). Finally, addition of the intercalating agent ethidium bromide to target DNA before treatment with AFB1 epoxide greatly reduces guanine reactivity (9). These experiments, coupled with stopped-flow kinetic analysis, support a model in which the intercalated intermediate provides a kinetic and entropic advantage for productive reaction with DNA over hydrolysis (15). Moreover, in the absence of this favorable interaction, AFB1 epoxide can be readily hydrolyzed to the inactive AFB1 diol.
Another agent that intercalates before its reaction with DNA is psoralen (16). Detailed structural analysis of psoralen thymidine adducts reveal that both the cis-syn furan-side monoadduct and the crosslink are intercalated in the DNA duplex (17). Previous work in our laboratory has focused on the total synthesis of a psoralen thymidine monoadduct and its solid-phase incorporation into oligonucleotides (18, 19). A structurally analogous model compound, a cis-syn thymidine benzofuran photoproduct, also was synthesized that possessed the same stereo- and regiochemistry as the cis-syn psoralen furan-side thymidine monoadduct (18); these constraints dictate that the benzofuran moiety occupies an intercalation site when situated in duplex DNA, raising the possibility that such a molecule could be used to inhibit subsequent intercalation by other molecular species.
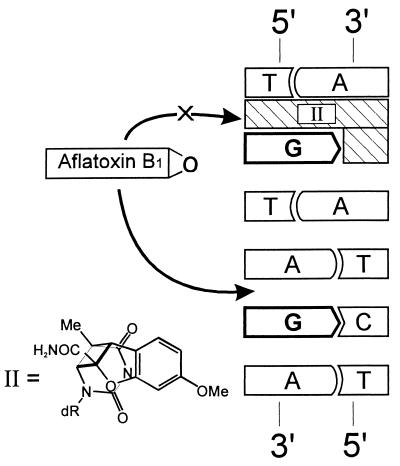
The goals of this work were to examine the dependence of AFB1 adduct formation on intercalation and to exploit this property to manipulate the sites of adduct formation. The construction of oligonucleotides and genomes containing AFB1-N7-Gua adducts at specific sites would facilitate understanding of the molecular origin of the observed p53 mutational hotspot in hepatocellular carcinoma. Study of the chemical basis of this hotspot has been hampered by the presence of numerous guanines in the nucleotide sequence surrounding codon 249. Due to the formidable obstacle of resolving and purifying highly labile AFB1 adducts from complex mixtures of multiple reaction products, the synthesis of site-specifically modified AFB1 adducts thus far has been limited to targets containing a single target guanine (4, 13). In light of the above observations suggesting that intercalation of AFB1 epoxide precedes covalent bond formation, we reasoned that physical occupation of an intercalation site with a thymidine-benzofuran photoproduct would inhibit intercalation by AFB1 epoxide and thereby prevent the formation of an AFB1 adduct at the proximal guanine (Fig. 2). Could this strategy be used to protect one or more reactive sites selectively to achieve an high yield of a given specific adduct? In this paper, we present evidence that the reaction of DNA with AFB1 epoxide is modulated by inhibiting intercalation. Moreover, judicious placement of intercalation inhibitors in a human p53 gene-derived sequence results in simplification of the local adduct spectrum and improves the yield of individual adducts.
Figure 2.
Experimental scheme. Occupation of an intercalation site 5′ to a guanine should prevent intercalation by AFB1 epoxide and reduce reactivity at the protected site. The covalently attached intercalation inhibitor (designated II), a cis-syn thymidine-benzofuran photoproduct, is positioned in the complementary strand. The benzofuran moiety is positioned on the 3′ side of the T component of the cis-syn thymidine-benzofuran adduct.
MATERIALS AND METHODS
Synthesis of Intercalation Inhibitor.
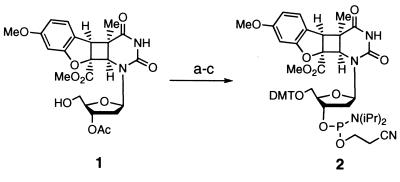
The synthesis of the suitably protected thymidine-benzofuran phosphoramidite is shown in Fig. 3. The synthesis of deoxynucleoside 1 has been previously described (18). Treatment of deoxynucleoside 1 with 4,4′-dimethoxytrityl chloride in the presence of silver nitrate led to rapid protection of the 5′ hydroxyl. Removal of the acetate protecting group with 5% 1,8-diazabicyclo[5.4.0]undec-7-ene in freshly distilled methanol afforded the free 3′ hydroxyl. Phosphitylation using standard conditions gave phosphoramidite 2. All reactions were performed under an argon atmosphere and at room temperature. The compounds were characterized by 1H, 13C, and 31P NMR (when applicable) and high-resolution MS.
Figure 3.
Conditions for synthesis of the cis-syn thymidine-benzofuran phosphoramidite with yields in parenthesis: (a) 4,4′-dimethoxytrityl chloride, AgNO3, collidine, DMF (84%); (b) 5% 1,8-diazabicyclo[5.4.0]undec-7-ene in methanol (98%); (c) bis(isopropylamine)-2-cyanoethyl phosphoramidite, diisopropylammonium tetrazolide, CH2Cl2 (74%).
Oligonucleotides.
All oligonucleotides were synthesized on an Applied Biosystems model 391 DNA synthesizer. Standard conditions were used except for oligonucleotides containing the intercalation inhibitor or 7-deaza-dG. For the thymidine-benzofuran phosphoramidite, the coupling time was extended to 15 min. The coupling yield for the modified phosphoramidite 2 was 85–92% as determined by trityl release. Oligonucleotides containing 7-deaza-dG were oxidized by using a 0.5 M solution of (1S)-(+)-(10-camphorsulfonyl)oxaziridine in anhydrous acetonitrile to prevent degradation of the 7-deazaguanine residues. The oxidation step was extended to 5 min to ensure complete oxidation. All oligonucleotides were deprotected with NH4OH for 18 hr at 55°C and subsequently purified by C18 reversed-phase HPLC before use. Oligonucleotides were annealed by heating for 5 min at 80°C in 100 mM NaHPO4 (pH 7.0), 100 mM NaCl. The samples were allowed to cool at room temperature for 1 hr and then at 4°C for 1 hr.
AFB1 Epoxide Reaction with DNA.
AFB1 epoxide was generated as reported (13). Reactions contained 10 nmol duplex DNA in a total volume of 20 μl (0.5 mM DNA). In a 4°C room, samples were treated with 1–2.5 molar equivalents of AFB1 epoxide, mixed by vortexing for 5 min, and diluted with buffer, and then the AFB1-diol was removed by extraction with CH2Cl2. Note: Everything contacting AFB1 or AFB1 epoxide solutions was treated with bleach to inactivate any residual toxin.
HPLC Purification.
Samples were loaded onto a C18 reversed-phase analytical column (Beckmann ODS) and eluted with a gradient of 10–30% B over 60 min (A = 0.1 M NH4OAc in H2O; B = 0.1 M NH4OAc in 50% H2O/acetonitrile) with UV monitoring at both 260 nm and 360 nm. Samples were desalted on a Sep-Pak C18 cartridge at 4°C and eluted with 50% acetonitrile.
32P-Labeling and Cleavage of AFB1-Treated Oligonucleotides.
Purified oligonucleotides were 5′-end labeled with T4 polynucleotide kinase and γ-32P ATP (New England Nuclear, 6,000 Ci/mmol) in 70 mM Tris⋅HCl (pH 7.6) for 5 min at 37°C. Unincorporated label was removed by centrifugation of the sample through a Sephadex G25 column. Assays to identify the position of adducts formed were based upon the known lability of AFB1 adducts to conditions of alkali and heat (20). One hundred microliters of 10% piperidine was added and heated for 30 min at 90°C. Piperidine was removed by lyophilization, and samples subsequently were electrophoresed through 20% 7 M urea polyacrylamide gels and visualized using a Molecular Dynamics PhosphorImager. The mobilities of bands from samples were compared with those of marker bands generated by the Maxam–Gilbert G-specific reaction (21).
RESULTS
Evidence accumulated over the past several years suggests that intercalation of AFB1 epoxide greatly enhances the reactivity of the potent carcinogen with DNA (9–13). Accordingly, if it were possible to reduce the likelihood of intercalation at a given site, one would expect to diminish substantially the adduct yield at that site (Fig. 2). A series of duplex oligonucleotides was designed to examine this possibility (Table 1). In each case, one strand contained two or more guanines that were potential adduction sites. In the complementary strand, a cis-syn thymidine-benzofuran intercalation inhibitor was situated to occupy the intercalation site immediately 5′ to one of the guanines. Chemical synthesis of the phosphoramidite of the intercalation inhibitor (Fig. 3) enabled us to control precisely both the number and position of the intercalation inhibitors in the target duplexes. Upon reacting the various DNA duplexes with AFB1 epoxide, HPLC analysis was used to separate and isolate the reaction products. The standard elution conditions were sufficient to denature the duplexes, resolving the target strands from their complements. By this method, the complementary strands, which contained the unnatural thymidine-benzofuran moieties, could be removed, and the single-stranded site-specifically AFB1-modified oligonucleotides could be isolated.
Table 1.
Oligonucleotides used in this work
| H | 5′-ATAGATTGTA-3′ |
| QC | 3′-TATCTAACAT-5′ |
| QT | 3′-TATTTAACAT-5′ |
| Q7 | 3′-TATXTAACAT-5′ |
| Q3 | 3′-TATCTAAXAT-5′ |
| GG | 5′-TATAGGTTAT-3′ |
| R0 | 3′-ATATCCAATA-5′ |
| R6 | 3′-ATATXCAATA-5′ |
| R5 | 3′-ATATCXAATA-5′ |
| P53 | 5′-CCGGAGGCC-3′ |
| S0 | 3′-T77TCTCC7-5′ |
| S6 | 3′-T77XCTCC7-5′ |
| S5 | 3′-T77CXTCC7-5′ |
7, 7-deaza-dG; X, thymidine-benzofuran photoproduct (intercalation inhibitor).
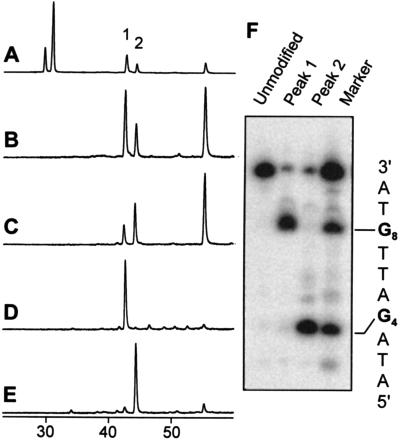
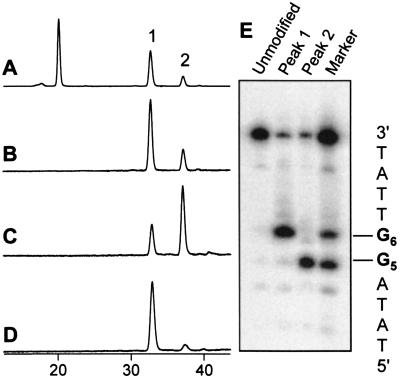
The simplest model system consisted of a 10-bp oligonucleotide duplex containing only two G residues in one strand separated by three nucleotides (Table 1, H/QC). In the control sequence, reaction with AFB1 epoxide afforded two peaks corresponding to AFB1 monoadducts and one peak representing the diadduct, as determined by the ratio of their UV absorbances at 360 nm and 260 nm (Fig. 4 A and B). The two monoadduct peaks were isolated, desalted, and then 5′-32P end-labeled by using polynucleotide kinase. The known alkaline lability of AFB1-N7-Gua adducts (20) was used to determine the identity of each peak. Electrophoresis of the piperidine-treated samples revealed that peak 1 was the G8 AFB1 adduct while peak 2 was the G4 adduct (Fig. 4F). It is known that the reaction of AFB1 epoxide with guanines in duplex DNA is not random, but rather displays sequence specificity. Although the basis for this discrimination is not well understood, a systematic investigation by Loechler and coworkers (22) empirically has determined reactivity rules based on the immediate 5′ and 3′ neighboring bases. In the first model system, the two adducts formed in a ratio of 2.4:1, in close agreement to the predicted 2:1 ratio.
Figure 4.
Intercalation inhibition in duplex (H/Q) series. Only molecules containing AFB1 have UV absorbance at 360 nm. (A) 260 nm HPLC trace of the AFB1 reaction with H/QC. The peaks near 30 min are the unreacted H and QC oligonucleotides, respectively. Peaks 1 and 2 are AFB1 monoadducts, and the peak near 55 min is an AFB1 diadduct. (B) 360 nm trace of H/QC. (C) 360 nm trace of H/QT reaction. (D) 360 nm trace of H/Q7. (E) 360 nm trace of H/Q3. (F) Electrophoretic analysis: piperidine cleavage of purified peaks 1 and 2 and comparison to markers generated by the Maxam–Gilbert G-reaction. Piperidine cleavage of the untreated duplex is shown in the lane labeled unmodified.
Upon altering the complementary strand to position an intercalation inhibitor in the 5′ intercalation site of G4 (H/Q7), the ratio of observed products was dramatically shifted; essentially no monoadducts derived from G4 or diadducts were observed (Fig. 4D). Conversely, placement of the intercalation inhibitor opposite G8 (H/Q3) greatly reduced the extent of reaction at G8 such that only 11% of the monoadducts arose from G8 (Fig. 4E). The asymmetry in the protection afforded is likely due to the inherently higher reactivity of G8, making total abolition of reactivity at that site more difficult.
Because positioning the intercalation inhibitor to occupy the intercalation site 5′ to a target guanine necessarily forms a G-T mismatch at that site, the effect of a mismatch alone (H/QT) on adduct distribution was examined. Interestingly, substitution of a G-T base pair for a G-C actually resulted in increased reactivity of the mismatched G (compare Fig. 4 B and C). Thus, the observed diminished reactivity upon introduction of an intercalation inhibitor can be attributed to the presence of an intercalated moiety and not merely distortions resulting from a mismatched base pair.
In the second model system, we examined reactivity and inhibition in oligonucleotides containing two adjacent guanines (GG). In the absence of an intercalation inhibitor (GG/R0), two peaks were observed in the HPLC trace, reflecting the two different monoadducts (Fig. 5 A and B). Loechler’s rules (22) predict that the two adducts should form in a 3.25:1 ratio, which agrees well with the observed product ratio of 3.3:1 (Fig. 5B). Interestingly, there were no diadduct peaks present in the trace (data not shown). This observation is consistent with the neighbor-exclusion principle that has been postulated to govern the ability of molecules to intercalate at adjacent sites (23, 24). Initial intercalation and reaction of an AFB1 epoxide molecule at one guanine results in inhibition of intercalation at the other adjacent guanine, thereby preventing the formation of a second adduct. Nevertheless, placement of the intercalation inhibitor at either of the two guanine sites (GG/R5 and GG/R6) resulted in an increase in the yield of adduct at the nonprotected guanine (Fig. 5 C and D).
Figure 5.
Intercalation inhibition in duplexes GG/R. (A) 260 nm HPLC trace of reaction with GG/R0. Large peak near 20 min is R0. Peaks 1 and 2 are AFB1 monoadducts. (B) 360 nm trace of GG/R0. (C) 360 nm trace of GG/R5. (D) 360 nm trace of GG/R6. (E) Piperidine cleavage of purified peaks 1 and 2.
Because the AFB1 adducts seemed to exhibit neighbor-exclusion properties, the benzofuran moiety was tested to see if it possessed similar characteristics. Duplexes were prepared with an intercalation inhibitor occupying the intercalation sites either 3′ to or 5′ to the actual target intercalation site (data not shown). Positioning the intercalation inhibitor 3′ to the target site had approximately the same effect as occupying the target site itself, whereas placement of the intercalation inhibitor to the 5′ side had no effect.
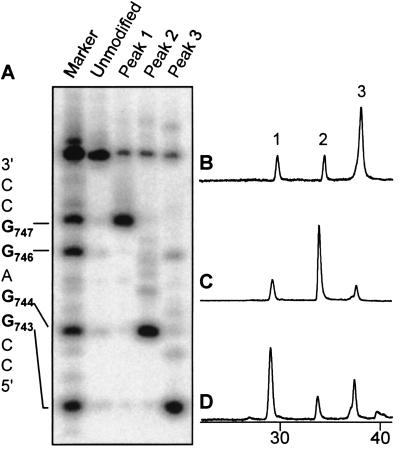
Finally, a sequence representing nucleotides 741–749 of the human p53 cDNA, 5′ CCGGAGGCC (P53), was tested (Fig. 6). In the complementary strands, guanine bases were substituted with 7-deaza-dG to prevent formation of additional AFB1 adducts in this strand; such adducts would complicate analysis of the adduct spectrum. Treatment of a control duplex (P53/S0) revealed three HPLC peaks that corresponded to AFB1 monoadducts (Fig. 6B). After 32P labeling and alkali cleavage, it was determined that peak 1 contained exclusively the G747 adduct, peak 2 contained the G744 adduct, and peak 3 was a mixture of the G743 and G746 (Fig. 6A). Under certain conditions, peak 3 actually could be resolved to afford two peaks (data not shown).
Figure 6.
Effects of introducing an intercalation inhibitor into duplexes derived from the human p53 sequence. For clarity, all HPLC traces are at 360 nm and show only the monoadduct region. (A) Electrophoretic analysis. Piperidine cleavage of peaks 1, 2, and 3 and comparison to markers. The sequence of oligonucleotide P53 is shown (Left). Numbers refer to the nucleotide position of each base in the human p53 cDNA. (B) AFB1 reaction with P53/S0. Peaks 1, 2, and 3 are monoadducts. (C) Reaction with duplex P53/S6. (D) Reaction with duplex P53/S5.
Experiments carried out with two intercalation inhibitors present within several bp of each other yielded reaction profiles consistent with single-stranded DNA, indicating that too much structural distortion was imposed by multiple intercalators or mismatches to maintain duplex structure (data not shown). Thus, we examined the effects of placing a single intercalation inhibitor at different positions in the DNA. The first experiment examined protection of G743 (P53/S6), the nucleotide predicted to have the greatest inherent reactivity. Protection of that site altered the product profile significantly: peak 3 was greatly reduced; peak 2 increased by approximately 4-fold; and peak 1 was relatively unchanged (Fig. 6C). In contrast, protection of G744 (P53/S5) resulted in diminished yield of peak 3 while enhancing peak 1 by 3-fold (Fig. 6D). The presence of intact AFB1 monoadduct in each sample was confirmed by electrospray MS.
DISCUSSION
Given that the half-life of AFB1 epoxide in water is approximately 1 sec (25), it has been proposed that intercalation of AFB1 epoxide provides a kinetic and entropic advantage for productive reaction with DNA versus unproductive hydrolysis (14, 15). To examine directly the effects of intercalation on AFB1 epoxide reactivity with DNA, a means was devised to occupy intercalation sites with covalently linked benzofuran derivatives (Fig. 2). Our results support the hypothesis that intercalation in DNA by AFB1 epoxide facilitates adduct formation, as evidenced by the observation that abolishing intercalation at a given site greatly diminished reactivity at that site. Furthermore, we have applied this principle to the problem of synthesizing specific AFB1-N7-Gua adducts within a region of the p53 gene and increased the yield of isolatable adducts by several-fold.
The first set of experiments clearly revealed that intercalation contributes significantly to the ability of AFB1 epoxide to react with DNA (Fig. 4). The relative adduct yields could be shifted from almost exclusively one adduct to almost exclusively the other depending upon the location of the intercalation inhibitor. In addition, diadduct formation was almost completely abolished. However, some residual adduction remained at the protected site (Fig. 4 D and E). This observation was not unexpected, because both single-stranded DNA and mononucleotides are known to react, albeit to a much lesser extent, with AFB1 epoxide (10), indicating that intercalation is not an obligate step for reaction with guanine.
The susceptibility of AFB1 adducts to piperidine cleavage enabled us to assign the HPLC peaks to specific guanine adducts by comparison to Maxam–Gilbert G-reaction markers (Fig. 4F). No cross-contamination of the two different adducts was observed. We note that a minor band (10%) of similar mobility to the unmodified target oligonucleotide was observed in the piperidine-treated samples for every monoadduct (Figs. 4F, 5E, and 6A). This band most likely arises during manipulation of the samples from chemical reversal of the AFB1-N7-Gua adduct to give free AFB1-diol and guanine, as previously has been observed in vitro (26, 27). It is unlikely to be a contamination from the HPLC purification process as the band persisted, even after multiple HPLC purifications.
An intriguing finding is the increased reactivity of DNA with AFB1 epoxide upon substituting T for C in the complementary strand (Fig. 4C), suggesting that the diminished reactivity observed after introduction of an intercalation inhibitor is due to the presence of the intercalated benzofuran moiety and not merely to distortions induced by a G-T mismatch. One possible explanation is that the guanine is more favorably positioned for nucleophilic attack on the epoxide when it is engaged in a wobble pairing characteristic of G-T base pairs (28). Alternatively, the mismatch itself, or the presence of the additional hydrophobic methyl group of T, might facilitate intercalation. As CpG groups in mammalian DNA often are methylated, methylation status could play a significant role in the reaction of AFB1 epoxide with DNA in vivo. Of particular relevance is recent evidence that CpG methylation significantly alters the distribution of DNA adducts of another intercalative carcinogen, benzo(a)pyrene diol epoxide (29).
The second model system tested the reactivity of AFB1 epoxide with a target DNA containing two adjacent guanines (Fig. 5). One interesting observation was that essentially no diadducts were observed with this duplex (data not shown). The absence of diadducts can be rationalized on the basis of the nearest neighbor-exclusion principle, which postulates that occupation of a single intercalation site prevents subsequent intercalation both immediately 5′ and 3′ to the initial site (23, 24). In this scenario, initial modification at one guanine results in occupancy of an intercalation site, effectively blocking intercalation at the adjacent guanine target site and thus diminishing reactivity of that guanine. Nevertheless, addition of intercalation inhibitors in the complementary strand had the expected effects. In both cases (GG/R5 and GG/R6), reaction at the protected guanine diminished whereas the yield of the other adduct increased (Fig. 5 C and D). From these experiments, it is unclear whether the benzofuran moiety possesses neighbor-excluding properties and to what extent this principle affects the reaction. If the neighbor-exclusion principle is equally applicable to all intercalators, then the expectation upon placing an intercalation inhibitor at one of the two sites would be that all reactivity should be abolished. This was not the case, however, suggesting that the neighbor-exclusion principle is not strictly applicable. Indeed, other violations of the neighbor-exclusion principle have been reported (30, 31).
Based on the observations with the GG/R0 duplex, we sought to test whether the intercalation inhibitor would exhibit neighbor-exclusion properties in a simpler system using oligonucleotide H (data not shown). Duplexes containing an intercalation inhibitor shifted one base to either the 5′ or 3′ side of the target were treated with AFB1 epoxide. These experiments revealed an asymmetry in the protection afforded by these constructs. Placing the intercalation inhibitor immediately 3′ to the target resulted in almost the same extent of protection as placing the intercalation inhibitor in the target intercalation site. However, when placed to the 5′ side, the reactivity at the target was indistinguishable from that of the entirely unprotected construct. The lack of consistently observable neighbor exclusion by this particular intercalation inhibitor may stem from the size of the intercalation inhibitor or the specific helix-distortion imposed. Expansion of the benzofuran moiety to a bulkier molecule that completely spans the helix may fill the entire intercalation space and serve as a more efficient inhibitor of intercalation. In any case, examination of the basis of the neighbor-exclusion principle warrants further investigation.
Having established the feasibility of altering chemical reactivities by intercalation inhibition, we sought to apply our methodology to help understand the conundrum presented by an observed AFB1-related p53 mutational hotspot. In greater than 50% of hepatocellular carcinomas in regions of the world with exposure to AFB1 and hepatitis B, a G→T mutation at G747 (the third position of codon 249) is observed (5–7). Based on Loechler’s predictions, G747 should not be exceptionally prone to adduction; the four potential target guanines in the p53 mutational hotspot sequence (Fig. 5E) are expected to have relative reactivities of 3:1.4:1:1.4 (22). Why, then, are so many mutations observed at G747? Although selective pressures certainly play a role in shaping the mutations observed in end-stage tumors, selection alone is unlikely to explain why G747 mutations are so abundant. One approach toward understanding the factors and processes subsequent to adduct formation that lead to a mutation is the use of oligonucleotides containing adducts at specific sites (32). By synthesizing a series of oligonucleotides each containing an AFB1 adduct at a different site, one can begin to study the relative ability of each adduct to induce a mutation. In addition, site-specific methods can address the identity of the actual mutagenic species. AFB1-N7-Gua adducts are highly labile, undergoing either depurination to afford an abasic site or imidazole ring opening to form the formamidopyrimidine derivative. Insertion of defined substrates into an appropriate vector will enable the first site-specific examination of the ability of AFB1 adducts or their decomposition products to induce mutations in the p53 sequence. However, to date, no study of the synthesis of site-specific AFB1 adducts in such a sequence or any oligonucleotide containing multiple guanines has been reported. It was our goal to use intercalation inhibitors to facilitate the synthesis and purification of specific AFB1 adducts in such a sequence.
In studies of the GC-rich p53 sequence between nucleotides 741–749 (Fig. 6), 7-deaza-dG, which lacks a nitrogen at position 7, was used in the place of dG in the complementary strand to simplify the reaction products. In the absence of an intercalation inhibitor (P53/S0), three peaks corresponding to monoadducts were obtained with peak 3 the dominant reaction product (Fig. 6B). Electrospray MS confirmed that the isolated products were of the expected molecular weight corresponding to intact AFB1 monoadducts. As anticipated, placement of two intercalation inhibitors within several bases of each other in a short oligonucleotide appeared to prevent hybridization of the duplex (data not shown). Thus, the effects of a single intercalation inhibitor on the product profile were examined. Upon protection of the theoretically most reactive site, G743, the yield of the other peaks increased significantly, with the G744 adduct comprising greater than 50% of the monoadducted species (Fig. 6C). Similarly, protection of G744 leads to a large increase in the yield of the G747 adduct (Fig. 6D). This is consistent with the observed asymmetry of protection afforded by the intercalation inhibitor, which appears to inhibit intercalation at both the site itself and the 5′ neighboring intercalation site. Thus, depending upon the location of the intercalation inhibitor, it was possible to alter the adduct distribution such that any of the three monoadduct peaks represented the major reaction product (Table 2).
Table 2.
Relative monadduct yield as a percentage of total monadducts
| Duplex | % Peak 1 | % Peak 2 | % Peak 3 |
|---|---|---|---|
| H/QC | 70 | 30 | N/A |
| H/QT | 34 | 66 | N/A |
| H/Q7 | >95 | <5 | N/A |
| H/Q3 | 11 | 89 | N/A |
| GG/R0 | 77 | 23 | N/A |
| GG/R6 | 93 | 7 | N/A |
| GG/R5 | 33 | 67 | N/A |
| P53/S0 | 17 | 14 | 69 |
| P53/S6 | 21 | 60 | 18 |
| P53/S5 | 51 | 14 | 35 |
Peak areas determined by integration of A360. N/A, not applicable.
This work demonstrates clearly that inhibiting intercalation of AFB1 epoxide can significantly alter the resulting adduct spectrum and that introduction of intercalation inhibitors into DNA duplexes provides a facile means of manipulating product yields (Table 2). Even in complex sequence environments, such as the p53 mutational hotspot sequence surrounding codon 249, a significant effect on adduct distribution with a single intercalation inhibitor can be demonstrated (Table 2). In addition, it should be possible to use these novel molecules to gain a better understanding of the phenomenon of intercalation. Structure-function investigations with intercalation inhibitors may shed more light on factors that contribute to the neighbor-exclusion principle, and second-generation intercalation inhibitors can be designed with superior inhibitory or neighbor-excluding capabilities. Moreover, intercalation inhibitors can be used to probe the intercalation requirements of other carcinogens, such as benzo(a)pyrene and aromatic amines, and facilitate the site-specific synthesis of carcinogen-DNA adducts.
Acknowledgments
We thank L. Marzilli and P. Vouros (Northeastern University) for electrospray MS of the oligonucleotides and L. Bailey for helpful discussions. This work was supported by National Institutes of Health Grant CA52127 to J.M.E., a National Science Foundation predoctoral fellowship to D.W., and National Institutes of Health Training Grant ES07020 and a predoctoral fellowship from Abbott Laboratories and the American Chemical Society Division of Medicinal Chemistry to W.R.K.
ABBREVIATIONS
- AFB1
aflatoxin B1
- AFB1-N7-Gua
8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 adduct
References
- 1.Qian G S, Ross R K, Yu M C, Yuan J M, Gao Y T, Henderson B E, Wogan G N, Groopman J D. Cancer Epidemiol Biomarkers Prev. 1994;3:3–10. [PubMed] [Google Scholar]
- 2.Croy R G, Essigmann J M, Reinhold V N, Wogan G N. Proc Natl Acad Sci USA. 1978;75:1745–1749. doi: 10.1073/pnas.75.4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster P L, Eisenstadt E, Miller J H. Proc Natl Acad Sci USA. 1983;80:2695–2698. doi: 10.1073/pnas.80.9.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey E A, Iyer R S, Stone M P, Harris T M, Essigmann J M. Proc Natl Acad Sci USA. 1996;93:1535–1539. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 6.Hsu I C, Metcalf R A, Sun T, Welsh J A, Wang N J, Harris C C. Nature (London) 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 7.Bressac B, Kew M, Wands J, Ozturk M. Nature (London) 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 8.Baertschi S W, Raney K D, Stone M P, Harris T M. J Am Chem Soc. 1988;110:7929–7931. [Google Scholar]
- 9.Misra R P, Muench K F, Humayun M Z. Biochemistry. 1983;22:3351–3359. doi: 10.1021/bi00283a008. [DOI] [PubMed] [Google Scholar]
- 10.Raney V M, Harris T M, Stone M P. Chem Res Toxicol. 1993;6:64–68. doi: 10.1021/tx00031a010. [DOI] [PubMed] [Google Scholar]
- 11.Gopalakrishnan S, Harris T M, Stone M P. Biochemistry. 1990;29:10438–10448. doi: 10.1021/bi00498a002. [DOI] [PubMed] [Google Scholar]
- 12.Iyer R S, Coles B F, Raney K D, Thier R, Guengerich F P, Harris T M. J Am Chem Soc. 1994;116:1603–1609. [Google Scholar]
- 13.Gopalakrishnan S, Stone M P, Harris T M. J Am Chem Soc. 1989;111:7232–7239. [Google Scholar]
- 14.Raney K D, Gopalakrishnan S, Byrd S, Stone M P, Harris T M. Chem Res Toxicol. 1990;3:254–261. doi: 10.1021/tx00015a011. [DOI] [PubMed] [Google Scholar]
- 15.Johnson W W, Guengerich F P. Proc Natl Acad Sci USA. 1997;94:6121–6125. doi: 10.1073/pnas.94.12.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hearst J E. Chem Res Toxicol. 1989;2:69–75. doi: 10.1021/tx00008a001. [DOI] [PubMed] [Google Scholar]
- 17.Spielmann H P, Dwyer T J, Sastry S S, Hearst J E, Wemmer D E. Proc Natl Acad Sci USA. 1995;92:2345–2349. doi: 10.1073/pnas.92.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobertz W R, Essigmann J M. J Am Chem Soc. 1996;118:7101–7107. [Google Scholar]
- 19.Kobertz W R, Essigmann J M. J Am Chem Soc. 1997;119:5960–5961. [Google Scholar]
- 20.D’Andrea A D, Haseltine W A. Proc Natl Acad Sci USA. 1978;75:4120–4124. doi: 10.1073/pnas.75.9.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Benasutti M, Ejadi S, Whitlow M D, Loechler E L. Biochemistry. 1988;27:472–481. doi: 10.1021/bi00401a068. [DOI] [PubMed] [Google Scholar]
- 23.Crothers D M. Biopolymers. 1968;6:575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- 24.Wilson W D. In: Nucleic Acids in Chemistry and Biology. Blackburn G M, Gait M J, editors. New York: Oxford Univ. Press; 1990. pp. 295–336. [Google Scholar]
- 25.Johnson W W, Harris T M, Guengerich F P. J Am Chem Soc. 1996;118:8213–8220. [Google Scholar]
- 26.Hertzog P J, Smith J R L, Garner R C. Carcinogenesis. 1980;1:787–793. doi: 10.1093/carcin/1.9.787. [DOI] [PubMed] [Google Scholar]
- 27.Groopman J D, Croy R G, Wogan G N. Proc Natl Acad Sci USA. 1981;78:5445–5449. doi: 10.1073/pnas.78.9.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel D J, Kozlowski S A, Marky L A, Rice J A, Broka C, Dallas J, Itakura K, Breslauer K J. Biochemistry. 1982;21:437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- 29.Denissenko M F, Chen J X, Tang M, Pfeifer G P. Proc Natl Acad Sci USA. 1997;94:3893–3898. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wakelin L P G, Romanos M, Chen T K, Glaubiger D, Canellakis E S, Waring M J. Biochemistry. 1978;17:5057–5063. doi: 10.1021/bi00616a031. [DOI] [PubMed] [Google Scholar]
- 31.Robledo-Luiggi C, Wilson W D, Pares E, Vera M, Martinez C S, Santiago D. Biopolymers. 1991;31:907–917. doi: 10.1002/bip.360310710. [DOI] [PubMed] [Google Scholar]
- 32.Singer B, Essigmann J M. Carcinogenesis. 1991;12:949–955. doi: 10.1093/carcin/12.6.949. [DOI] [PubMed] [Google Scholar]