Abstract
Background
Hybridization, or the interbreeding of two species, is now recognized as an important process in the evolution of many organisms. However, the extent to which hybridization results in the transfer of genetic material across the species boundary (introgression) remains unknown in many systems, as does the length of time after initial divergence that the species boundary remains porous to such gene flow.
Results
Here I use genome-wide genotypic and DNA sequence data to show that there is introgression and admixture between the melpomene/cydno and silvaniform clades of the butterfly genus Heliconius, groups that separated from one another as many as 30 million generations ago. Estimates of historical migration based on 523 DNA sequences from 14 genes suggest unidirectional gene flow from the melpomene/cydno clade into the silvaniform clade. Furthermore, genetic clustering based on 520 amplified fragment length polymorphisms (AFLPs) identified multiple individuals of mixed ancestry showing that introgression is on-going.
Conclusion
These results demonstrate that genomes can remain porous to gene flow very long after initial divergence. This, in turn, greatly expands the evolutionary potential afforded by introgression. Phenotypic and species diversity in a wide variety of organisms, including Heliconius, have likely arisen from introgressive hybridization. Evidence for continuous gene flow over millions of years points to introgression as a potentially important source of genetic variation to fuel the evolution of novel forms.
Background
Hybridization has long been recognized as an important mechanism of diversification in plants [1,2], and the exchange of genetic material via horizontal gene transfer has played a significant role in the evolution of many prokaryotic genomes [3]. In animals however, hybridization has historically been viewed as rare and evolutionarily inconsequential [4]. Despite this bias in opinion, we now know that hybridization is relatively wide-spread among animal species [5], and in some instances, it has likely had important evolutionary ramifications, such as in the origin of new species [6-10]. Surveys of hybridization in animals show that it occurs predominantly between closely-related sister species [5], and well-characterized examples of interspecific gene flow generally involve species that diverged very recently [10-13]. These observations are consistent with theory and data which show that genetic incompatibilities that result in hybrid sterility and inviability accumulate as species diverge [14]. Hybrid sterility and inviability, in turn, reduce or eliminate the opportunity for gene exchange. Despite these general trends, there are occasional examples of hybridization between distantly-related non-sister species in various animal groups [15-20]. Do these cases result in the long-term sharing of genetic material or are they simply evolutionary dead-ends?
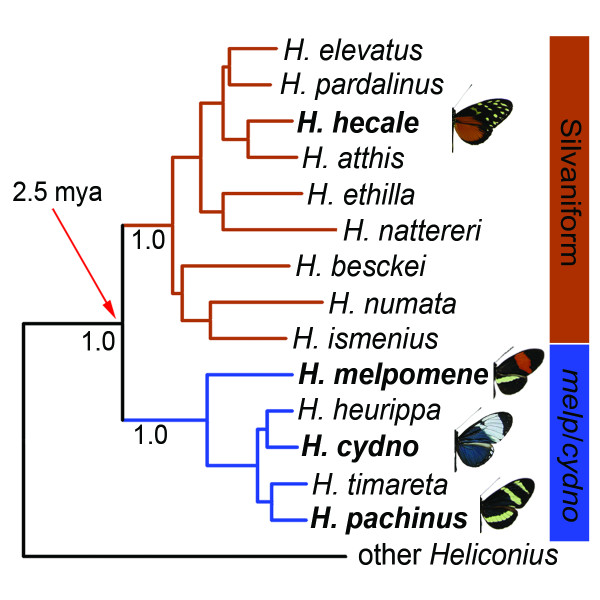
To address this question I focused on the Neotropical butterfly genus Heliconius, a group well known for its diversity of mimetic wing patterns and for extensive hybridization [21]. As in other organisms, most hybridization in Heliconius occurs between closely-related species and subspecies [21]. Among these groups hybridization is known to result in gene flow [22-24]. However, members of two ecologically and morphologically distinct Heliconius subgroups, the melpomene/cydno clade and the silvaniform clade (Figure 1), also occasionally hybridize with one another [21,25]. In captivity, first-generation and backcross hybrids have resulted from multiple crosses between the two clades [21,26], and there are at least eleven suspected hybrids that have been collected in the field [21,25,27]. Based on phenotype and collection location, four of these field-caught specimens are believed to be hybrids between H. melpomene and H. numata, five are believed to be hybrids between H. melpomene and H. ethilla, and two are believed to be hybrids between H. melpomene and H. hecale [27]. Recent genetic data demonstrated that one of these suspected H. melpomene/H. ethilla hybrids was indeed an F1 hybrid [25].
Figure 1.
Bayesian phylogeny showing the relationship between the distantly-related silvaniform and melpomene/cydno clades of the genus Heliconius. Posterior probabilities for three major nodes are shown (1.0), as are the wing pattern phenotypes for the four analyzed species. The age of the split between the melpomene/cydno and silvaniform clades was estimated assuming an evolutionary rate of 1.15% per lineage per million years [32].
While the existence of rare hybrids between the melpomene/cydno and silvaniform clades provides a potential avenue for gene flow between them, it is unknown whether introgression occurs over these large phylogenetic distances. To determine whether these distantly related groups continue to exchange genes, I used two complementary population genetic datasets to measure the extent of historical gene flow and contemporary admixture between sympatric populations of the two clades.
Results and Discussion
Historical migration inferred from DNA sequence data
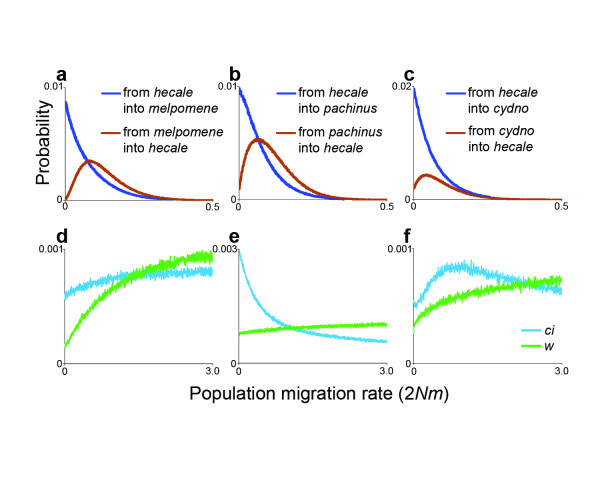
In order to examine rates of gene flow between the two clades, I sequenced multiple haplotypes for one mitochondrial and 13 nuclear genes from three species in the melpomene/cydno clade (H. cydno, H. pachinus, H. melpomene) and one species in the silvaniform clade (H. hecale). I then used these data and the coalescent-based approach implemented in IM [28] to estimate the population migration rate (2Nm) between H. hecale and each of the three melpomene/cydno clade species. All three pairwise analyses revealed non-zero peaks for the marginal posterior probability distributions corresponding to the rate of historical migration from the melpomene/cydno clade into H. hecale (Figure 2a–c). Furthermore, for the comparisons involving H. pachinus and H. melpomene, the 90% highest posterior density interval for this function did not include zero, allowing rejection of the no gene flow hypothesis. These results, which are suggestive of unidirectional gene flow from the melpomene/cydno clade into the silvaniform clade, are consistent with the observation that of the two suspected backcross hybrids that have been collected in the field, both are believed to have resulted from backcrossing in the direction of the silvaniform clade parent (H. melpomene × H. ethilla backcrossed to H. ethilla) [25,27].
Figure 2.
Posterior probability distributions for between-species population migration rates. Three pairwise comparisons based on DNA sequence data for 14 genes revealed non-zero rates of historical gene flow from the melpomene/cydno clade into the silvaniform clade (a, b, c). Locus specific estimates of gene flow into H. hecale from H. melpomene, H. pachinus, and H. cydno, respectively, for the two loci that exhibit evidence of between-clade introgression, cubitus interruptus and white (d, e, f).
It is important to note that the evolutionary history of the DNA sequences studied here violates one of the underlying assumptions of the Isolation with Migration model. The IM model assumes that the two populations being examined are sister-taxa, each being more closely related to the other than either is to any other population. That is clearly not the situation with these inter-clade comparisons. However, if the sampled species provide an unbiased approximation of the genetic divergence between the two groups under study (the silvaniform and melpomene/cydno clades in this case), then it seems reasonable to model the system under the Isolation with Migration framework. The fact that analyses based on independent data (AFLPs, see below) are consistent with the IM results lends support to both the approach and the results.
Gene flow has a variable influence across the genome
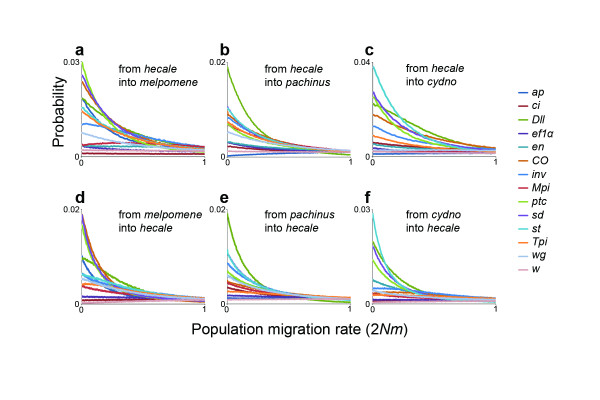
A prediction of speciation models that permit gene flow during the process of divergence is that the influence of introgression should vary throughout the genome [29]. For instance, regions of the genome linked to loci that are under divergent selection between species should be prevented from crossing the species boundary even while other portions of the genome remain interchangeable. To test this prediction, I performed a second series of IM analyses, this time estimating separate population migration rates for each locus. Consistent with the prediction, the shapes of the migration rate posterior probability distributions varied substantially across loci. Most genes had probability distributions that peaked at or near zero indicating little or no historical gene flow (Figure 3). However, two loci, cubitus interruptus (ci) and white (w), had probability distributions consistent with gene flow from the melpomene/cydno clade into H. hecale (Figure 2d–f). While the migration rate probability distributions for ci and w were not well-defined, their shapes suggest that some amount of historical migration fit the data better than no migration.
Figure 3.
Marginal posterior probability distributions for locus-specific migration rates. Probability distributions for gene flow from H. hecale into H. melpomene, H. pachinus, and H. cydno, respectively (a, b, c). Probability distributions for gene flow into H. hecale from H. melpomene, H. pachinus, and H. cydno, respectively (d, e, f). Despite substantial variation in distribution shapes, most peak at or near zero suggestive of little or no gene flow.
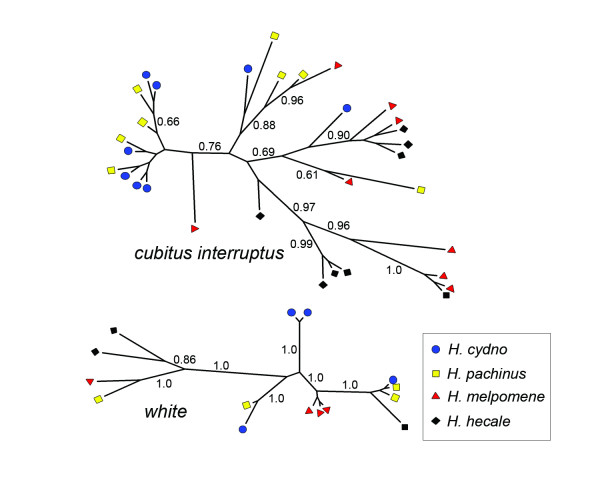
Gene genealogies for these two loci revealed the extent of shared genetic variation between the two clades. For the other 12 loci, H. hecale haplotypes formed a well-supported clade that was distinct from melpomene/cydno clade haplotypes [24], but for both ci and w, H. hecale haplotypes were distributed across the tree (Figure 4). In addition, one identical ci haplotype was shared between H. melpomene and H. hecale. This haplotype was 281 bp long, 139 bp of which consisted of an otherwise highly variable intron. The sharing of an identical haplotype between these two clades is difficult to explain without recent gene flow. Previous work has found evidence of gene flow at ci among species within the melpomene/cydno clade [23,24] but interestingly, the locus with the clearest signature of gene flow between closely-related Heliconius species, Mannose phosphate isomerase [22,23], does not exhibit evidence of introgression over the larger phylogenetic distances examined here.
Figure 4.
Gene genealogies for cubitus interruptus and white. Across loci, haplotypes from H. hecale and the melpomene/cydno clade were reciprocally monophyletic except for at cubitus interruptus and white [24]. The lack of association between species and tree topology for these two loci is indicative of interspecific gene flow. Nodes with posterior probabilities > 0.60 are labeled.
Contemporary admixture
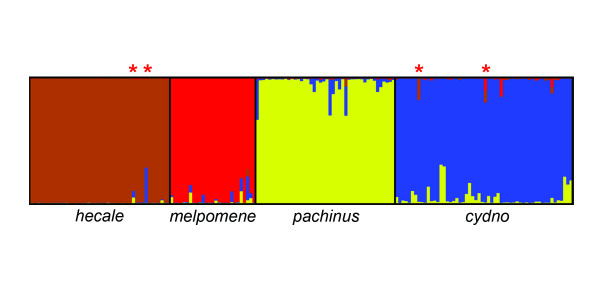
While the analyses of the DNA sequence data are consistent with a low but detectable level of gene flow between the two clades, these results do not reveal its timing. Perhaps gene flow persisted for some time after the initial divergence of the two clades but has since ceased. Alternatively, current hybridization could continue to provide a bridge between the gene pools. To test for evidence of contemporary gene flow, I genotyped 56 H. cydno, 44 H. pachinus, 27 H. melpomene and 44 H. hecale individuals at 520 polymorphic AFLP loci. Using these data and the Bayesian clustering method implemented in STRUCTURE 2.2 [30,31], I performed genetic clustering assuming four populations and allowing individuals to be of mixed ancestry. This analysis correctly delineated the four species and identified two H. cydno individuals and two H. hecale individuals with ancestry from the opposite clade (Figure 5). For each of these individuals, the 95% posterior probability interval for the genome proportion derived from the population of origin did not include one, and for three of the four individuals, the interval for the introgressed genome proportion did not include zero (Table 1). To further assess the statistical confidence for these suspected instances of admixture, I performed a second clustering analysis, this time estimating the posterior probability that each individual had pure ancestry after first indicating the population of origin for each individual and setting the prior probability of pure ancestry to 0.95. All four of the individuals identified in the first analysis had low probabilities of pure ancestry and high probabilities of mixed ancestry with the other clade (Table 1), supporting the hypothesis of contemporary admixture. Interestingly, the four individuals with recently-mixed ancestry based on the AFLP data were not included in the DNA sequence analyses so the signatures of gene flow in the two datasets are independent of one another.
Figure 5.
Genetic clustering based on 520 AFLP loci. Each individual is represented by a narrow vertical column with the proportion of the four colors indicating the genome proportion derived from each of the four populations. Two H. hecale and two H. cydno individuals exhibited evidence of mixed ancestry with the opposite clade (asterisks).
Table 1.
AFLP based estimates of mixed ancestry. Admixture proportions (with 95% confidence intervals) and ancestry probabilities for the four individuals that exhibited evidence of between-clade mixed ancestry.
| Individual | Proportion of genome derived from: | Probability of pure ancestry | Probability of mixed ancestry with the opposite clade | |||
|---|---|---|---|---|---|---|
| H. hecale | H. melpomene | H. pachinus | H. cydno | |||
| hecale #33 | 0.898 (0.791–0.992) | 0.005 (0.000–0.064) | 0.045 (0.000–0.159) | 0.052 (0.000–0.199) | 0.211 | 0.789 |
| hecale #37 | 0.709 (0.594–0.818) | 0.006 (0.000–0.072) | 0.002 (0.000–0.024) | 0.283 (0.167–0.400) | 0.000 | 1.000 |
| cydno #8 | 0.172 (0.092–0.262) | 0.001 (0.000–0.015) | 0.011 (0.000–0.143) | 0.816 (0.674–0.905) | 0.000 | 1.000 |
| cydno #29 | 0.096 (0.009–0.186) | 0.098 (0.000–0.224) | 0.005 (0.000–0.060) | 0.801 (0.653–0.937) | 0.169 | 0.626 |
Gene flow has persisted for millions of years after speciation
Together, these population genetic data are consistent with a history of divergence with gene flow between the melpomene/cydno and silvaniform clades. Average pairwise mtDNA divergence between these two groups is 5.7% (SE = 0.5%). Using the estimate of 1.1 – 1.2% divergence per lineage per million years [32], this equates to approximately 2.5 million years of divergence. With a minimal Heliconius generation time of one month, this represents as many as 30 million generations of evolution along each lineage since the speciation event that precipitated cladogenesis.
Conclusion
These data suggest that the process of divergence that ultimately results in reproductively isolated species can be prolonged. The fact that genomes can remain open to gene flow very long after the speciation process is initiated greatly expands the evolutionary novelty that can be generated from introgression. Some portion of the phenotypic and species diversity in Heliconius has very likely arisen from introgressive hybridization [6,26]. The results presented here suggest that the melpomene/cydno and silvaniform clades of Heliconius have experienced continuous gene flow over millions of years. Thus, introgression has had the potential to provide a ready source of genetic variation to fuel this expansive adaptive radiation. For many organisms, even rare hybridization with distantly related species may allow for the continued exchange of genetic material which may serve as a long-term source of variation for adaptive change.
Methods
Samples
I collected 56 H. cydno, 44 H. pachinus, 27 H. melpomene, and 44 H. hecale individuals from various locations throughout Costa Rica. None of the sampled individuals exhibited phenotypic evidence of introgression from the opposite clade. All specimens were collected as adults in the summer of 2000 and 2002. Tissue was preserved in 95% ethanol and DNA was extracted with a DNeasy Tissue Kit (Qiagen) or using a phenol/chloroform extraction protocol.
DNA sequencing and analysis
I sequenced multiple haplotypes for one mitochondrial locus and 13 nuclear loci from the four species using primers and methods described previously [24,33]. The loci were apterous (ap), cubitus interruptus (ci), cytochrome oxidase I &II (CO, mtDNA), Distal-less (Dll), elongation factor 1α (ef1α), engrailed (en), invected (unv), Mannose phosphate isomerase (Mpi), patched (ptc), scalloped (sd), scarlet (st), Triose phosphate isomerase (Tpi), white (w), and wingless (wg). I also sequenced portions of the genes cinnabar [24] and decapentaplegic [34] but these loci were excluded from the analyses because only one H. hecale sequence was obtained for each. All sequences have been deposited in GenBank under accession numbers AY744577–AY744672, AY745254–AY745278, AY745315–AY745335, AY745356–AY745490, DQ448305–DQ448516 and EF041105–EF041122. For analysis, the datasets for CO,ef1α,Mpi, and Tpi were supplemented with published sequences from GenBank [24].
I used the comparative DNA sequence data and the Isolation with Migration model implemented in IM [28] to infer historical rates of between-species gene flow. IM cannot handle alignment gaps or evidence of recombination in DNA sequence data. Therefore, I removed gaps and searched for evidence of recombination using the four-gamete test in DnaSP 3.5 [35]. For those loci that exhibited evidence of recombination, the sequence alignment was divided into portions that showed no evidence of recombination and only one portion was included in the analysis. In an effort to preserve as much genealogical information as possible, the portion with the most polymorphisms was chosen. The size of these non-recombining portions ranged from 60 bp for st in the H. cydno/H. hecale comparison to 1111 bp for wg in the H. melpomene/H. hecale comparison.
For each comparison between H. hecale and one of the three melpomene/cydno clade species, I ran IM in two different ways. First, I used the data for all loci to estimate a single pair of bidirectional population migration rates (Figure 2a–c). Then, using the same dataset, I allowed each locus to have a separate pair of population migration rates (Figure 3). For each analysis, IM was run with 10 Metropolis-coupled chains for 300,000 burn-in steps followed by 2 × 107steps of data collection. Following other implementations of IM [28], I used the HKY model for the mitochondrial locus and the infinite-sites model for the nuclear loci.
Phylogeny estimation
I used published DNA sequences from six genes [36] and MrBayes [37] to estimate a phylogeny of the silvaniform and melpomene/cydno clades of Heliconius. Four Metropolis-coupled Markov chains were run for 250,000 burn-in generations followed by 1.75 × 106 generations of data collection. The age of the melpomene/cydno and silvaniform split was estimated based on 1606 base pairs of mtDNA from ref. 36, assuming an evolutionary rate of 1.15% per lineage per million years [32]. Gene genealogies for ci and w were also estimated using MrBayes, based on the GTR+I+Γ model of molecular evolution.
AFLP genotyping and analysis
Using the ABI Plant Mapping Kit (PE Applied Biosystems), I genotyped each individual with four selective AFLP primer combinations; EcoRI-ACT/MseI-CAT, EcoRI-ACT/MseI-CTG, EcoRI-ACA/MseI-CAT, and EcoRI-ACA/MseI-CTG. AFLP fragments were separated using an ABI 3100 automated genotyper and then scored using ABI GENEMAPPER software version 3.7. In total, each individual was typed for the presence or absence of a fragment at 925 AFLP loci. In an effort to focus on the most informative and reliable markers, only the 520 AFLPs that had a minor allele frequency ≥ 0.05 were used in further analyses.
I used the AFLP data and the Bayesian-based genetic clustering program STRUCTURE 2.2 [30,31] to define populations and estimate admixture among them. The data were analyzed in two ways. First, I performed naïve admixture clustering assuming four populations. As part of this analysis, I estimated the 95% posterior probability interval around each individual admixture proportion. Second, I performed an additional round of admixture clustering after first indicating the population of origin for each individual and setting the prior probability of pure ancestry to 0.95. As part of this analysis, I estimated the posterior probability that each individual was misclassified or had an ancestor from each of the other species within the last three generations. For both analyses, STRUCTURE was run for 10,000 burn-in generations followed by 106 generations of data collection.
Authors' contributions
MRK conceived of the study, performed the research, analyzed the data, and wrote the manuscript.
Acknowledgements
I thank Larry Gilbert, Ulrich Mueller, Joan Strassmann, Dave Queller, Lauren Blume, Laura Young, Andres Vega, and Kenny Kronforst for facilitating this research. I also thank reviewers for comments on the manuscript. This work was funded by National Science Foundation Grants DEB 0415718 & DEB 0640512.
References
- Rieseberg L. Hybrid origins of plant species. Annu Rev Ecol Syst. 1997;28:359–389. doi: 10.1146/annurev.ecolsys.28.1.359. [DOI] [Google Scholar]
- Grant V. Plant Speciation. New York, Columbia Univ. Press; 1981. [Google Scholar]
- Jain R, Rivera MC, Moore JE, Lake JA. Horizontal gene transfer in microbial genome evolution. Theor Popul Biol. 2002;61:489–495. doi: 10.1006/tpbi.2002.1596. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Cambridge, Harvard Univ. Press; 1963. [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Mavarez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Syst. 1997;28:593–619. doi: 10.1146/annurev.ecolsys.28.1.593. [DOI] [Google Scholar]
- Schwarz D, Matta BM, Shakir-Botteri NL, McPheron BA. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature. 2005;436:546–549. doi: 10.1038/nature03800. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Fordyce JA, Forister ML, Shapiro AM, Nice CC. Homoploid hybrid speciation in an extreme habitat. Science. 2006;314:1923–1925. doi: 10.1126/science.1135875. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Machado CA, Hey J. The causes of phylogenetic conflict in a classic Drosophila species group. Proc R Soc Lond B. 2003;270:1193–1202. doi: 10.1098/rspb.2003.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Touré Y, Sagnon NF. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci USA. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PR, Grant BR, Markert JA, Keller LF, Petren K. Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution. 2004;58:1588–1599. doi: 10.1111/j.0014-3820.2004.tb01738.x. [DOI] [PubMed] [Google Scholar]
- Edmands S. Does parental divergence predict reproductive compatability? Trends Ecol Evol. 2002;17:520–527. doi: 10.1016/S0169-5347(02)02585-5. [DOI] [Google Scholar]
- Hubbs CL. Hybridization between fish species in nature. Syst Zool. 1955;4:1–20. doi: 10.2307/2411933. [DOI] [Google Scholar]
- Hubbs CL, Laritz CM. Occurrence of a natural intergeneric Etheostomatine fish hybrid. Copeia. 1961;2:231–232. doi: 10.2307/1440009. [DOI] [Google Scholar]
- Smith GR. Analysis of several hybrid cyprinid fishes from western North America. Copeia. 1973. pp. 395–410. [DOI]
- Smith DG. Evidence for hybridization between two crayfish species (Decapoda: Cambaridae: Orconectes) with a comment on the phenomenon in Cambarid crayfish. Am Midl Nat. 1981;105:405–407. doi: 10.2307/2424764. [DOI] [Google Scholar]
- Aspinwall N, Carpenter D, Bramble J. The ecology of hybrids between the peamouth, Mylocheilus caurinus, and the redside shiner, Richardsonius balteatus, at Stave Lake, British Columbia, Canada. Can J Zool. 1993;71:83–90. [Google Scholar]
- Gergus EWA, Malmos KB, Sullivan BK. Natural hybridization among distantly related toads (Bufo alvarius, Bufo cognatus, Bufo woodhousii) in central Arizona. Copeia. 1999. pp. 281–286. [DOI]
- Mallet J, Beltrán M, Neukirchen W, Linares M. Natural hybridization in heliconiine butterflies: the species boundary as a continuum. BMC Evol Biol. 2007;7:28. doi: 10.1186/1471-2148-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán M, Jiggins CD, Bull V, Linares M, Mallet J, McMillan WO, Bermingham E. Phylogenetic discordance at the species boundary: comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol Biol Evol. 2002;19:2176–2190. doi: 10.1093/oxfordjournals.molbev.a004042. [DOI] [PubMed] [Google Scholar]
- Bull V, Beltrán M, Jiggins CD, McMillan WO, Bermingham E, Mallet J. Polyphyly and gene flow between non-sibling Heliconius species. BMC Biol. 2006;4:11. doi: 10.1186/1741-7007-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst MR, Young LG, Blume LM, Gilbert LE. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006;60:1254–1268. [PubMed] [Google Scholar]
- Dasmahapatra KK, Silva-Vásquez A, Chung JW, Mallet J. Genetic analysis of a wild-caught hybrid between non-sister Heliconius butterfly species. Biol Lett. 2007;3:660–663. doi: 10.1098/rsbl.2007.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LE. In: Ecology and Evolution Taking Flight: Butterflies as Model Systems. Boggs, CL, Watt WB, Ehrlich PR, editor. Chicago, Univ. Chicago Press; 2003. Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic "tool box" from synthetic hybrid zones and a theory of diversification; pp. 281–318. [Google Scholar]
- Mallet J, Neukirchen W, Linares M. Hybrids between species of Heliconius and Eueides butterflies: a database. 2006. http://www.ucl.ac.uk/taxome/hyb/
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelianov I, Marec F, Mallet J. Genomic evidence for divergence with gene flow in host races of the larch budmoth. Proc R Soc Lond B. 2004;271:97–105. doi: 10.1098/rspb.2003.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst MR. Primers for the amplification of nuclear introns in Heliconius butterflies. Mol Ecol Notes. 2005;5:158–162. doi: 10.1111/j.1471-8286.2004.00873.x. [DOI] [Google Scholar]
- Kronforst MR, Salazar C, Linares M, Gilbert LE. No genomic mosaicism in a putative hybrid butterfly species. Proc R Soc Lond B. 2007;274:1255–1264. doi: 10.1098/rspb.2006.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Beltrán M, Jiggins CD, Brower AVZ, Bermingham E, Mallet J. Do pollen feeding, pupal-mating and larval gregariousness have a single origin in Heliconius butterflies? Inferences from multilocus DNA sequence data. Biol J Linn Soc. 2007;92:221–239. doi: 10.1111/j.1095-8312.2007.00830.x. [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]