Abstract
Xeroderma pigmentosum (XP) patients fail to remove pyrimidine dimers caused by sunlight and, as a consequence, develop multiple cancers in areas exposed to light. The second most common sign, present in 20–30% of XP patients, is a set of neurological abnormalities caused by neuronal death in the central and peripheral nervous systems. Neural tissue is shielded from sunlight-induced DNA damage, so the cause of neurodegeneration in XP patients remains unexplained. In this study, we show that two major oxidative DNA lesions, 8-oxoguanine and thymine glycol, are excised from DNA in vitro by the same enzyme system responsible for removing pyrimidine dimers and other bulky DNA adducts. Our results suggest that XP neurological disease may be caused by defective repair of lesions that are produced in nerve cells by reactive oxygen species generated as by-products of an active oxidative metabolism.
Xeroderma pigmentosum (XP) is an hereditary disease characterized clinically by extreme sensitivity to sunlight, a predisposition to skin cancer and other dermatological changes in exposed areas, and progressive neurological abnormalities. The underlying cause of solar sensitivity is an inability to repair UV-induced DNA damage (1, 2), and some investigators have suggested that XP neurological disease, which is particularly prevalent in groups A and D, is caused by DNA damage in nerve cells (3, 4). XP can be caused by mutations in any one of seven genes, XPA through XPG (1). With the exception of XPE, whose gene product has not been unambiguously identified, the proteins encoded by each of these genes are essential for nucleotide excision repair, the primary enzyme system for removal of bulky DNA lesions (2, 5, 6). Recent studies have revealed that some of these genes, in addition to DNA repair, play a role in transcription, thus raising the possibility that XP neurological abnormalities are caused by defects in transcription (7–9). The XPB and XPD gene products are subunits of the general transcription factor TFIIH (10, 11), so it is conceivable that the neurological disease in complementation groups (CG) B and D is caused by defective transcription. Similarly, the XPG protein binds tightly to TFIIH (12–14) and has been found in some RNA polymerase II holoenzyme preparations (15); hence, the neurological disease in the XPG CG may also be ascribed to a transcription defect. However, neurological abnormalities also are observed in CG A, but there is no evidence that the XPA protein has any function other than DNA repair.
Neurons consume great amounts of molecular oxygen, and the reactive oxygen species that are by-products of cellular respiration can cause considerable damage to DNA; if not repaired, such damage may be the cause of neurodegeneration in XP patients (3, 4). The major DNA lesions produced by oxidative damage, 8-oxoguanine (8-OxoG), thymine glycol (Tg), pyrimidine hydrates, and urea, are nonbulky lesions thought to be repaired mostly, if not exclusively, by the base excision pathway that does not depend on XP proteins (16). There is no evidence that nucleotide excision repair, the repair system for bulky adducts, contributes substantially to the repair of these nonbulky lesions. Therefore, it has been proposed (17) that neuronal death in XP patients is caused by rare bulky lesions known to be induced by reactive oxygen species (18). Considering the rarity of such lesions and knowing that the functionally homologous Escherichia coli (A)BC excinuclease system removes Tg lesions (19), we reasoned that the more common, nonbulky lesions induced by oxidative damage (20) may be the cause of XP neurological disease if the mammalian nucleotide excision repair system contributes significantly to the removal of such damaged bases in unaffected individuals. Hence, we decided to use the XP protein-dependent excision assay (12, 21, 22) to examine the effect of nucleotide excision repair on the major DNA lesions caused by oxidative stress (Fig. 1a).
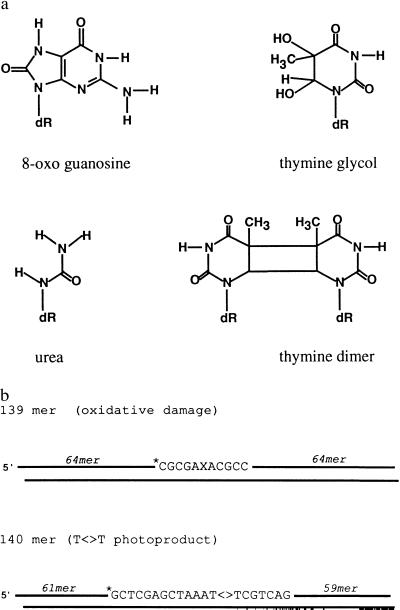
Figure 1.
Substrates used in this study. (a) Structures of the base lesions; dR, deoxyribose. (b) Schematic of duplexes used in the excision assay. Linear duplex molecules, 139 or 140 bp in length, were prepared by phosphorylation, annealing, and ligation of six overlapping oligonucleotides; ∗, position of the radiolabel. (Upper) X denotes 8-OxoG, Tg, or urea.
Tg and 8-OxoG are generated directly by reactions of the corresponding bases with hydroxyl radicals; urea is a secondary product generated in substantial quantities by spontaneous hydrolytic cleavage of Tg. The classical substrate for nucleotide excision repair, the bulky cis, syn-cyclobutane thymine dimer (T<>T), was used as a reference lesion. Mammalian nucleotide excision repair is initiated by dual incisions, one on each side of the lesion, resulting in release of oligonucleotides 24- to 32-nt in length (21). To test for removal of oxidative base lesions by nucleotide excision, we constructed substrates (Fig. 1b) in which a 139-bp duplex contained the damaged base at nucleotide 70; for comparison, we used a 140-bp duplex with T<>T at nucleotides 74–75.
MATERIALS AND METHODS
Cell Lines, Cell-Free Extracts (CFEs), and Purified Repair Factors.
HeLa S3 cells were from the stock of Lineberger Comprehensive Cancer Center (Chapel Hill, NC). The following Chinese hamster ovary (CHO) cell lines were obtained from the American Type Culture Collection Repository (Rockville, MD): CRL 1859 (AA8, parental cell line), CRL 1862 (UV20, ERCC1), CRL 1865 (UV5, ERCC2, XP-D), CRL 1866 (UV24, ERCC3, XP-B), CRL 1860 (UV41, ERCC4, XP-F), and CRL 1867 (UV135, ERCC5, XP-G). The National Institute of General Medical Sciences Human Mutant Cell Repository (Coriell Institute, Camden, NJ) provided these human cell lines: XP-A (XP20S, GM02345), XP-B (XP11BE, GM02252), XP-C (XP1BE, GM02246; XP3BE, GM02248; XP21RO, GM00709), XP-D (XP17BE, GM02253; XP7BE, GM02485), XP-F (XP2YO, GM08437), and XP-G (XP2BI, GM03021). Cells were cultured as described (22), and CFEs were prepared from cells in exponential growth phase according to well established methods (23) and stored at −80°C as described (24). The six factors that reconstitute human excision nuclease were purified from HeLa cells (RPA, TFIIH) or as recombinant proteins (XPA, XPC•HHR23B, XPF•ERCC1, XPG) as described (12, 25–28).
Preparation of Substrates.
Double-stranded DNA molecules were prepared by phosphorylation, annealing, and ligation of six partially overlapping oligonucleotides as described (29), using T4 DNA polynucleotide kinase and T4 DNA ligase (New England BioLabs) to generate duplexes with 1-nt overhangs at the 5′ ends. The oligomers containing centrally located Tg, urea, and T<>T lesions were prepared as described (30, 31), and the oligomer containing 8-OxoG was obtained from the Midland Certified Reagent Company (Midland, TX). All oligomers used for substrates were purified by HPLC; NMR was used to characterize the oligomer containing Tg (30), and the composition of the 8-OxoG oligomer was verified by mass spectral analysis (Midland Certified Reagent Company). The damaged oligomers were phosphorylated with [γ-32P]ATP (7000 Ci/mmol; 1Ci = 37 GBq) and used for substrate preparation. For DNA containing oxidative base lesions, the substrate was a 139-bp duplex with the adduct at nucleotide 70 of one strand and 32P label at the 6th phosphodiester bond 5′ to the lesion. For comparison, we used a 140-bp duplex with T<>T at nucleotides 74–75 and 32P label at the 13th bond 5′ to the photoproduct. Because the manipulations used for substrate purification may induce abasic (AP) sites in the DNA, we tested the integrity of the Tg and 8-OxoG substrates by incubating the DNA with human AP endonuclease as described (32). We found that ≈1% of the 8-OxoG and 8% of the Tg substrate molecules were incised under conditions in which a bona fide AP site was digested to completion.
Excision Assays.
The excision reactions with either CFE or purified repair factors were conducted under substrate-limiting conditions (data not shown). For assays with extracts based on a single time point, 50 μg of CFE (25 μg each for complementation assays) and 12.6 fmol of pBR322 were mixed in 25 μl of reaction buffer as described (24). The repair reaction was at 30°C for 60 min and was initiated by addition of substrate DNA (40 fmol of 8-OxoG or 15 fmol of Tg). For the time course with HeLa CFE, 150 μg of extract and 60 fmol of pBR322 were mixed with substrate DNA (90 fmol of 8-OxoG or 120 fmol of Tg) in 75 μl of reaction buffer, and 18-μl aliquots were removed at the indicated time points. For the kinetic experiments with CHO AA8 extract, substrate DNA (100 fmol) was incubated at 30°C with 330 μg of CFE and 60 fmol of pBR322 in 125 μl of reaction buffer, and 18-μl aliquots were removed at the indicated time points. For the excision reactions with purified repair factors, substrate DNA (30 fmol) was incubated at 30°C for 2.5 h with the indicated repair factors in 25 μl of reaction buffer similar to that described (25). Processing of DNA, PAGE, autoradiography, and quantitation of repair with an AMBIS Systems Scanner have been described (24). The level of repair for each reaction was determined from the radioactivity migrating as 20–30 mers as a percentage of the total radioactivity in the lane. We have shown previously that, after incubation with CFE and resolution by gel electrophoresis, fragments in this size range contain the excised thymine dimer (33), psoralen monoadduct (T.B., D. Mu, and A.S., unpublished work), ABPD (2-aminobutyl-1, 3-propanediol) synthetic analog of an AP site (J.T.R. and A.S., unpublished work), ABPD site with a cholesterol side chain (29), and cisplatin GG diadduct (J. C. Huang and A.S., unpublished work).
RESULTS
Removal of Oxidative DNA Damage by Extracts.
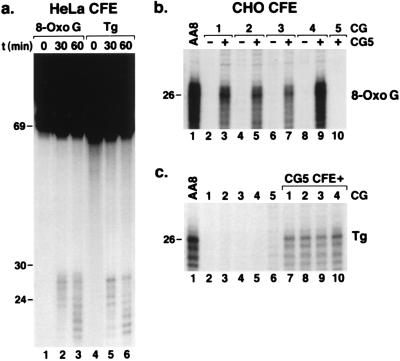
The 8-OxoG- and Tg-containing DNAs were tested for removal by nucleotide excision repair using HeLa CFE known to be capable of excising thymine dimers (21). Fig. 2a shows that, at early time points, both lesions were excised by HeLa CFE primarily as 26-nt long oligomers; at later time points, smaller species appeared, in part because of exonucleolytic digestion of excised fragments (35). CFE prepared from human XP cell lines (XP-A, XP-B, XP-C, XP-D, XP-F, and XP-G) failed to excise these lesions in oligonucleotides (data not shown), consistent with the requirement of all XP proteins for nucleotide excision repair. However, some of the CFE from XP lymphoblastoid and fibroblast cell lines gave high background caused by degradation of the DNA by nonspecific endonucleases. Hence, we decided to investigate the requirement for XP proteins and to characterize the excision reaction further by using either CFE from CHO cell lines mutated in XP genes or the reconstituted human excision nuclease system, the dual incision activity that requires all XP proteins. We used the CHO AA8 cell line and its mutant derivatives for experiments with CFE because of the ease of growing these cells and obtaining high quality extracts. Furthermore, both genetic (36) and biochemical (22) data show that the human and hamster nucleotide excision repair proteins are interchangeable and, hence, any result obtained with CFE from these cell lines should be applicable to human excision repair.
Figure 2.
Excision of oxidative DNA lesions by human and rodent CFEs. (a) Autoradiograph of sequencing gel showing time course of excision by HeLa CFE. In lanes 1 and 4, substrate DNA was resolved without incubation in reaction buffer. (b) Excision of 8-OxoG by CFE from CHO AA8 and its excision repair mutant derivatives. The mutant cell lines used were UV20, UV5, UV24, UV41, and UV135 representative of CG 1–5, respectively. (c) Excision of Tg by CHO AA8 CFE and the same set of mutant cell lines. In a, the entire gel is shown; in b and c, only the region encompassing the excision products is shown. For the 60-min time points with HeLa CFE, the excision products were 0.1% of input DNA (a, lanes 3 and 6) whereas the level of excision with AA8 CFE was ≈2% for 8-OxoG (b, lane 1) and 0.7% for Tg (panel c, lane 1). As with the T<>T substrate (22), the mixture of two mutant CFEs typically gave ≈50% of the wild-type signal. The faint bands visible in lane 6 (c) are due to degradation of the substrate by nucleases in the UV135 CFE.
Results presented in Fig. 2 b and c demonstrate that the CHO AA8 CFE excised both lesions and that CFEs from cell lines (CGs) with mutations in ERCC1 (CG1), XPD (CG2), XPB (CG3), XPF (CG4), and XPG (CG5) genes were as defective in removing 8-OxoG and Tg as they are in removing T<>T (22). Because one of the hallmarks of nucleotide excision repair is an essentially infinite substrate range (5, 35), it was not surprising to find that nucleotide excision repair removes nonbulky lesions caused by oxidative damage. The important question is whether these nonbulky lesions are removed at a physiologically relevant rate. To assess the potential significance of excision of oxidative lesions by nucleotide excision repair, we compared the rates of removal of these lesions to that of T<>T, the classical substrate for nucleotide excision repair used in the vast majority of in vivo assays that measure nucleotide excision repair on the basis of the disappearance of pyrimidine dimers from cellular DNA (2, 36).
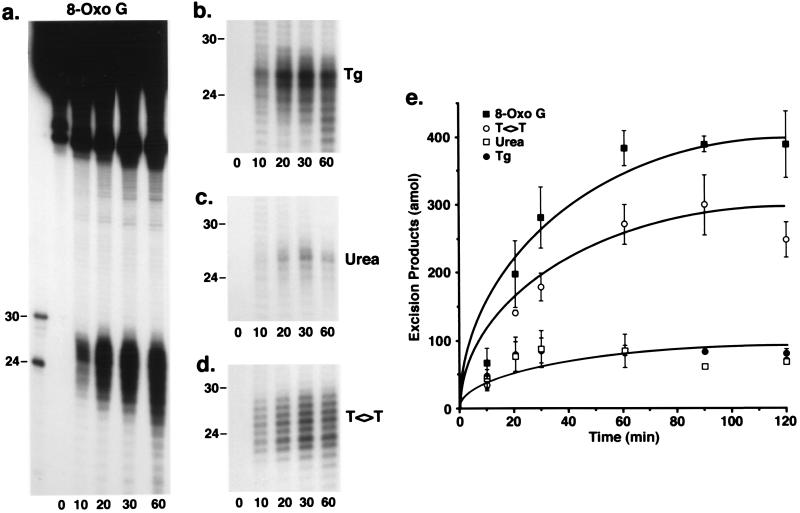
Fig. 3 shows the results of kinetic assays conducted with CHO AA8 CFE and the four substrates illustrated in Fig. 1. All four lesions were released primarily as oligonucleotides 20–30 nt in length, with the shorter species being more abundant with the oxidative damage substrates. At later time points, the smaller species predominated with all four substrates, similar to the results obtained with HeLa CFE. A quantitative analysis of the results shown in Fig. 3 a–d, combined with data from other experiments performed under identical conditions, is presented in Fig. 3e. As is apparent, 8-OxoG was excised at a 1.5-fold faster rate than T<>T, which was excised at about a 3-fold faster rate than Tg or urea. With either the Tg or urea substrates, the excision reaction was essentially complete at 20 min, and excision of 8-OxoG and T<>T continued to increase during the first 60 min of incubation. Although we have no explanation for these different kinetics, these results clearly show that oxidative DNA lesions are repaired in vitro at rates comparable to that of T<>T, the reference bulky lesion. Therefore, we consider the removal of 8-OxoG, Tg, and urea by the nucleotide excision repair system to be physiologically relevant.
Figure 3.
Time course of excision of oxidative base damage by mammalian excision nuclease using CHO CFEs. Sequencing gels show the results of kinetic experiments conducted as described in the text. (a) The entire gel is shown; (b–d) only the region encompassing the excision products is shown. In the time 0 lanes, substrate DNA was resolved without incubation in reaction buffer. (e) Quantitative analyses of the data shown in a–d and of additional experiments conducted under identical conditions; n = 2–5 experiments except the 90- and 120-min time points with the urea substrate. The bars indicate SEs; in these experiments, 200 amol of excision products represents ≈1.4% excision of input DNA.
The results shown in Figs. 2 and 3 suggest that the nucleotide excision system, as defined by the requirement for all proteins encoded by the XP genes and the dual incision mode, removes the major oxidative base lesions. However, data obtained with CFE have certain limitations. For instance, these extracts contain proteins that specifically bind to oxidative base lesions (37–41), and recognition by such proteins could be necessary for the subsequent action of the mammalian excision nuclease. It is also possible that nucleotide excision repair enzymes act on a secondary lesion, a glycosylase-generated AP site, known to be a weak substrate for HeLa CFE in our in vitro system (5, 35). However, the possibility that the repair signal was generated by an AP site was eliminated by the following observations. The 8-OxoG substrate contains at most 1% AP site, and we have reported that, under optimal conditions, only 4% of AP substrate can be excised in our system (35). Hence, the maximum excision that may arise from an AP contaminant would be 0.04% of the substrate. In fact, we obtain ≈2% excision with the 8-OxoG substrate, indicating that the excised lesion is 8-OxoG itself and not an AP site. This is further supported by the finding that the Tg substrate, which contains more AP sites than the 8-OxoG substrate, is in fact a weaker substrate in vitro for the excision nuclease. Alternatively, it can be argued that cleavage by glycosylase/AP endonuclease is a prerequisite for excision nuclease activity. Cleavage of these substrates by the joint actions of glycosylase and AP endonuclease is expected to give rise to a 69-nt fragment, and such a band is seen in our assays (Figs. 2a and 3a). However, even incubation of 8-OxoG, Tg, and urea substrates in reaction buffer without CFE generated this fragment; upon incubation with CFE, there was no or only a modest increase in the level of this cleavage product, which varied from 1 to 5% of input substrate in different preparations (data not shown). Hence, we conclude that only a minor fraction of the 69 mer seen in our excision experiments is produced by glycosylase/AP endonuclease action. To ascertain that the excision of oxidative base damage that we detect in CFE is bona fide nucleotide excision repair, which requires all excision repair factors, we conducted experiments with purified repair proteins.
Excision of Oxidative Damage by Purified Repair Factors.
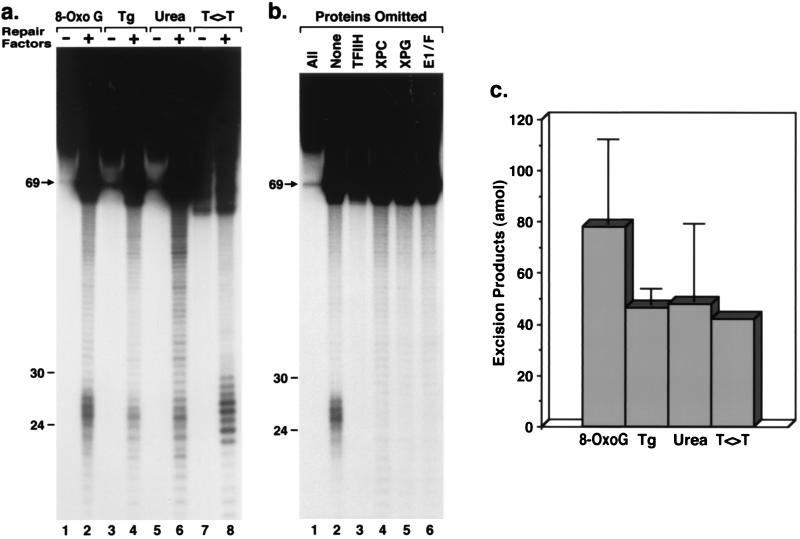
Six repair factors consisting of XPA, RPA, TFIIH (XPB and XPD plus other polypeptides), XPC•HHR23B, XPG, and XPF•ERCC1 are necessary and sufficient to reconstitute the human excision nuclease (12, 25). Hence, the four substrates that were used with CFE also were tested with the reconstituted system. All four lesions were excised by the reconstituted system (Fig. 4a), and omission of individual repair factors abolished excision of 8-OxoG (Fig. 4b) as well as T<>T (25), Tg, and urea (data not shown). Quantitative analysis of the data with the reconstituted system revealed that levels of excision for all four lesions were within a factor of 2 of one another (Fig. 4c). These results demonstrate that 8-OxoG, Tg, and urea can be excised in vitro by the human excision nuclease. We believe that these findings are relevant to the progression of neurological disease in XP patients.
Figure 4.
Excision of oxidative DNA lesions by reconstituted human excision nuclease. (a) Autoradiograph of sequencing gel showing excision by the XP-dependent reconstituted system. (b) Repair factor omission experiment (25) with 8-OxoG. For these experiments, substrate DNA was resolved in the lanes designated “− Repair Factors” or “All Proteins Omitted” without prior incubation in reaction buffer. (c) For quantitative analysis of excision of oxidative damage with the reconstituted system, the averages of two experiments are plotted. For these experiments, 60 amol of excision products corresponds to ≈0.2% excision of input DNA.
DISCUSSION
Two hereditary diseases in which patients present signs of neurological abnormalities are known to be caused by mutations in genes that affect DNA repair: XP and Cockayne’s syndrome (CS) (1, 2, 7, 8). In light of our results and of recent reports on the possible cause of neurological disease in these patients (7, 42, 43), we wish to reconsider the models proposed for the neurodegeneration associated with CS and XP. CS patients suffer acute sun sensitivity, severe cachetic dwarfism, and developmental neurological abnormalities caused primarily by demyelination. Cultured CS cells are defective in transcription-coupled repair (TCR) of pyrimidine dimers (44) and Tgs (43). Mutations in two genes exclusively associated with CS, CSA and CSB, and in three genes essential for excision repair, XPB, XPD, and XPG, can cause CS. Of interest, although CSA and CSB mutations affect only TCR of pyrimidine dimers and other bulky adducts, mutations in XPB, XPD, and XPG greatly reduce or abolish overall repair as well (1, 2). A finding with potential implications for the pathogenesis of CS is that not only nucleotide excision repair of T<>T, but also base excision repair of Tg, is coupled to transcription (43). However, in contrast to TCR of T<>T, which requires all six excision repair factors, TCR of Tg depends on the XPG polypeptide but not on XPA or XPF and does not require the 3′ nuclease function of XPG (43). Thus, certain XPG mutations that completely abolished excision repair of T<>T did not affect the rate of either overall or TCR of Tg (43). In contrast, XPG null mutations completely eliminated TCR of both T<>T and Tg, totally abolished excision of T<>T, and moderately reduced the rate of transcription-independent repair of the Tg lesion (43).
Based on these data, it was proposed that Tg is removed from human chromosomes by two pathways (43). The first pathway (fast rate) requires the physical presence of XPG (but not its nuclease activity) + CSB protein + (glycosylase/AP endonuclease) but not the XPA or XPF proteins; this is TCR of Tgs. The second pathway (intermediate rate) requires XPG (but not its nuclease activity) + (glycosylase/AP endonuclease) but not the XPA, XPF, or CSB proteins; this is XPG-dependent genome overall repair of Tg lesions. The requirement for glycosylase/AP endonuclease activity in these two pathways is quite likely but has not been experimentally demonstrated. Similarly, it has not been shown whether these two pathways remove oxidative damage other than Tg. The third pathway (slow rate) that we describe in this study is nucleotide excision repair, which requires the entire set of excision repair proteins including XPA, XPB, XPC, XPD, XPF, and XPG as well as the other polypeptides necessary for excision of T<>T (12, 25). The properties of the three pathways may help explain the different neuropathologies in the two diseases. Although there is no absolute demarcation between the neuropathology of XP and CS, degenerative changes predominate in XP and developmental abnormalities predominate in CS (45, 46). It is conceivable that the two fast pathways, which are dependent on XPG but independent of XPA and XPF, play an important role in maintaining genomic integrity during neuroskeletal development. The recent findings that null mutations of genes essential for base excision, AP endonuclease and DNA polymerase β, cause embryonic lethality (47, 48) underscore the importance of base excision repair during development.
In contrast, the comparatively slower nucleotide excision repair pathway for the removal of the most common oxidative lesions, such as 8-OxoG and Tg, may play a more significant role in helping to maintain the long term genetic integrity of fully differentiated neurons, and defects in this pathway may lead to XP neurological disease. There are three caveats to this model for neurodegeneration in XP: (i) Our data simply show that the human nucleotide excision repair system can remove 8-OxoG and Tg in vitro; the rate may be too slow to be of relevance in vivo. However, the fact that the classical T<>T substrate for excision repair is removed at an intermediate rate compared with 8-OxoG and Tg suggests that the removal of oxidative base damage by nucleotide excision repair is fast enough to be physiologically significant. (ii) If nucleotide excision repair is essential to prevent neurodegeneration, one would expect all XP patients to exhibit neurological disease; yet it is reported that only 20–30% of patients exhibit neurological signs and some CGs such as XP-C are, as a rule, free of neurological disease (1, 49). However, a careful analysis of available case histories of XP patients and, where available, the nature of the mutation, suggests that, if they do not succumb to skin cancers, most XP patients will eventually develop neurological disease. Patients with leaky mutations in any XP gene tend to develop the disease later in life compared with patients with null mutations; XP-C individuals carry out normal TCR (50, 51) and hence develop neurological symptoms only at a relatively advanced age compared with other XP patients (46, 52). (iii) If nucleotide excision repair plays a significant role in repairing the most abundant lesions of oxidative DNA damage, cultured XP cells would be expected to be hypersensitive to ionizing radiation, which causes DNA damage by generating hydroxyl radicals that produce 8-OxoG and Tg in DNA. Yet, as a rule, XP cell lines are not hypersensitive to x-ray killing (1). However, killing by x-rays is largely due to double-strand breaks induced directly as evidenced by the extreme sensitivity of scid mutants to killing by ionizing radiation (53).
The experiments reported in this paper were conducted under substrate limiting conditions and hence the rates contain both Km and kcat components and may be taken as approximate values for catalytic efficiency (kcat/Km). Looked at in this light, clearly, the major lesions of oxidative damage are repaired with efficiency comparable to that for the major UV photoproduct. From a cellular perspective, the two types of lesions differ in that Tg and other oxidative lesions can also be repaired by glycosylases whereas, in humans, only nucleotide excision repair can remove pyrimidine dimers, which are the causative lesions of XP dermatological symptoms. However, it is quite possible that the level and activities of glycosylases in neurons are insufficient to deal with the large amount of oxidative damage generated in these cells and that nucleotide excision repair plays a major role in defending neural cells against oxidative damage. Further research on relative activities of base excision and nucleotide excision repair systems in various types of cells and tissues are needed to examine this possibility.
In summary, our results suggest that the defective repair of nonbulky lesions, such as 8-OxoG and Tg, in XP patients may be the cause of neurodegeneration in XP. Furthermore, these results lend support to DNA damage models for other neurodegenerative diseases (54). Although such diseases have not been correlated with overt defects in known DNA repair pathways, it is possible that, under certain conditions, oxidative base damage is so extensive that, even in repair proficient individuals, all three pathways discussed above are saturated and the remaining lesions cause neuron death. There is abundant experimental as well as clinical data suggesting that oxidative DNA damage contributes to tissue damage in stroke, trauma, and some chronic neurological disorders (34).
Acknowledgments
We thank Drs. P. K. Cooper, A. Kazantsev, K. H. Kraemer, and J. H. Robbins for helpful discussions and critical reading of the manuscript, and we thank J.-S. Taylor, Washington University, for the T<>T-containing oligomer and B. Demple, Harvard University, for human Ape protein. This work was supported by National Institutes of Health Grant GM3283 to A.S. and by a grant from the American Cancer Society to P.H.B.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: XP, xeroderma pigmentosum; 8-OxoG, 8-oxoguanine; Tg, thymine glycol; T<>T, cis, syn-cyclobutane thymine dimer; CHO, Chinese hamster ovary; CFE, cell-free extract; CS, Cockayne’s Syndrome; AP, abasic; TCR, transcription-coupled repair; CG, complementation group.
References
- 1.Cleaver J E, Kraemer K H. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. Vol. 2. New York: McGraw–Hill; 1989. pp. 2949–2971. [Google Scholar]
- 2.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 3.Robbins J H. J Child Neurol. 1989;4:143–146. doi: 10.1177/088307388900400215. [DOI] [PubMed] [Google Scholar]
- 4.Robbins J H, Brumback R A, Mendiones M, Barrett S F, Carl J R, Cho S, Denckla M B, Ganges M B, Gerber L H, Guthrie R A, Meer J, Moshell A N, Polinsky R J, Ravin P D, Sonies B C, Tarone R E. Brain. 1991;114:1335–1361. doi: 10.1093/brain/114.3.1335. [DOI] [PubMed] [Google Scholar]
- 5.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 6.Wood R D. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 7.Hanawalt P C. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg E C. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- 9.Chu G, Mayne L. Trends Genet. 1996;12:187–192. doi: 10.1016/0168-9525(96)10021-4. [DOI] [PubMed] [Google Scholar]
- 10.Drapkin R, Reardon J T, Ansari A, Huang J C, Zawel L, Ahn K, Sancar A, Reinberg D. Nature (London) 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 11.Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers J H J, Egly J-M. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mu D, Park C-H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 13.Habraken Y, Sung P, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1996;93:10718–10722. doi: 10.1073/pnas.93.20.10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer N, Reagan M S, Wu K J, Canagarajah B, Friedberg E C. Biochemistry. 1996;35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 15.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 16.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 17.Satoh M S, Jones C J, Wood R D, Lindahl T. Proc Natl Acad Sci USA. 1993;90:6335–6339. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmichael P L, Shé M N, Phillips D H. Carcinogenesis. 1992;13:1127–1135. doi: 10.1093/carcin/13.7.1127. [DOI] [PubMed] [Google Scholar]
- 19.Lin J-J, Sancar A. Biochemistry. 1989;28:7979–7984. doi: 10.1021/bi00446a002. [DOI] [PubMed] [Google Scholar]
- 20.Dizdaroglu M. Int J Radiat Biol. 1992;61:175–183. doi: 10.1080/09553009214550791. [DOI] [PubMed] [Google Scholar]
- 21.Huang J C, Svoboda D L, Reardon J T, Sancar A. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reardon J T, Thompson L H, Sancar A. Cold Spring Harbor Symp Quant Biol. 1993;58:605–617. doi: 10.1101/sqb.1993.058.01.067. [DOI] [PubMed] [Google Scholar]
- 23.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reardon J T, Thompson L H, Sancar A. Nucleic Acids Res. 1997;25:1015–1021. doi: 10.1093/nar/25.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu D, Hsu D S, Sancar A. J Biol Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 26.Matsunaga T, Park C-H, Bessho T, Mu D, Sancar A. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 27.Reardon J T, Mu D, Sancar A. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 28.Bessho T, Sancar A, Thompson L H, Thelen M P. J Biol Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- 29.Matsunaga T, Mu D, Park C-H, Reardon J T, Sancar A. J Biol Chem. 1995;270:20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- 30.Kung H C, Bolton P H. J Biol Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 31.Taylor J-S, Brockie I R, O’Day C L. J Am Chem Soc. 1987;109:6735–6742. [Google Scholar]
- 32.Wilson D M, III, Takeshita M, Grollman A P, Demple B. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 33.Svoboda D L, Taylor J-S, Hearst J E, Sancar A. J Biol Chem. 1993;268:1931–1936. [PubMed] [Google Scholar]
- 34.Chopp M, Chan P H, Hsu C Y, Cheung M E, Jacobs T P. Stroke. 1996;27:363–369. doi: 10.1161/01.str.27.3.363. [DOI] [PubMed] [Google Scholar]
- 35.Huang J C, Hsu D S, Kazantsev A, Sancar A. Proc Natl Acad Sci USA. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson L H. In: DNA Damage and Repair: Biochemistry, Genetics, and Cell Biology. Nickoloff J A, Hoekstra M, editors. Totowa, NJ: Humana; 1997. , in press. [Google Scholar]
- 37.Bessho T, Tano K, Kasai H, Ohtsuka E, Nishimura S. J Biol Chem. 1993;268:19416–19421. [PubMed] [Google Scholar]
- 38.Hilbert T P, Boorstein R J, Kung H C, Bolton P H, Xing D, Cunningham R P, Teebor G W. Biochemistry. 1996;35:2505–2511. doi: 10.1021/bi952516e. [DOI] [PubMed] [Google Scholar]
- 39.Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Mutation Res. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- 40.Strauss P R, Beard W A, Patterson T A, Wilson S H. J Biol Chem. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 41.Bennett, R. A. O., Wilson, D. M., Wong, D. & Demple, B. (1997) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 42.Leadon S A, Cooper P K. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 44.Venema J, Mullenders L H F, Natarajan A T, van Zeeland A A, Mayne L V. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otsuka F, Robbins J H. Am J Dermatopathol. 1985;7:387–392. doi: 10.1097/00000372-198508000-00013. [DOI] [PubMed] [Google Scholar]
- 46.Robbins J H. J Am Med Assoc. 1988;260:384–388. [Google Scholar]
- 47.Xanthoudakis S, Smeyne R J, Wallace J D, Curran T. Proc Natl Acad Sci USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 49.Kraemer K H, Lee M M, Scotto J. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 50.Venema J, van Hoffen A, Natarajan A T, van Zeeland A A, Mullenders L H F. Nucleic Acids Res. 1990;18:443–448. doi: 10.1093/nar/18.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Hoffen A, Venema J, Meschini R, van Zeeland A A, Mullenders L H F. EMBO J. 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robbins J H, Brumback R A, Moshell A N. Eur Neurol. 1993;33:188–190. doi: 10.1159/000116932. [DOI] [PubMed] [Google Scholar]
- 53.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 54.Robbins J H. Arch Neurol. 1987;44:579. doi: 10.1001/archneur.1987.00520180005004. [DOI] [PubMed] [Google Scholar]