Abstract
Infection of primates by HIV-1 and SIV induces multiple hematological abnormalities of central hematopoietic origin. Although these defects greatly contribute to the pathophysiology of HIV-1 infection, the molecular basis for altered BM function remains unknown. Here we show that when cynomolgus macaques were infected with SIV, the multipotent potential of their hematopoietic progenitor cells was lost, and this correlated with downregulation of STAT5A and STAT5B expression. However, forced expression of STAT5B entirely rescued the multipotent potential of the hematopoietic progenitor cells. In addition, an accessory viral protein required for efficient SIV and HIV replication and pathogenicity, “Negative factor” (Nef), was essential for SIV-mediated impairment of the multipotent potential of hematopoietic progenitors ex vivo and in vivo. This newly uncovered property of Nef was both conserved between HIV-1 and SIV strains and entirely dependent upon the presence of PPARγ in targeted cells. Further, PPARγ agonists mimicked Nef activity by inhibiting STAT5A and STAT5B expression and hampering the functionality of hematopoietic progenitors both ex vivo and in vivo. These findings have extended the role of Nef in the pathogenicity of HIV-1 and SIV and reveal a pivotal role for the PPARγ/STAT5 pathway in the regulation of early hematopoiesis. This study may provide a basis for investigating the potential therapeutic benefits of PPARγ antagonists in both patients with AIDS and individuals with hematopoietic disorders.
Introduction
Patients with AIDS exhibit multiple hematopoietic abnormalities, including anemia, granulocytopenia, and thrombocytopenia (1, 2). These reflect central hematopoietic deficiency (3, 4). Long-term BM cultures from HIV-infected patients have low CD34+ progenitor cell growth and differentiation (5, 6), indicating impaired functionality of early hematopoietic progenitors. Such hematopoietic failure affects T cell production and should contribute to the immunodeficiency characteristic of AIDS patients (7). Also, abnormalities in fetal hematopoiesis have been reported in aborted fetuses from seropositive women (8). However, CD34+ BM progenitors from HIV-infected patients are devoid of proviral DNA (9–12). This suggests that HIV-1 infection hampers hematopoiesis indirectly. Ex vivo, HIV-1 alters the hematopoietic microenvironment (1, 13, 14); affects hematopoietic progenitors through the viral envelope gp120, through Gag p24, or through Negative factor (Nef) (15–19); and enhances secretion of inhibitory cytokines, including tumor necrosis factor alpha (20). However, the way HIV affects early hematopoiesis in vivo is still unknown.
SIV-infected macaques are a very good animal model to study how immunodeficiency viruses affect early hematopoiesis in vivo. SIV-infected macaques present immunodeficiency syndrome and hematological changes — including impaired clonogenic growth of CD34+ BM progenitors — that mimic those of human AIDS (21, 22).
To study the effects of immunodeficiency virus on early hematopoiesis, we tested BM progenitors from SIVmac251-infected macaques for deregulated genes possibly responsible for the observed defects. We report that the hematopoietic defects of infected animals correlated with downregulation of STAT5A and STAT5B gene expression in CD34+ progenitors. These defects were corrected by STAT5B overexpression in CD34+ cells. We show for the first time to our knowledge that Nef was responsible for these defects, both ex vivo and in vivo, and relied on the presence and activation of the PPARγ signaling pathway. These data reveal what we believe to be a previously unsuspected inhibitory role of the PPARγ signaling pathway in early hematopoietic progenitors and suggest its involvement in hematopoietic dysfunction in infected patients.
Results
STAT5 is responsible for SIV-dependent loss of functional multipotent hematopoietic progenitors.
Blood and BM samples from 5 macaques were collected at various times before and after animals were intravenously inoculated with fifty 50% animal infectious dose (AID50) of the pathogenic SIVmac251 strain. Upon SIV inoculation, animals developed an infection with typical plasma viral load profiles, as previously described (7) (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI33037DS1). To number and assess the functionality of multipotent hematopoietic progenitors, CD34+ cells were purified from BM samples, and well-known short-term colony-forming assays (23) were first performed. The proportion of CD34+ BM cells was constant along infection (7% ± 2% before infection, 6% ± 2% on day 35 after SIV injection, 6% ± 1% on day 127 after SIV injection, and 9% ± 2% on day 260 after SIV injection). Also, as reported (7) and confirmed in the present samples (data not shown), the collected CD34+ cells lacked any provirus and viral RNA at any time following SIV injection, as assayed by sensitive nested PCR and real time RT-PCR assays, respectively. However, for all tested animals, total CFCs in these CD34+ progenitors decreased from 4 weeks after injection and remained at low levels during chronic infection (Figure 1A). By day 260 after injection, the total CFC number was 26% ± 3% of that before infection. These results are consistent with the CD34+ progenitor clonogenic defects reported in human HIV-seropositive patients (6, 10, 12).
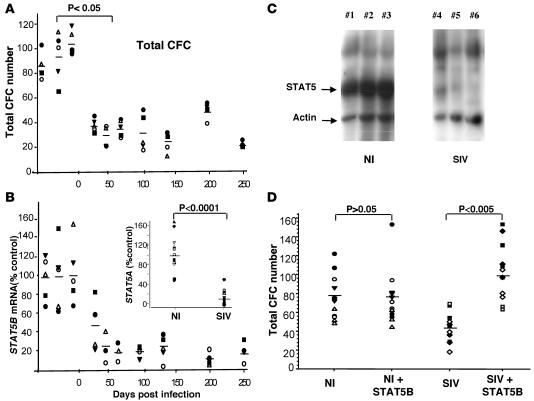
Figure 1. SIV inhibits the clonogenic potential of hematopoietic progenitors through downregulation of STAT5.
(A) Evaluation of total progenitor cell counts in semisolid cultures of CD34+ BM cells collected from 5 animals before and after infection with SIV. Day 0 is the day of SIV injection. Horizontal lines indicate mean CFC numbers scored from all cell cultures from the 5 animals analyzed at the indicated time. Each symbol represents samples from a single animal. (B) STAT5B mRNA was evaluated by RT-PCR in CD34+ BM cells at various times following animal infection with SIV. Levels were normalized to GAPDH mRNA. Results were expressed relative to the average level of STAT5B mRNA in samples collected before SIV injection. Horizontal lines indicate the mean values for STAT5B mRNA in 5 animal samples at the indicated time. STAT5A mRNA was similarly evaluated and compared in noninfected control (NI) and chronically SIV-infected animals (SIV) (inset). (C) CD34+ BM cells from 3 noninfected control and 3 SIVmac251-infected macaques at 93 days after injection were lysed. Lysates were subjected to SDS-PAGE and western blotting. STAT5 and actin proteins were detected by coincubation with pan STAT5 and anti-actin antibodies. (D) CD34+ BM cells from 4 control or 4 chronically SIV-infected macaques were transduced with or without a lentiviral vector encoding simian STAT5B. They were processed for CFC assays. Experiments were repeated twice. Horizontal lines indicate the mean number of colonies scored in each condition. Each symbol represents samples from a single animal.
We investigated the origin of the hematopoietic defect in SIV-infected animals. HSC renewal and determination are tightly controlled by intrinsic and extrinsic factors. A few transcription factors have been suggested to maintain HSC pluripotency, including HoxB4, Notch1, and Bmi1 among others (24). Similarly, various cytokines and growth factors play crucial roles in HSC renewal through activation of a discrete number of signaling pathways. Among these are the STAT5A and STAT5B (25, 26), 2 very similar proteins that have been reported to be required for early hematopoiesis and long-term multilineage reconstitution following HSC transplantation in mice (27–32). We used quantitative RT-PCR to evaluate the expression of genes important for HSC functions in hematopoietic progenitors isolated from infected animals. We observed a progressive decrease in STAT5B mRNA in CD34+ progenitors following SIV injection (Figure 1B). This decrease correlated with the loss of CFCs in infected animals (P = 0.0002). The lowest level was observed 50 days after injection and remained constant during chronic infection. STAT5B mRNA levels were only 15% of those observed in progenitors from uninfected macaques. STAT5A mRNA levels were also substantially lower in CD34+ cells from chronically infected macaques than in CD34+ cells from uninfected animals (Figure 1B, inset). Moreover, STAT5A/B proteins were nearly undetectable in CD34+ cells from infected animals (Figure 1C). This suggests that downregulation of STAT5 gene expression in hematopoietic progenitors is responsible for the SIV-dependent clonogenic defect of CD34+ progenitors. To address this, we evaluated the functionality of CD34+ cells following transduction with a lentiviral expression vector encoding STAT5B. The SIN-PGK-WHV lentiviral vector used allowed expression of a Tg in 70%–75% of the normal simian CD34+ as observed with control GFP vector (Supplemental Figure 2A). In these normal progenitors, the levels of STAT5B expression obtained in STAT5B-vector transduced cells were 5 times greater than levels found in untransduced cells (Supplemental Figure 2B), but clonogenic capacities were unaffected (Figure 1D). However, similar transduction of CD34+ cells collected from infected macaques strongly enhanced their clonogenic capacities. Overexpressing STAT5B rescued the clonogenic capacity of BM progenitors collected from infected animals. This indicates that SIV-related hematopoietic failure is STAT5 dependent.
Nef mediates hematopoietic defects induced by SIV infection.
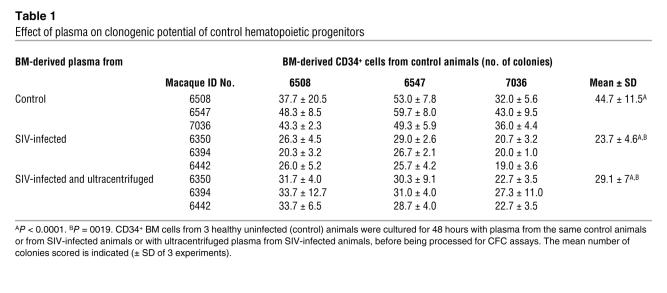
To delineate the origin of the SIV-dependent hematopoietic defects, CD34+ BM cells from control (noninfected) macaques were incubated for 2 days with plasma from healthy or chronically SIV-infected animals and then processed for CFC assays as described above (see Figure 1A). Progenitor clonogenicity was lower in normal CD34+ cells incubated with SIV-containing plasma than in those incubated with uninfected plasma (Table 1). If infected plasma were initially ultracentrifuged to remove any detectable viral particles (less than 60 copies, as determined by real-time RT-PCR), they still significantly decreased clonogenicity of CD34+ cells (Table 1), suggesting that a soluble factor from infected plasma directly affected the clonogenic potential of normal hematopoietic progenitors ex vivo. To determine whether such a factor was of viral origin, we added purified SIVmac251 infectious particles to plasma from control animals and performed CFC assays on normal CD34+ cells. CFC number decreased significantly in the presence of 102 SIV infectious particles/ml (Figure 2A). However, the infectious titer for reasonably packaged lentiviral vector is often 102–104 lower than the lentiviral physical titer as active viral stocks contain large amounts of defective or partially denaturated particles, which all still express SIV proteins (33). To assess the actual amounts of physical lentiviral particles added to culture medium, we quantified the p27 core protein present and evaluated that 102 SIV infectious particles/ml actually contained 160 pg/ml SIV p27. Because there are roughly 2,000 molecules of p27 per lentiviral particle, 160 pg/ml SIV p27 corresponded to 3.2 105/ml SIV physical particles. This inhibitory activity was maintained when SIV particles were subjected to a brief heat-inactivation process that suppressed their capacity to infect cells, indicating that it was independent of viral infectivity. Altogether these data suggested that a soluble factor present in viral particles and also in infected but virus-depleted plasma affects the biological properties of CD34+ progenitors.
Table 1 .
Effect of plasma on clonogenic potential of control hematopoietic progenitors
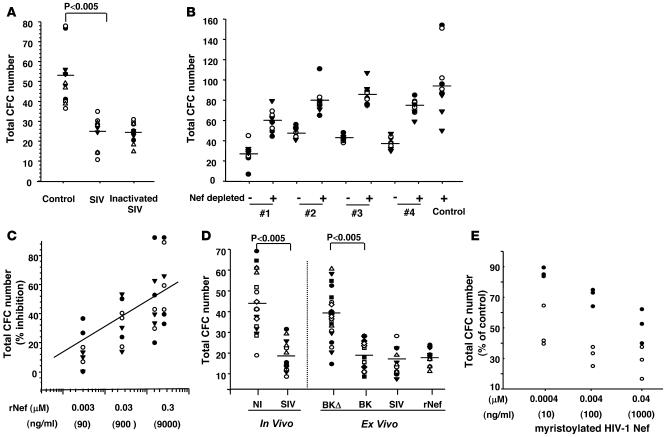
Figure 2. Nef mimics SIV actions on hematopoietic progenitors.
(A–C) CFC assays were performed with CD34+ cells isolated from 3–4 control animals following incubation for 48 hours with (A) plasma from noninfected animals in the absence (control) or presence of infectious or heat-inactivated SIVmac251 particles (1 × 102 particles/ml), (B) plasma from noninfected animal (control) or plasma from 4 chronically infected macaques without (–) or with (+) Nef immunodepletion, or (C) with various concentrations of recombinant SIVmac251 Nef. (D) CFC assays were performed with CD34+ BM cells isolated from SIVmac251-infected or noninfected macaques. Progenitors from uninfected animals were either left untreated or incubated for 48 hours with the viral isolate SIVmac251, molecular clones BK28-41 (BK) or BK28-41ΔNef (BKΔ) (1 × 102 infectious particles/ml), or with rNef (0.15 μM) before being processed for CFC assays. (E) Inhibitory activity of recombinant myristoylated HIV-1 Nef was assayed on CD34+ BM cells isolated from 2 healthy macaques. CD34+ BM cells were preincubated for 48 hours with the indicated concentration of myristoylated HIV-1 Nef before CFC assays. Horizontal lines and the diagonal line in C indicate mean of CFC numbers scored from all cell cultures from the animals analyzed. Each kind of symbol represents samples from a single animal.
We investigated whether any SIV protein known to be present in infected plasma and also packaged in substantial quantities into viral particles can be responsible for the defect. The Nef gene encodes a 206–/292–amino acid myristoylated protein required for efficient in vivo viral replication and pathogenicity (34). It is produced in large amounts early after HIV/SIV infection, is present in viral particles (35), and is released into the serum at concentrations up to 10 ng/ml (36). We thus tested whether Nef is involved in SIV-dependent hematopoietic failure by first depleting Nef from infected plasma before performing CFC assays. Addition of anti-Nef antibodies to plasma collected from chronically infected macaques and removal of the immune complex that had formed, depleted Nef from plasma (Supplemental Figure 3A) and prevented plasma inhibitory activity (Figure 2B). We next added E. coli–recombinant SIVmac251 Nef protein (rNef) to culture medium of CD34+ progenitors isolated from uninfected animals and processed for CFC assays as described above. rNef (0.15 μM) reduced the clonogenic potential of normal CD34+ BM progenitors as much as 1 × 102 infectious particles/ml of SIVmac251 (or 160 pg/ml SIV p27) (Figure 2, C and D). To avoid possible artefacts from E. coli contaminants, we further tested various sources of recombinant Nef. Similar results were obtained with baculovirus-recombinant SIV J5 Nef, and with E. coli–recombinant HIV-1 Nef Laï (data not shown). Moreover, fully myristoylated recombinant HIV-1 Nef was also tested and shown to exhibit enhanced inhibitory activity (Figure 2E). These data indicate that soluble HIV-1 and SIV Nef exhibit a new conserved activity that impairs the functionality of normal hematopoietic progenitors when added to culture medium.
To strengthen the importance of Nef in SIV-dependent hematopoietic defects, we made use of another well-studied SIV strain, BK28-41, which allows synthesis of all SIV (viral) proteins with Nef truncated in the C-terminal domain, while its reported variant BK28-41ΔNef produces all viral proteins except Nef due to mutation of the Nef initiation codon and deletion of 174 nt located 200 nt further downstream (37). As shown in Figure 2D, BK28-41 induced a severe clonogenic defect when added ex vivo to normal CD34+ BM progenitors in CFC assays. This defect was comparable to that observed with SIVmac251 ex vivo and also in vivo following SIV infection (Figure 2D, left panel). However BK28-41ΔNef did not affect the clonogenic potential of CD34+ cells (Figure 2D, right panel). Nef is known to be located within virus particles, therefore the inhibitory activities of exogenously added virions looked quite puzzling. We hypothesized the presence of soluble Nef in the virus preparations used. These stocks were thus incubated directly with anti-Nef antibodies and the immune complexes formed were analyzed by SDS-PAGE and immunoblotting to assess for the presence of soluble Nef. These experiments indicated that rather large amounts of soluble Nef were present in infectious virus stocks, which even exceeded those present within virions themselves (see Supplemental Figure 3B).
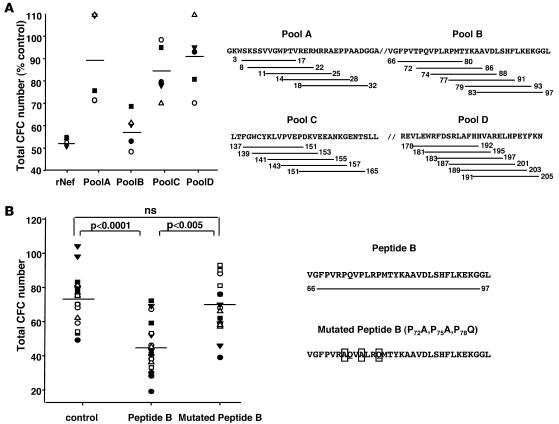
We investigated which domain(s) of Nef is responsible for this conserved inhibitory activity. Lentiviral Nef peptide sequences can be roughly divided into 4 functional regions. Region A covers the amino-terminus of Nef and includes a myristoylation site; region B includes a putative Src homology 3 (SH3) binding motif; region C overlaps an SH2 domain; and region D includes the carboxy-terminus of the molecule. We used CFC assays to test pools of overlapping peptides (15 amino acids long), spanning the 4 functional domains of the HIV-1 Nef Laï strain (Figure 3A, right panel). The pool of peptides covering region B mimicked Nef action and caused a major clonogenic defect (P < 0.05) in CD34+ cells; the other pools were inactive (Figure 3A, left panel). We further observed that a single peptide that covers region B entirely (amino acids 66–97) was sufficient to affect CD34+ BM cell clonogenicity substantially (Figure 3B, left panel). Interestingly, the sequence of region B is the only 1 conserved (>80%; Los Alamos Database; http://hiv-web.lanl.gov) within SIV and HIV genomes; it is moreover absent in the inactive BK28-41ΔNef strain (37). This region contains a Pro rich motif (P72XXP75XXP78), whose mutation to A72XXA75XXQ78 completely abolished the pathogenic potential of Nef for the induction of a severe AIDS-like disease in CD4C/HIV Tg mice (38). This motif is conserved between the majority of SIV strains, including SIVmac251 (39). We therefore assessed the role of this motif. We observed that mutation to A72XXA75XXQ78 similarly abolished Nef inhibitory actions on CD34+ BM cells (Figure 3B, left panel). Overall, these data indicate that the conserved Pro-rich peptide sequence from the central region of HIV/SIV Nef can impair the hematopoietic potential of normal progenitors ex vivo.
Figure 3. HIV/SIV-related hematopoietic failure is dependent on a central Pro-rich motif of Nef.
(A) CD34+ cells from control animals were incubated for 48 hours with rNef (0.15 μM) or the indicated pools of Nef peptides (100 nM). (B) Alternatively, CD34+ cells were incubated with either wild-type or mutated (P72, P75, P78) peptide B (100 nM). Cells were then processed for CFC assays.
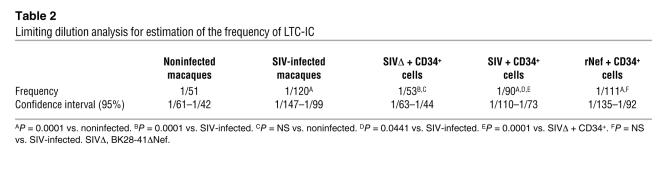
We assessed the impact of Nef on more immature progenitor cells and tested functionality of the long-term colony-initiating cell (LTC-IC) compartment (40). Normal CD34+ BM progenitors incubated with either SIVmac251 particles or rNef had many fewer LTC-ICs than untreated samples (Figure 4). This decrease was similar to that observed for CD34+ cells isolated from chronically infected macaques (Figure 4, compare left and right panels). Limiting dilution analysis showed that CD34+ BM cells from macaques infected with SIV and normal CD34+ BM cells incubated with either rNef or SIVmac251 particles had similar LTC-IC frequencies (Table 2). However, there was no significant difference in LTC-IC frequency between progenitors incubated with control plasma in the presence or absence of BK28-41ΔNef particles. These data confirm the role of Nef in mediating HIV/SIV-related hematopoietic defects.
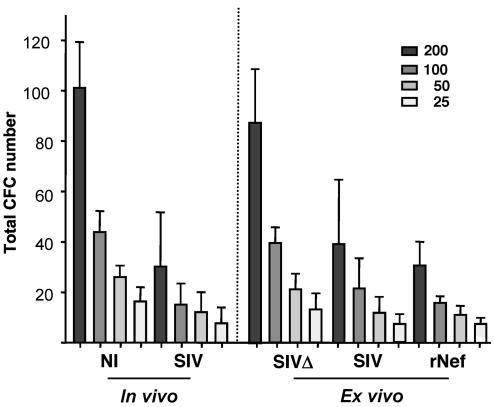
Figure 4. SIV impairs LTC-IC.
The left panel shows LTC-IC assay of CD34+ BM cells from 5 uninfected and 5 SIV-infected macaques. Various numbers of progenitor cells were initially seeded as indicated. The right panel shows LTC-IC assay of CD34+ BM cells from 5 control animals that were preincubated for 48 hours with plasma from noninfected animals supplemented with SIVmac251 (SIV), BK28-41ΔNef (SIVΔ) (1 × 102 infectious particles/ml), or rNef (0.15 μM).
Table 2 .
Limiting dilution analysis for estimation of the frequency of LTC-IC
PPARγ mediates Nef activities and inhibits hematopoietic progenitors.
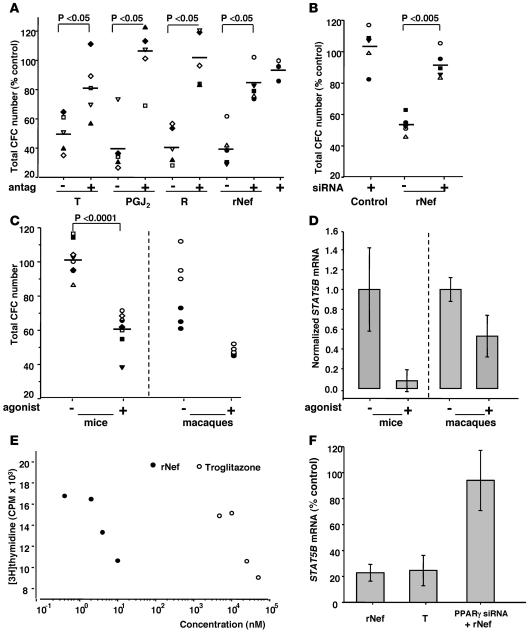
PPARγ is a nuclear receptor that has antiproliferative effects in preadipocytes and mammary epithelial cells (41). Because Nef binds to PPARγ (42) and suppresses adipogenesis, we tested whether PPARγ mediates the hematopoietic actions of Nef. We assayed clonogenic properties of normal CD34+ BM cells in response to 6 potent and specific PPARγ ligands (PGJ2, troglitazone, ciglitazone, rosiglitazone, pioglitazone, and MCC-555) and to the PPARα, -γ, -δ agonist, GW501516. Similar to Nef, all PPARγ agonists inhibited CD34+ clonogenicity in CFC assays (Figure 5A and data not shown). We preincubated the irreversible low-affinity PPARγ antagonist, GW9662, before adding each of the agonists to assess the specificity of PPARγ action. GW9662 counteracted the action of all PPARγ agonists tested. Surprisingly, it also counteracted SIV rNef action in the same CFC assays.
Figure 5. Nef action on hematopoietic cells depends on the presence of PPARγ and its downregulation of STAT5.
(A) Clonogenic capacities of CD34+ BM cells were assayed following preincubation for 2 days with culture medium alone (control) or supplemented with SIV-rNef (rNef, 0.15 μM) or PPARγ agonists troglitazone (T) or rosiglitazone (R) or PGJ2 (25 μM) in the presence or absence of PPARγ antagonist GW9662 (antag, 10 μM). Samples from 5 macaques were analyzed, each represented by 1 symbol. The number of colonies scored was expressed as percentage of control values. (B) Clonogenic capacities of CD34+ BM cells were assayed on cells transfected with or without PPARγ siRNA or irrelevant siRNA (ir) and incubated for 2 days in culture medium in the absence or presence of rNef before CFC assays. Samples from 5 macaques were analyzed, each represented by 1 symbol. CFC numbers were expressed relative to the CFC scored in the absence of siRNA PPARγ and rNef. (C) Clonogenic capacities of hematopoietic progenitors isolated from mice (n = 8, left) or macaques (n = 2, right), pretreated with or without rosiglitazone (agonist) during 2 weeks. (D) Real-time RT-PCR of STAT5B mRNA from murine BM sca-1+ c-kit+ CD3– cells or macaques CD34+ BM cells, treated with or without rosiglitazone (agonist) during 2 weeks. STAT5B mRNA were normalized to GAPDH mRNA. (E) K562 cell proliferation, evaluated by [3H]thymidine incorporation, in cells incubated for 7 days in the presence of various concentrations of rNef or troglitazone. (F) Real-time RT-PCR of STAT5B mRNA from K562 cells incubated with or without siRNA directed against PPARγ and troglitazone (25 μM) or rNef (0.15 μM). STAT5B mRNA were normalized to GAPDH mRNA and were expressed relative to untreated cells. Efficiency of the siRNA transfection has assessed with a fluorescent control siRNA (25 nM) was over 98%; inhibition of PPARγ mRNA was 98% ± 2% (n = 5).
To extend these results, we introduced siRNA directed against PPARγ into CD34+ cells. Liposome-based reagents allowed highly efficient introduction of siRNAs in simian CD34+ BM cells (96% ± 2%; n = 5) as observed with fluorescent control siRNA (Supplemental Figure 4A). Transfection of the PPARγ-directed siRNA nearly abolished the expression of PPARγ transcripts in the progenitors as opposed to control siRNA (Supplemental Figure 4B). The transfected cells were maintained in culture for 2 days in the presence or absence of rNef and then processed for CFC assays. In the absence of rNef, the highly efficient PPARγ-directed siRNA did not change the clonogenic potential of CD34+ cells (Figure 5B). However, the same siRNA abolished Nef-dependent clonogenic defects of the progenitors. As expected, control siRNA had no action (data not shown). These data demonstrate that Nef requires the presence of PPARγ to impair the functionality of early hematopoietic progenitors.
We tested whether PPARγ agonists as such still modulate hematopoiesis in vivo. Either mice or macaques were treated with rosiglitazone for 15 days, BM hematopoietic progenitors were then selected, and CFC assays were performed. Sca-1+ c-kit+ CD3– and CD34+ BM cells from treated mice and macaques, respectively, showed a strong clonogenic failure as compared with untreated animals (Figure 5C). Taken together, these data indicate that activation of PPARγ modulates the functionality of early myeloid progenitors in vivo.
Activation of Nef/PPARγ pathway inhibits hematopoietic cell growth and STAT5 expression.
To more thoroughly investigate links between Nef, PPARγ, and STAT5 expression in hematopoietic cells, we made use of the human hematopoietic K562 cell line that exhibits cytokine-independent cell growth and constitutive STAT5 activation (43). We observed that both HIV-1 rNef and the PPARγ agonist troglitazone inhibited K562 cell growth in a dose-dependent manner, with HIV-1 rNef exhibiting much stronger potency (Figure 5E). This inhibition correlated with downregulation of STAT5A (data not shown) and STAT5B transcripts in the treated cells (Figure 5F). In addition, suppression of PPARγ expression through siRNA-based approaches abolished both actions of Nef, i.e., cell-growth inhibition (data not shown) and STAT5 mRNA downregulation (Figure 5F). These data indicated that active PPARγ deregulated cell growth and STAT5 expression in hematopoietic K562 cells ex vivo. To confirm these activities in vivo, we further analyzed the expression of STAT5A/B in the hematopoietic progenitors isolated from rosiglitazone-treated animals (see above). We observed that activating PPARγ inhibited STAT5 mRNA expression in murine and simian hematopoietic progenitors in vivo (Figure 5D).
Discussion
Hematopoiesis is the highly regulated process of continuous production of blood cells from early progenitors. HIV/SIV induces large hematopoietic defects that reflect progenitor failure and contribute to weak lymphopoiesis and immunodeficiency in infected individuals. Our study showed that SIV infection affected the functionality of early hematopoietic progenitors, including LTC-IC and CFC progenitors, by downregulating STAT5A and STAT5B expression in vivo. Under similar conditions, SIV did not affect expression of HoxB4, Notch1, GATA2, GATA3, RbTn2, or Tal1 genes (data not shown). Similarly, neither STAT3 nor STAT4 expression was impaired (data not shown). These observations reveal for what we believe to be the first time the crucial role of STAT5 genes in the maintenance of functional multipotent hematopoietic progenitors in primates in vivo. They further indicate that STAT3 cannot compensate for such STAT5 biological activities.
STAT5A/B factors are essential for the generation of functional murine HSCs (30, 44). BM cells isolated from mice bearing partial deletion of both genes, which actually expressed partially active, amino-terminal truncated STAT5A/B proteins (renamed STAT5ABΔn/Δn mice; ref. 43), are unable to contribute to hematopoiesis after competitive repopulation against wild-type BM progenitors in irradiated animals (45, 46). However, these mice do reach birth at expected mendelian frequency and exhibit rather mild lymphoid phenotypes as compared with the recently described STAT5A/Bnull/null mice that have a complete deletion of the Stat5a and Stat5b gene locus and exhibit more than 99% perinatal lethality (32, 43). Furthermore, lymphopoiesis is also highly abnormal in these mice, and STAT5ABΔn/Δn mice have fewer CD8+ T cells and a lower T cell response to interleukin-2 than wild-type animals; similar T cell alterations are observed in infected individuals upon HIV/SIV infection (47). HIV-1 mediates selective decrease in STAT5 levels in purified T cells from infected patients (48). Our data further indicate that SIV also suppresses STAT5 expression in early hematopoietic progenitors, this suppression being responsible for the observed hematopoietic defects. Such STAT5 inhibition should similarly occur in patients and contribute to their hematopoietic dysfunction, severe lymphopoietic defects, and immunodeficiency.
We further demonstrated that SIV inhibitory activity on hematopoietic progenitors entirely depends on the presence of Nef based on the following observations: (a) immunodepletion of Nef from plasma from SIV-infected animals suppressed their inhibitory activity; (b) SIV rNef and HIV-1 rNef, as well as peptide B from HIV-1 Nef, recapitulated SIV inhibitory activities on early progenitors ex vivo, in both CFC and LTC-IC assays; (c) the variant BK28-41ΔNef SIV strain, which only differs from the BK28-41 strain by mutations abolishing Nef production, was inactive on early hematopoietic progenitors. These data indicated that lentiviral Nef is not only an immunosuppressor but also a strong inhibitor of early hematopoiesis in primates. These data suggest that, similarly, Nef is responsible for the central hematopoietic defects observed in HIV seropositive and AIDS patients.
Nef is a small, myristoylated, and highly abundant early viral protein that is required for efficient HIV/SIV replication and pathogenicity. It impairs expression of the MHC-1 class I and affects numerous signaling pathways in infected lymphoid cells. It was recently shown that Nef is also involved in a variety of bystander effects. It protects HIV-1 infected cells from apoptotic signals while simultaneously promoting killing of bystander cells through the induction of FasL (49); it can penetrate in primary uninfected human lymphoid B cells (50) as well as in various hematopoietic and nonhematopoietic cell lines (51–54) in the presence of neighboring infected macrophages and/or Nef-supplemented culture medium (33, 52). In these uninfected B cells, Nef suppresses CD40-dependent immunoglobulin class switching. Our data extend these observations and indicate that Nef can similarly alter biological functions of healthy uninfected myeloid progenitors. Such a capacity resembles that described for another early HIV-1 protein, Tat. However, we didn’t observe any inhibitory activities of recombinant Tat on CD34+ BM progenitors in similar CFC assays, thus assessing the specific function of Nef (data not shown). The question of how Nef interacts with CD34+ cells remains to be known. Nef was recently shown to freely enter human primary lymphoid cells when added to the culture medium (50). However, we have been unable to detect Nef in a large number of CD34+ cells by immunofluorescence microscopy, suggesting that either only small amounts of Nef interact/enter hematopoietic progenitors or that Nef interacts quite transiently with the progenitors.
The amounts of soluble Nef detected in serum from HIV-infected patients are rather high, around 10 ng/ml, but may be even higher when taking into account the presence of immune Nef/anti-Nef complexes (36). Higher amounts may also be present in the hematopoietic niche, in which potentially infected stromal cells may express large amounts of the myristoylated membrane-anchored Nef, while tightly interacting with hematopoietic progenitors. Our data indicated that 10 ng/ml recombinant myristoylated Nef are sufficient to impair CD34+ functionality. This form of Nef was much more active than recombinant Nef from E. coli, but may still be much less active than native properly folded and membrane-anchored Nef expressed by infected cells.
We demonstrated that a peptide from a central region of the HIV Laï Nef directly impaired the clonogenic potential of progenitors and recapitulated, ex vivo, the hematopoietic defects observed in SIV-infected animals (our present data) and in HIV seropositive– or AIDS-patients (1, 2, 4, 6, 10, 12). These results indicate that peptide B contains all information(s) to act on hematopoietic progenitors. This region is uniquely conserved among lentiviral Nef sequences (34). Its contains a well-known Pro-rich motif that is reported to interact in vitro with numerous SH3-containing signaling molecules, including Hck, NAK, Vav, and Src-related kinase (reviewed by Geyer et al.; ref. 34); these interactions were reported to be essential for Nef activities in human lymphoid cells and for the induction of a severe AIDS-like disease in CD4C/HIV Tg mice (38). Our data extend these observations and indicate that this same Pro-rich motif is still essential for Nef inhibitory activity on CD34+ hematopoietic progenitors. They provide the first evidence to our knowledge that this central Nef region can also mimic PPARγ agonist activities, at least in hematopoietic progenitors. Whether this Pro-rich motif functions as a whole and active SH3 binding motif that allows association with an SH3 factor regulating PPARγ activity or instead works independently of any kind of SH3 binding through a single selective Pro crucial for Nef inhibitory activities warrants further investigation. It is worth noting that some viral isolates, such as SIVmac239, express a PxxP motif instead of the conserved PxxPxxP in their Nef sequence, while still inducing central hematopoietic defects. Such observations would lend support to an SH3 independent activity for Nef on hematopoietic progenitors. Also, whether other conserved actions of Nef, such as those affecting T cell functions, are also mediated by PPARγ-dependent signaling pathways remains to be determined.
Finally, our data further reveal that SIV, Nef, or PPARγ agonists all inactivate/deregulate hematopoietic progenitors, through a PPARγ-dependent signaling pathway, that inhibit the expression of STAT5A/B genes, both ex vivo and in vivo, in mice and macaques. STAT5A promoter was recently shown to contain numerous PPAR-responsive elements that mediate PPARγ regulatory activity on Stat5a gene expression in rat mammary cells (55). Some of these PPAR-responsive elements are conserved in human STAT5A and STAT5B genes and may similarly mediate PPAR regulatory activities in CD34+ hematopoietic progenitors. Although PPARγ was initially thought mainly to regulate adipogenesis (41), 1 report indicated that PPARγ agonists regulate proliferation and differentiation of erythroid precursors (56). In addition, recent reports have mentioned unexplained hematopoietic abnormalities in a large cohort of patients with type 2 diabetes mellitus who participated in clinical trials with the PPARγ agonist pioglitazone (57). PPARγ agonists have also been recently used in clinical trials in the prevention of tumor development with contradictory effects (58). Our data therefore suggest that side effects on hematopoietic progenitors favoring anemia and pancytopenia may hamper the benefit of PPARγ agonists as therapeutic agents. They strongly suggest that abnormal PPARγ-dependent signaling pathways may be responsible for various hematologic disorders and hemopathies.
Overall, we demonstrated that SIV profoundly affects hematopoiesis through expression of a soluble or membrane-anchored, virally-encoded protein, Nef, which mimics a highly potent PPARγ agonist. We further demonstrated that Nef/PPARγ regulates early hematopoiesis through deregulation of STAT5 genes. This study provides new clues for the development of novel drugs targeting PPARγ activity to cure hematopoietic disorders, including those affecting seropositive/AIDS patients.
Methods
Animals and virus.
Adult male cynomolgus macaques were imported from Mauritius and housed in single cages within level 3 biosafety facilities. Animals were handled under ketamine chlorhydrate anesthesia (Rhone-Mérieux). All experimental procedures were conducted in accordance with European guidelines for animal care and approved by Direction Départementale des Services Vétérinaires. The pathogenic SIVmac251 strain was produced and used for infection of macaques as previously described (7). Briefly, the virus was isolated from macaque PBMCs cocultured with a spleen homogenate from a rhesus macaque infected with SIVmac251 (provided by R.C. Desrosiers, New England Regional Primate Center, Southborough, Massachusetts, USA). The cell-free stock was obtained by a second passage of the isolate in macaque PBMCs. This SIVmac251 resulting stock contained 1.22 × 108 viral RNA copies/ml and 3.6 × 104 50% tissue culture infection doses/ml (TCID50/ml) when the titer was determined on CEMx174 cells. The amount of viral antigen (p27) was 57 ng/ml and the in vivo titer after intravenous inoculation was 40,000 AID50/ml. We injected 50 AID50 into the saphenous vein of the monkey. SIVmac BK28-41 (2.8 × 104 TCID50/ml) and BK28-41ΔNef (4.1 × 104 TCID50/ml) were described previously (37). Female-specific pathogen-free C3H mice were obtained from Charles River Laboratories. During a period of 15 days, PPARγ agonist (Rosiglitazone; AVANDIA; GlaxoSmithKline) was administered to mice (10 mg/kg/day; intragastric bolus; n = 8) and for cynomolgus macaques (0.5 mg/kg/day; soft in banana; n = 2) during a period of 15 days.
Reagents.
Recombinant SIVmac251 Nef proteins and SIVmac251 Nef antibody (17.2) were provided by the National Institutes of Health (AIDS Research and Reference Reagent Program; catalog no. 2999). Recombinant HIV-1 Laï Nef and SIVmac251 Nef antibody (ARP3201) were provided by the National Institute for Biological Standards and Control (NIBSC) Centralized Facility for AIDS Reagents (repository reference, EVA650) or CEA/DIEP. Recombinant myristoylated HIV-1 Nef from strain SF2 (Jena Bioscience) was generated as reported (59). Nef-derived peptides (15AA) were synthesized using standard Fmoc chemistry; peptide B and mutated peptide B were produced by NeoMPS. The purity of all peptide preparations was more than 80%. PPARγ agonists and antagonist were provided by Cayman Chemical (PPARγ-PAK; SPI-BIO).
Isolation of macaques CD34+ BM mononuclear cells.
BM mononuclear cells were obtained from the iliac crest by aspiration and isolated by standard Ficoll density-gradient centrifugation (Eurobio). Cells were enriched in CD34+ cells by positive immunomagnetic selection (clone 561; Dynabeads M-450 CD34; Dynal) according to the manufacturer’s instructions. Enrichment for CD34+ cells was ascertained by flow cytometry using anti-CD34 monoclonal antibody (clone 563; BD Pharmingen) and always exceeded 95%.
Isolation of mice sca-1+ c-kit+ CD3– BM cells.
BM mononuclear cells were filtered on a cell strainer (40 μm; BD Falcon and BD Biosciences Europe), stained with PE-conjugated anti–sca-1 (BD Pharmingen) and anti-PE microbeads (Miltenyi Biotec). Enriched sca-1 cells were immuno-selected on MS columns (Miltenyi Biotec) and then stained with FITC-conjugated anti–c-kit and APC-conjugated anti-CD3 (BD Pharmingen). FACS analysis and cell sorting of sca-1+ c-kit+ CD3– cells were performed on a FACSAria (Becton Dickinson).
Culture of hematopoietic myeloid BM cells.
For ex vivo assays, simian CD34+ or murine sca-1+ c-kit+ CD3– mononuclear BM cells were suspended (1 × 104) in 3 ml of MethoCult H4434 medium or MethoCult M3434, respectively (StemCell Technologies). Cells were incubated and were scored as previously described (23).
LTC-IC were obtained as previously described (23). The following limiting dilution coculture method was used: CD34+ cells were added to MS-5 cultures, with 200, 100, 50, or 25 cells per well added to 20 wells for each dilution point.
For in vitro assays, CD34+ BM cells from healthy animals were incubated for 2 days in the presence of plasma from either infected or noninfected macaques. Plasma from noninfected macaques was added either with or without the following reagents: SIV or SIV-derived virus, HIV/SIV rNef, HIV-1 Nef-derived peptides, or PPARγ agonists and antagonist.
Plasma viral load.
Viral RNA was prepared from 200 μl of EDTA-treated, cell-free plasma using the High Pure Viral RNA kit (Roche Diagnostics). One-tube RT-PCR was performed in an iCycler real-time thermocycler (Bio-Rad) as described by Hofmann-Lehmann et al. (60).
RNA extraction and RT-PCR analysis.
RNA was extracted from 2 × 105 cells using RNAqueous-4PCR (Ambion). Reverse transcription was carried out for 1 hour at 42°C using 100 U MMLV reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed in an iCycler thermocycler (Bio-Rad) using iQ Supermix SYBR GRN (Bio-Rad). The following primer pairs were used: STAT5B forward, 5′-GGCAGAGTCGGTGACAGAAG-3′ and reverse, 5′-GGGTCTGCAAAAGCATTGTC-3′; STAT5A forward, 5′-CGAGTGCAGTGGTGAGATCC-3′ and reverse, 5′-TCCTCTGTCACGGACTCTGC-3′; murine STAT5B forward, 5′-GGCAGGGTCAGTAACGGAAG-3′ and reverse, 5′-GGCTCTGCAAAGGCGTTGTC-3′; PPARγ forward, 5′-AGCTCCGTGGATCTCTCCGT-3′ and reverse, 5′-CATGAGGGAGTTGGAAGGCTCT-3′; and GAPDH forward, 5′-TCGTGGAAGGACTCATGACC-3′ and reverse, 5′-TCAGCTCAGGGATGACCTTG-3′.
STAT5 protein analysis.
CD34+ BM cells (2.5 × 105) were lysed in RIPA lysis buffer on ice. Whole-cell extracts were boiled for 5 minutes in Laemmli sample buffer and subjected to SDS-PAGE in 10% acrylamide gels. Proteins were transferred to Hybond N+ filters (Amersham). Membranes were probed with the following antibodies: STAT5 (sc-1656), actin (sc-8432), and goat anti-mouse IgG-HRP (sc-2005) (Santa Cruz Biotechnology Inc.). Antibody binding was detected by ECL+ (Amersham).
Nef immunodepletion and immunoprecipitation.
Plasma from infected macaques or virus stocks were incubated for 12 hours at 4°C with a cocktail of 2 different monoclonal anti-SIVmac251 Nef antibodies (ARP3201 and 17.2). Immunocomplexes were removed on a magnet following a 4-hour incubation with Dynabeads Protein G (Dynal; Invitrogen) at 4°C. Virions from virus stocks were pelleted by ultracentrifugation (100,000 g, 4 hours at 4°C). Samples were checked by western blotting analysis using anti-SIVmac251 Nef 17.2.
Lentiviral vector production and transduction.
The cDNA encoding Macaca fascicularis STAT5B was cloned, sequenced (Genbank acquisition number DQ267926), and inserted into the SIN-cPPT-PGK-WHV lentiviral transfer vector. We used pCMVDR-8.92, pRSV-Rev, and pMD.G as packaging and vesicular stomatitis virus G protein (VSV-G) envelope constructs. Viral particles were produced by transient transfection of 293T cells with these 4 plasmids (61).
CD34+ cells were incubated for 48 hours (1 × 106/ml) in StemSpan (StemCell Technologies), supplemented with protamine sulfate (4 μg/ml), SCF (100 ng/ml), Flt-3-L (100 ng/ ml), IL-3 (20 ng/ml), and IL-6 (20 ng/ml) in a 96-well plate. Lentiviral particles were then added to cell suspensions for 12 hours before the cells were washed twice and suspended in MethoCult GF4434 medium (StemCell Technologies) for clonogenic analysis.
Proliferation assays.
K562 cells (2 × 105) were cultured in 96-well plates, in complete Dulbecco’s Modified Eagle medium alone and with or without variable concentration of HIV-1 rNef or troglitazone. Cells were cultured for 7 days in the presence of 1 μCi/well [3H]thymidine, collected by centrifugation, and counted on a plate reader. Eight replicates were used for each set of conditions.
PPARγ-specific siRNA.
siRNA targeting the human and macaque PPARγ sequence 5′-TGTTCCGTGACAATCTGTC-3′ were synthesized (Proligo). CD34+ BM cells were transfected with PPARγ-specific siRNA (25 nM) or control siRNA in the presence of Lipofectamine 2000 (Invitrogen) and maintained for 48 hours before CFC assay. Control siRNA was purchased from Invitrogen (BLOCK-iT). Transfection efficiency was assessed using a fluorescein-labeled, double-strand RNA duplex (BLOCK-iT Fluorescent Oligo; Invitrogen).
Statistics.
For culture assays and quantitative real-time PCR, values were calculated as mean ± SD for at least 3 separate experiments done in triplicate. Paired and unpaired comparisons were made, using the nonparametric Wilcoxon rank test and the Mann-Whitney test, respectively. P values of less than 0.05 were considered significant. Limiting dilution analysis was carried out with L-Calc software (StemCell Technologies). Correlations were analyzed with Spearman’s correlation coefficient test. All statistical analyses were carried out with StatView software (SAS Institute Inc.).
Supplementary Material
Acknowledgments
We thank J. Calvo, B. Delache, P. Brochard, N. Dufour, H. Felix, and J. Raguet for excellent technical assistance. We thank D. Gillet and S. Pichard (CEA/DIEP, Saclay, France) for supplying recombinant HIV LAI Nef protein and B. Delorme and P. Charbord (INSERM/ESPRI EA 3855, Tours, France) for providing PPARγ siRNA. We thank the NIH (Bethesda, Virginia, USA), European Program for Vaccine against AIDS and NIBSC (Potters Bar, United Kingdom) for supplying recombinant Nef protein. This work was supported by the Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (ANRS, Paris, France), the Association de Recherches contre le Cancer (to I. Dusanter-Fourt), the Association Française contre les Myopathies (AFM, Evry, France), the fondation de France, the Agence Nationale de la Recherche (to I. Dusanter-Fourt), and the Commissariat à l’Energie Atomique (CEA).
Footnotes
Nonstandard abbreviations used: LTC-IC, long-term colony-initiating cell; Nef, Negative factor; rNef, E. coli–recombinant SIVmac251 Nef protein; SH3, Src homology 3.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:1765–1775 (2008). doi:10.1172/JCI33037
Stéphane Prost and Mikael Le Dantec contributed equally to this work.
Isabelle Dusanter-Fourt and Marek Kirszenbaum contributed equally to this work.
See the related Commentary beginning on page 1622.
References
- 1.Moses A., Nelson J., Bagby G.C., Jr. The influence of human immunodeficiency virus-1 on hematopoiesis. Blood. 1998;91:1479–1495. [PubMed] [Google Scholar]
- 2.Zon L.I., Arkin C., Groopman J.E. Haematologic manifestations of the human immune deficiency virus (HIV). Br. J. Haematol. 1987;66:251–256. doi: 10.1111/j.1365-2141.1987.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 3.Koka P.S., Reddy S.T. Cytopenias in HIV infection: mechanisms and alleviation of hematopoietic inhibition. Curr. HIV Res. 2004;2:275–282. doi: 10.2174/1570162043351282. [DOI] [PubMed] [Google Scholar]
- 4.Levine A.M., Scadden D.T., Zaia J.A., Krishnan A. Hematologic aspects of HIV/AIDS. Hematology Am. Soc. Hematol. Educ. Program. 2001;2001:463–478. doi: 10.1182/asheducation-2001.1.463. [DOI] [PubMed] [Google Scholar]
- 5.Marandin A., et al. Loss of primitive hematopoietic progenitors in patients with human immunodeficiency virus infection. Blood. 1996;88:4568–4578. [PubMed] [Google Scholar]
- 6.Sloand E.M., et al. Secondary colony formation after long-term bone marrow culture using peripheral blood and bone marrow of HIV-infected patients. AIDS. 1997;11:1547–1553. doi: 10.1097/00002030-199713000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Thiebot H., et al. Impact of bone marrow hematopoiesis failure on T-cell generation during pathogenic simian immunodeficiency virus infection in macaques. Blood. 2005;105:2403–2409. doi: 10.1182/blood-2004-01-0025. [DOI] [PubMed] [Google Scholar]
- 8.Burstein Y., et al. Alterations in human fetal hematopoiesis are associated with maternal HIV infection. Pediatr. Res. 1992;32:155–159. doi: 10.1203/00006450-199208000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Bieniasz P.D. An intrinsic host defense against HIV-1 integration? J. Clin. Invest. 2007;117:302–304. doi: 10.1172/JCI31290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Luca A., et al. Haemopoietic CD34+ progenitor cells are not infected by HIV-1 in vivo but show impaired clonogenesis. Br. J. Haematol. 1993;85:20–24. doi: 10.1111/j.1365-2141.1993.tb08640.x. [DOI] [PubMed] [Google Scholar]
- 11.Koka P.S., Jamieson B.D., Brooks D.G., Zack J.A. Human immunodeficiency virus type 1-induced hematopoietic inhibition is independent of productive infection of progenitor cells in vivo. J. Virol. 1999;73:9089–9097. doi: 10.1128/jvi.73.11.9089-9097.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molina J.M., et al. Lack of evidence for infection of or effect on growth of hematopoietic progenitor cells after in vivo or in vitro exposure to human immunodeficiency virus. Blood. 1990;76:2476–2482. [PubMed] [Google Scholar]
- 13.Jenkins M., Hanley M.B., Moreno M.B., Wieder E., McCune J.M. Human immunodeficiency virus-1 infection interrupts thymopoiesis and multilineage hematopoiesis in vivo. Blood. 1998;91:2672–2678. [PubMed] [Google Scholar]
- 14.Moses A.V., et al. Human immunodeficiency virus infection of bone marrow endothelium reduces induction of stromal hematopoietic growth factors. Blood. 1996;87:919–925. [PubMed] [Google Scholar]
- 15.Calenda V., Graber P., Delamarter J.F., Chermann J.C. Involvement of HIV nef protein in abnormal hematopoiesis in AIDS: in vitro study on bone marrow progenitor cells. Eur. J. Haematol. 1994;52:103–107. doi: 10.1111/j.1600-0609.1994.tb01294.x. [DOI] [PubMed] [Google Scholar]
- 16.Rameshwar P., Denny T.N., Gascon P. Enhanced HIV-1 activity in bone marrow can lead to myelopoietic suppression partially contributed by gag p24. J. Immunol. 1996;157:4244–4250. [PubMed] [Google Scholar]
- 17.Reinhold D., Wrenger S., Kahne T., Ansorge S. HIV-1 Tat: immunosuppression via TGF-beta1 induction. Immunol. Today. 1999;20:384–385. doi: 10.1016/s0167-5699(99)01497-8. [DOI] [PubMed] [Google Scholar]
- 18.Sugiura K., Oyaizu N., Pahwa R., Kalyanaraman V.S., Pahwa S. Effect of human immunodeficiency virus-1 envelope glycoprotein on in vitro hematopoiesis of umbilical cord blood. Blood. 1992;80:1463–1469. [PubMed] [Google Scholar]
- 19.Zauli G., Re M.C., Furlini G., Giovannini M., La Placa M. Human immunodeficiency virus type 1 envelope glycoprotein gp120- mediated killing of human haematopoietic progenitors (CD34+ cells). J. Gen. Virol. 1992;73:417–421. doi: 10.1099/0022-1317-73-2-417. [DOI] [PubMed] [Google Scholar]
- 20.Maciejewski J.P., Weichold F.F., Young N.S. HIV-1 suppression of hematopoiesis in vitro mediated by envelope glycoprotein and TNF-alpha. J. Immunol. 1994;153:4303–4310. [PubMed] [Google Scholar]
- 21.Hillyer C.D., et al. CD34+ and CFU-GM progenitors are significantly decreased in SIVsmm9 infected rhesus macaques with minimal evidence of direct viral infection by polymerase chain reaction. Am. J. Hematol. 1993;43:274–278. doi: 10.1002/ajh.2830430409. [DOI] [PubMed] [Google Scholar]
- 22.Thiebot H., et al. Early and persistent bone marrow hematopoiesis defect in simian/human immunodeficiency virus-infected macaques despite efficient reduction of viremia by highly active antiretroviral therapy during primary infection. J. Virol. 2001;75:11594–11602. doi: 10.1128/JVI.75.23.11594-11602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradley T.R., Metcalf D. The growth of mouse bone marrow cells in vitro. Aust. J. Exp. Biol. Med. Sci. 1966;44:287–299. doi: 10.1038/icb.1966.28. [DOI] [PubMed] [Google Scholar]
- 24.Stein M.I., Zhu J., Emerson S.G. Molecular pathways regulating the self-renewal of hematopoietic stem cells. Exp. Hematol. 2004;32:1129–1136. doi: 10.1016/j.exphem.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Hou S.X., Zheng Z., Chen X., Perrimon N. The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev. Cell. 2002;3:765–778. doi: 10.1016/S1534-5807(02)00376-3. [DOI] [PubMed] [Google Scholar]
- 26.Ihle J.N., et al. The roles of Jaks and Stats in cytokine signaling. Cancer J. Sci. Am. 1998;4(Suppl. 1):S84–S91. [PubMed] [Google Scholar]
- 27.Baskiewicz-Masiuk M., Paczkowski M., Machalinski B. The influence of antisense oligonucleotides against STAT5 on the regulation of normal haematopoiesis in a bone marrow model. Cell Prolif. 2004;37:231–245. doi: 10.1111/j.1365-2184.2004.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradley H.L., Couldrey C., Bunting K.D. Hematopoietic-repopulating defects from STAT5-deficient bone marrow are not fully accounted for by loss of thrombopoietin responsiveness. Blood. 2004;103:2965–2972. doi: 10.1182/blood-2003-08-2963. [DOI] [PubMed] [Google Scholar]
- 29.Bradley H.L., Hawley T.S., Bunting K.D. Cell intrinsic defects in cytokine responsiveness of STAT5-deficient hematopoietic stem cells. Blood. 2002;100:3983–3989. doi: 10.1182/blood-2002-05-1602. [DOI] [PubMed] [Google Scholar]
- 30.Kyba M., et al. Enhanced hematopoietic differentiation of embryonic stem cells conditionally expressing Stat5. Proc. Natl. Acad. Sci. U. S. A. 2003;100(Suppl. 1):11904–11910. doi: 10.1073/pnas.1734140100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nosaka T., et al. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao Z., et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michael N.L., et al. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 1999;37:2557–2563. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geyer M., Fackler O.T., Peterlin B.M. Structure--function relationships in HIV-1 Nef. EMBO Rep. 2001;2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandori M.W., et al. Producer-cell modification of human immunodeficiency virus type 1: Nef is a virion protein. J. Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujii Y., Otake K., Tashiro M., Adachi A. Soluble Nef antigen of HIV-1 is cytotoxic for human CD4+ T cells. FEBS Lett. 1996;393:93–96. doi: 10.1016/0014-5793(96)00859-9. [DOI] [PubMed] [Google Scholar]
- 37.Chakrabarti L., et al. Limited viral spread and rapid immune response in lymph nodes of macaques inoculated with attenuated simian immunodeficiency virus. Virology. 1995;213:535–548. doi: 10.1006/viro.1995.0026. [DOI] [PubMed] [Google Scholar]
- 38.Hanna Z., et al. The pathogenicity of human immunodeficiency virus (HIV) type 1 Nef in CD4C/HIV transgenic mice is abolished by mutation of its SH3-binding domain, and disease development is delayed in the absence of Hck. J. Virol. 2001;75:9378–9392. doi: 10.1128/JVI.75.19.9378-9392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couillin I., Letourneur F., Lefebvre P., Guillet J.G., Martinon F. DNA vaccination of macaques with several different Nef sequences induces multispecific T cell responses. Virology. 2001;279:136–145. doi: 10.1006/viro.2000.0705. [DOI] [PubMed] [Google Scholar]
- 40.Sutherland H.J., Lansdorp P.M., Henkelman D.H., Eaves A.C., Eaves C.J. Functional characterization of individual human hematopoietic stem cells cultured at limiting dilution on supportive marrow stromal layers. Proc. Natl. Acad. Sci. U. S. A. 1990;87:3584–3588. doi: 10.1073/pnas.87.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehrke M., Lazar M.A. The many faces of PPARgamma. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 42.Otake K., et al. HIV-1 Nef protein in the nucleus influences adipogenesis as well as viral transcription through the peroxisome proliferator-activated receptors. AIDS. 2004;18:189–198. doi: 10.1097/00002030-200401230-00007. [DOI] [PubMed] [Google Scholar]
- 43.Hoelbl A., et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–4906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuringa J.J., Wu K., Morrone G., Moore M.A. Enforced activation of STAT5A facilitates the generation of embryonic stem-derived hematopoietic stem cells that contribute to hematopoiesis in vivo. Stem Cells. 2004;22:1191–1204. doi: 10.1634/stemcells.2004-0033. [DOI] [PubMed] [Google Scholar]
- 45.Bunting K.D., et al. Reduced lymphomyeloid repopulating activity from adult bone marrow and fetal liver of mice lacking expression of STAT5. Blood. 2002;99:479–487. doi: 10.1182/blood.V99.2.479. [DOI] [PubMed] [Google Scholar]
- 46.Snow J.W., et al. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood. 2002;99:95–101. doi: 10.1182/blood.V99.1.95. [DOI] [PubMed] [Google Scholar]
- 47.Kryworuchko M., et al. Defective interleukin-2-dependent STAT5 signalling in CD8 T lymphocytes from HIV-positive patients: restoration by antiretroviral therapy. AIDS. 2004;18:421–426. doi: 10.1097/00002030-200402200-00007. [DOI] [PubMed] [Google Scholar]
- 48.Pericle F., et al. HIV-1 infection induces a selective reduction in STAT5 protein expression. J. Immunol. 1998;160:28–31. [PubMed] [Google Scholar]
- 49.Geleziunas R., Xu W., Takeda K., Ichijo H., Greene W.C. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell. Nature. 2001;410:834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 50.Qiao X., et al. Human immunodeficiency virus 1 Nef suppresses CD40-dependent immunoglobulin class switching in bystander B cells. Nat. Immunol. 2006;7:302–310. doi: 10.1038/ni1302. [DOI] [PubMed] [Google Scholar]
- 51.Alessandrini L., et al. T-tropic human immunodeficiency virus (HIV) type 1 Nef protein enters human monocyte-macrophages and induces resistance to HIV replication: a possible mechanism of HIV T-tropic emergence in AIDS. J. Gen. Virol. 2000;81:2905–2917. doi: 10.1099/0022-1317-81-12-2905. [DOI] [PubMed] [Google Scholar]
- 52.Mangino G., et al. In vitro treatment of human monocytes/macrophages with myristoylated recombinant Nef of human immunodeficiency virus type 1 leads to the activation of mitogen-activated protein kinases, IkappaB kinases, and interferon regulatory factor 3 and to the release of beta interferon. J. Virol. 2007;81:2777–2791. doi: 10.1128/JVI.01640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olivetta E., et al. HIV-1 Nef induces the release of inflammatory factors from human monocyte/macrophages: involvement of Nef endocytotic signals and NF-kappa B activation. J. Immunol. 2003;170:1716–1727. doi: 10.4049/jimmunol.170.4.1716. [DOI] [PubMed] [Google Scholar]
- 54.Varin A., et al. Exogenous Nef protein activates NF-kappa B, AP-1, and c-Jun N-terminal kinase and stimulates HIV transcription in promonocytic cells. Role in AIDS pathogenesis. J. Biol. Chem. 2003;278:2219–2227. doi: 10.1074/jbc.M209622200. [DOI] [PubMed] [Google Scholar]
- 55.Olsen H., Haldosen L.A. Peroxisome proliferator-activated receptor gamma regulates expression of signal transducer and activator of transcription 5A. Exp. Cell Res. 2006;312:1371–1380. doi: 10.1016/j.yexcr.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 56.Nagasawa E., et al. Pivotal role of peroxisome proliferator-activated receptor gamma (PPARgamma) in regulation of erythroid progenitor cell proliferation and differentiation. Exp. Hematol. 2005;33:857–864. doi: 10.1016/j.exphem.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 57.Berria R., et al. Reduction in hematocrit and hemoglobin following pioglitazone treatment is not hemodilutional in type II diabetes mellitus. Clin. Pharmacol. Ther. 2007;82:275–281. doi: 10.1038/sj.clpt.6100146. [DOI] [PubMed] [Google Scholar]
- 58.Wang T., Xu J., Yu X., Yang R., Han Z.C. Peroxisome proliferator-activated receptor gamma in malignant diseases. Crit. Rev. Oncol. Hematol. 2005;58:1–14. doi: 10.1016/j.critrevonc.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 59.Geyer M., Peterlin B.M. Domain assembly, surface accessibility and sequence conservation in full length HIV-1 Nef. FEBS Lett. 2001;496:91–95. doi: 10.1016/S0014-5793(01)02394-8. [DOI] [PubMed] [Google Scholar]
- 60.Hofmann-Lehmann R., et al. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res. Hum. Retroviruses. 2000;16:1247–1257. doi: 10.1089/08892220050117014. [DOI] [PubMed] [Google Scholar]
- 61.Hottinger A.F., Azzouz M., Deglon N., Aebischer P., Zurn A.D. Complete and long-term rescue of lesioned adult motoneurons by lentiviral-mediated expression of glial cell line-derived neurotrophic factor in the facial nucleus. J. Neurosci. 2000;20:5587–5593. doi: 10.1523/JNEUROSCI.20-15-05587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.