Abstract
In fission yeast, mating-type switching involves replacing genetic information contained at the expressed mat1 locus by that of either the mat2P or mat3M donor loci. Donor selection is nonrandom, as mat1P cells preferentially use mat3M for switching, whereas mat1M cells use mat2P. Switching directionality is determined by the cell-type-specific distribution of the Swi2–Swi5 complex that, in mat1P cells, localises to mat3M and, only in mat1M cells, spreads to mat2P in a heterochromatin-dependent manner. Mechanisms regulating spreading of Swi2–Swi5 across heterochromatin are not fully understood. Here, we show that the fission yeast homologue of CENP-B, Abp1, binds to the silent domain of the mating-type locus and regulates directionality of switching. Deletion of abp1 prevents utilisation of mat2P, as when heterochromatin is disrupted and spreading of Swi2–Swi5 is impaired. Our results show that, indeed, deletion of abp1 abolishes spreading of Swi2–Swi5 to mat2P. However, in abp1Δ cells, heterochromatin organisation at the mating-type locus is preserved, indicating that Abp1 is actually required for efficient spreading of Swi2–Swi5 through heterochromatin. Cbh1 and Cbh2, which are also homologous to CENP-B, have only a minor contribution to the regulation of directionality of switching, which is in contrast with the strong effects observed for Abp1.
Keywords: Abp1, CENP-B, heterochromatin, mating type, S. pombe
Introduction
In the fission yeast Schizosaccharomyces pombe, haploid cells switch mating type by means of a tightly regulated gene conversion event that involves long distance interactions between an expressed locus (mat1) with either of two silent donor loci (mat2 and mat3), which are located 17 and 29 kb away from mat1, respectively (reviewed in Arcangioli and Thon, 2004). Mating-type information is contained in the silent mat2P (plus) and mat3M (minus) loci but it is expressed only after translocation to the mat1 locus giving rise to mat1P or mat1M cells, depending on whether mat2 or mat3 information is expressed at mat1. Silencing at the mat2 and mat3 loci is mediated by heterochromatin that, in the mating-type region, extends for a 20-kb long domain (Thon and Klar, 1993; Grewal and Klar, 1997; Noma et al, 2001). This silenced domain is flanked by two inverted repeats (IR-L and IR-R) and, in addition to mat2 and mat3, contains a repeated element (cenH) that is homologous to the centromeric dg/dh repeats and nucleates the formation of heterochromatin in an RNAi-dependent manner (Hall et al, 2002). At the mating-type region, heterochromatin formation is also induced through an additional RNAi-independent mechanism that involves binding of the transcription factors Atf1 and Pcr1 (Jia et al, 2004a).
Mating-type switching initiates during DNA replication with the introduction of a strand-specific single-strand break (SSB) imprint at mat1, which was proposed to result from the incorporation of two ribonucleotides (Vengrova and Dalgaard, 2004, 2006), and that, in the next round of DNA replication, is converted into a double-strand break (DSB) (reviewed in Arcangioli and Thon, 2004). This DSB is, then, healed by gene conversion using mat2 or mat3 as donors. Donor selection is, however, not random. In contrast, mat1P cells preferentially use mat3 as a donor, whereas mat1M cells use mat2 (reviewed in Arcangioli and Thon, 2004). Directionality of switching, therefore, ensures that cells switch to the opposite mating type with a very high frequency. Directionality of switching is determined by the cell-type specific distribution of the Swi2–Swi5 complex that promotes recombination (Jia et al, 2004b). In mat1P cells, Swi2–Swi5 localisation is restricted to a recombination-enhancer element (SRE) located adjacently to mat3 so that, under these circumstances, only mat3 is efficiently used as a donor. On the other hand, in mat1M cells, Swi2–Swi5 spreads across the entire mating-type region reaching the mat2 locus that becomes the preferred donor site due to the structural constraints imposed by heterochromatin. Spreading of Swi2–Swi5 in mat1M cells relies on heterochromatin, as it is abolished in a swi6 mutant without affecting its binding to SRE (Jia et al, 2004b). As a consequence, in a swi6 mutant background, mat3 is used as a donor at a much higher frequency than mat2, so that swi6 mutant cells are predominantly of the mat1M type. Mutations in several other heterochromatin assembly factors (i.e. crl4, rik1 and sir2) were also shown to affect directionality of switching (Ivanova et al, 1998; Nakayama et al, 2001; Shankaranarayana et al, 2003; Tuzon et al, 2004). These observations unveil the essential contribution of heterochromatin to spreading of Swi2–Swi5. Heterochromatin-dependent spreading was also reported for other multi-protein complexes such as SHREC (Sugiyama et al, 2007). The mechanisms that regulate spreading of Swi2–Swi5 across heterochromatin are, however, not fully understood. Direct interaction with Swi6 likely contributes to binding of Swi2–Swi5 to heterochromatin, as Swi2 selectively colocalises with Swi6 and the two proteins interact strongly in vitro (Akamatsu et al, 2003; Jia et al, 2004b). However, in vivo, only a small proportion of Swi2 is associated to Swi6 (Jia et al, 2004b), indicating that additional factors must contribute to binding of Swi2–Swi5 to heterochromatin.
In this paper, we show that Abp1 binds at the mating-type locus and regulates directionality of switching. In abp1Δ cells, mat3 is preferentially used as a donor as when, in swi6Δ or crl4Δ cells, spreading of Swi2–Swi5 to mat2 is abolished due to disruption of heterochromatin. Our results show that deletion of abp1 impairs spreading of Swi2–Swi5 to mat2 without disrupting heterochromatin organisation at the mating-type region, indicating that Abp1 is actually required for efficient spreading of Swi2–Swi5 across heterochromatin.
Abp1 is homologous to CENP-B, an evolutionarily conserved sequence-specific DNA-binding protein that associates to centromeric heterochromatin (Murakami et al, 1996; Halverson et al, 1997). In S. pombe, Cbh1 and Cbh2 are also homologous to CENP-B and, together with Abp1, have redundant functions in the regulation of various nuclear processes (Halverson et al, 1997; Lee et al, 1997; Baum and Clarke, 2000; Ireland et al, 2001; Nakagawa et al, 2002). However, the contribution to the regulation of directionality of mating-type switching is specific to Abp1, as deletion of cbh1 or cbh2 shows no effects on switching. Interestingly, Abp1 also appears to have an important function in DNA replication (Murakami et al, 1996; Locovei et al, 2006), suggesting a possible link between spreading of Swi2–Swi5 and DNA replication.
Results
Abp1 regulates directionality of mating-type switching
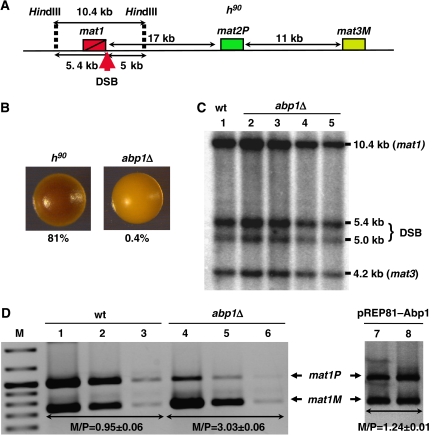
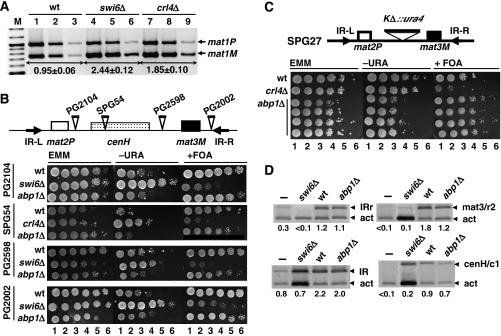
Efficiency of mating-type switching can be monitored by staining with iodine vapours (Bresch et al, 1968; Thon and Klar, 1993; Jia et al, 2004b). Efficient switching results into colonies containing an homogeneous mixture of cells of opposite mating types that, when grown in sporulation medium, can readily mate and sporulate giving rise to colonies staining dark with iodine vapours due to the accumulation of starch products occurring during sporulation. In contrast, cells showing poor mating-type switching form light-staining colonies. Deletion of abp1 in a switching-efficient homothallic h90 strain (Figure 1A) results in a strong decrease in its ability to mate and sporulate. abp1Δ cells form colonies staining much lighter with iodine than wild-type h90 colonies (Figure 1B). Actually, the frequency of spores observed in abp1Δ colonies is as low as 0.4% of total cells, whereas in wild-type colonies is approximately 81%. These results indicate that mating-type switching is very inefficient in abp1Δ cells.
Figure 1.
Analysis of the effects on mating-type switching of deleting abp1 in a homothallic h90 strain. (A) Schematic representation of the structural organisation of the mating-type region in an h90 strain. The positions of mat1, mat2P and mat3M loci are indicated. The position of the SSB/DSB imprint, contained within the 10.4-kb HindIII fragment spanning the mat1 locus, is also indicated. (B) Iodine staining of wild-type (h90) and abp1Δ colonies. Before staining, colonies were grown in sporulation medium at 25°C for 3 days. Numbers below each panel indicate the frequencies of sporulation. Similar results were obtained with several independent abp1Δ colonies. (C) Presence of the SSB/DSB imprint was determined by Southern blot analysis of a wild-type (lane 1) and four independent abp1Δ strains (lanes 2–5). Genomic DNA was digested with HindIII and probed with a 9-kb DNA fragment, which spans the mat1 locus and contains mat3M information at mat1, so that it also detects a 4.2-kb HindIII band corresponding to the mat3M locus. Bands arising from cleavage at the SSB/DBS imprint of the 10.4-kb HindIII fragment are indicated (DSB). (D) Quantitative multiplex PCR determination of the predominant mating type adopted by wild-type (wt, lanes 1–3) and abp1Δ cells (lanes 4–6). PCR reactions were performed using appropriate primers to amplify mat1M and mat1P sequences simultaneously. For each strain, three 10-fold dilutions of the PCR products were analysed. Similar results were obtained with several independent abp1Δ colonies. Lanes 7 and 8, correspond to abp1Δ cells transformed with plasmid pREP81–Abp1 to express Abp1. In this case, single dilutions of the PCR products obtained from two independent isolates are shown. The positions of the bands corresponding to mat1P and mat1M are indicated. The M/P ratios (±s.d.) are indicated below the corresponding lanes.
As discussed above, a first step in mating-type switching is the generation of a strand-specific SSB imprint that, latter, is converted into a DSB. A number of factors (swi1, swi3 and swi7) are known to participate in the formation of this SSB/DSB imprint (reviewed in Arcangioli and Thon, 2004). Abp1 could also participate in this process, as it contains a DDE domain with significant homology to the catalytic domains of the pogo family of transposases (Ireland et al, 2001). To test this possibility, we performed Southern blotting analysis to determine the presence of the SSB/DSB imprint in abp1Δ cells (Figure 1C). In these experiments, genomic DNA was digested with HindIII, which generates a 10.4-kb fragment spanning the mat1 locus (Figure 1A). Presence of the SSB/DSB imprint results in cleavage of this HindIII fragment into two fragments of 5.4 and 5.0 kb, respectively. It must be noticed that, though many cells contain only a SSB at this position, it is converted into a DSB during DNA isolation due to sharing forces (Arcangioli, 1998). Bands reflecting formation of the SSB/DBS imprint are readily detected in abp1Δ (Figure 1C, lanes 2–5) as well as in wild-type h90 cells (Figure 1C, lane 1), indicating that deletion of abp1 does not affect generation of the imprint.
Next, we analysed whether inefficient switching of abp1Δ cells is the consequence of a deregulation on directionality of switching, so that they are unable to use either mat2 or mat3 as a donor locus. To test this possibility, we carried out quantitative multiplex PCR analysis using primers that specifically determine the presence at the expressed mat1 locus of either mat2 (mat1P) or mat3 (mat1M) information (Jia et al, 2004b) (Figure 1D). In wild-type h90 cells, efficient mating-type switching results in equal utilisation of mat2 or mat3 as a donor, so that bands representing mat1P and mat1M cells are roughly of the same intensity (M/P ratio of 0.95±0.06) (Figure 1D, lanes 1–3). In contrast, abp1Δ cells are predominantly of the mat1M type, which contains mat3 information at mat1 (M/P=3.03±0.06) (Figure 1D, lanes 4–6). Ectopic expression of Abp1 from an episomal vector reverts the switching defect of abp1Δ cells (M/P=1.24±0.01) (Figure 1D, lanes 7 and 8). These results indicate that Abp1 regulates directionality of switching, so that abp1Δ cells preferentially use mat3 as a donor.
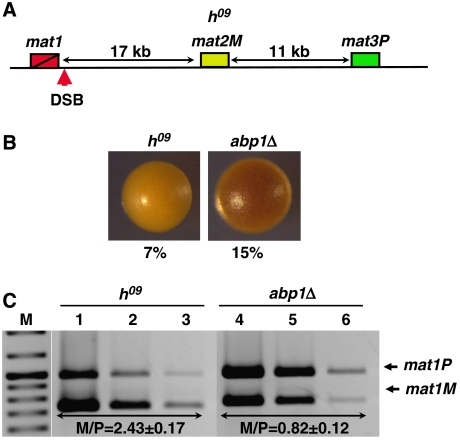
To further confirm the contribution of Abp1 to the regulation of directionality of mating-type switching, we analysed the effects of deleting abp1 in an h09 background. In h09 cells, mating-type information contained at the donor mat2 and mat3 loci is swapped, so that mat3 contains plus (P) information (mat3P), whereas mat2 contains minus (M) information (mat2M) (Figure 2A) (Thon and Klar, 1993). Switching is determined by the chromosomal position of the donor loci with respect to mat1 rather than by the actual genetic information contained at mat1 (Thon and Klar, 1993). As a consequence, in h09 cells, switching does not replace mating information at mat1, as mat1M cells preferentially use mat2M as a donor and, vice versa, mat1P cells use mat3P, resulting in non-productive switching. Consistent with that, h09 colonies stain lightly with iodine and show a low frequency of sporulation (Figure 2B). As shown in Figure 2C, the h09 strain used in these experiments is predominantly of the mat1M type (M/P=2.43±0.17) (Figure 2C, lanes 1–3). In this h09 strain, deletion of abp1 results also in a preferred utilisation of mat3P, so that abp1Δ cells become predominantly mat1P (M/P=0.82±0.12) (Figure 2C, lanes 4–6). Concomitantly, abp1Δ cells stain darker with iodine than wild-type h09 cells and sporulation is partially rescued, from 7 to 15% (Figure 2B).
Figure 2.
Analysis of the effects on mating-type switching of deleting abp1 in h09 cells. (A) Schematic representation of the structural organisation of the mating-type region in an h09 strain. The positions of mat1, mat2M and mat3P loci are indicated. The position of the SSB/DSB imprint is also indicated. (B) Iodine staining of wild-type (h09) and abp1Δ colonies. Numbers below each panel correspond to the frequencies of sporulation. (C) Quantitative multiplex PCR determination of the predominant mating type adopted by wild-type h09 (lanes 1–3) and abp1Δ cells (lanes 4–6). PCR reactions were performed as in Figure 1D. The M/P ratios (±s.d.) are indicated below the corresponding lanes.
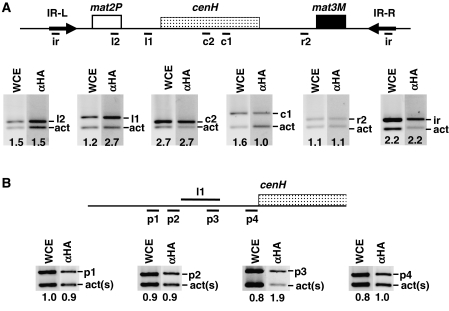
Next, we asked whether Abp1 actually binds to the mating-type locus. To address this question, we performed chromatin immunoprecipitation (ChIP) experiments from cells carrying HA-tagged Abp1 (Figure 3). In these experiments, the presence of Abp1 was determined at several regions spanning the entire silent domain of the mating-type locus (Figure 3A). Crosslinked material was immunoprecipitated with αHA antibodies and analysed by PCR using appropriate primers to amplify specific bands of the indicated mating-type regions (Figure 3A, bands ir, r2, c1, c2, l1 and l2), and, in the same reaction, with a second set of primers to amplify a control DNA fragment of similar length corresponding to the act1 gene (Figure 3A, bands act). As shown in Figure 3A, only one of the regions analysed (l1) was found significantly enriched in the immunoprecipitated material, indicating binding of Abp1 to this region. Region l1, which spans approximately 500 bp, maps immediately upstream to the cenH element. To confirm binding of Abp1 to this region, and to narrow down its binding site, we analysed presence of Abp1 at different regions flanking l1 (Figure 3B, regions p1, p2 and p4), as well as within it (Figure 3B, region p3). As shown in Figure 3B, immunoprecipitation with αHA antibodies results in enrichment of region p3, but not the rest. Altogether, these results indicate that Abp1 binds to the silent domain of the mating-type locus, with a major binding-site centred around region l1, close to the cenH element. Similar results have been recently reported by others (Cam et al, 2007).
Figure 3.
Abp1 localises to the silent domain of the mating-type locus. (A) Binding of Abp1–HA at the silent domain of the mating-type locus was determined by ChIP-analysis using αHA antibodies. Immunoprecipitated material was analysed by multiplex PCR using primers specific for the indicated regions of the mating-type locus (bands ir, r2, c1, c2, l1 and l2) (see Supplementary Table SII for a description of the primers used) and, in the same PCR reaction, primers that amplify a fragment of similar length of the act1 locus (bands act), used as control. (B) Similar experiments as those described in (A) but with primers designed to amplify shorter DNA fragments flanking region l1 (bands p1, p2 and p4) or contained within it (bands p3). In this case, a shorter fragment from the act1 gene was used as control (bands act(s)). Lanes WCE correspond to PCR products obtained from the input material before immunoprecipitation. Lanes αHA correspond to the products obtained from the immunoprecipitated material. Numbers below each lane correspond to the ratio of the corresponding mating-type-specific band with respect to the control act1 band.
Directionality of mating-type switching is not compromised by deletion of cbh1 and/or cbh2
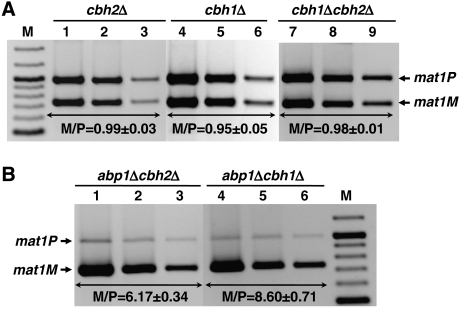
In S. pombe, Cbh1 and Cbh2 are closely related to Abp1 having redundant functions in the regulation of several cellular processes (Halverson et al, 1997; Lee et al, 1997; Baum and Clarke, 2000; Ireland et al, 2001; Nakagawa et al, 2002). In particular, all three proteins show synergistic effects on the regulation of centromeric silencing and chromosome segregation (Nakagawa et al, 2002). Therefore, it is possible that Cbh1 and Cbh2 would also contribute to the regulation of directionality of mating-type switching. To address this possibility, we examined the effects on mating-type switching of deleting cbh1 and cbh2 in h90 cells (Figure 4). cbh1Δ and cbh2Δ cells stain dark with iodine (not shown) and show high frequencies of sporulation (∼80%), indicating that they do not have major defects in directionality of mating-type switching. Quantitative multiplex PCR analysis confirmed these results (Figure 4A). In cbh1Δ and cbh2Δ cells, the bands corresponding to mat1P and mat1M are of almost equal intensity (M/P=0.95±0.05 and 0.99±0.03) (Figure 4A, lanes 1–6), indicating that, in these mutant backgrounds, both mat2 and mat3 are efficiently used as donor locus during mating-type switching, which is in contrast to the strong donor preference observed in abp1Δ cells (M/P=3.03±0.06). Similarly, cbh1Δcbh2Δ double mutants show no significant defects on directionality of switching (M/P=0.98±0.01) (Figure 4A, lanes 7–9). On the other hand, in abp1Δcbh1Δ and abp1Δcbh2Δ double mutants, mat3 is also the preferred donor locus, as in abp1Δ cells (Figure 4B). It must be noticed, however, that abp1Δcbh1Δ and abp1Δcbh2Δ double mutants show significantly higher M/P ratios than abp1Δ cells, suggesting a synergistic cooperation to regulate switching directionality. Actually, it was recently shown that Abp1 interacts both with Cbh1 and Cbh2 and that, in the mating-type locus, Cbh1 colocalises with Abp1 (Cam et al, 2007). Moreover, binding of Cbh1 to chromatin appears to depend on Abp1 but not vice versa (Cam et al, 2007). Altogether, these observations indicate that Abp1 has a major function in the regulation of mating-type switching, whereas Cbh1 and Cbh2 have only minor contributions that, most likely, depend on Abp1.
Figure 4.
Analysis of the effects on mating-type switching of deleting cbh1 and cbh2 in h90 cells. (A) Quantitative multiplex PCR determination of the predominant mating type adopted by cbh2Δ (lanes 1–3), cbh1Δ (lanes 4–6) and cbh1Δcbh2Δ cells (lanes 7–9). (B) Quantitative multiplex PCR determination of the predominant mating type adopted by abp1Δcbh2Δ (lanes 1–3) and abp1Δcbh1Δ cells (lanes 4–6). PCR reactions were performed as in Figure 1D. The M/P ratios (±s.d.) are indicated below the corresponding lanes.
Heterochromatin organisation of the mating-type region is preserved in abp1Δ cells
Heterochromatin organisation of the mating-type region is essential for the efficient utilisation of mat2 as a donor (Jia et al, 2004b). On the one hand, in mat1M cells, heterochromatin mediates spreading of the Swi2–Swi5 complex to mat2, so that it can be used as template for gene conversion. In addition, although in mat1M cells Swi2–Swi5 is also present at mat3, heterochromatin provides a higher order structure that makes mat2 the preferred donor loci. As a consequence, mutations that disrupt heterochromatin organisation prevent efficient utilisation of mat2 as a donor and, therefore, alter directionality of switching in a similar way as deletion of abp1 does (Figure 5A) (Jia et al, 2004b).
Figure 5.
Heterochromatin organisation of the mating-type region is not disrupted in abp1Δ cells. (A) Quantitative multiplex PCR determination of the predominant mating type adopted by wild-type (lanes 1–3), swi6Δ (lanes 4–6) and crl4Δ cells (lanes 7–9). PCR reactions were performed as in Figure 1D. The M/P ratios (±s.d.) are indicated below the corresponding lanes. (B) The effect of deleting abp1 on heterochromatin-mediated silencing was determined in several strains carrying ura4+ insertions at different positions of the silent domain of the mating-type locus. Sites of insertion are schematically indicated on top. Exponentially growing cells were plated as serial 10-fold dilutions (lanes 1–6) onto selective media with (panels EMM) or without uracil (panels −URA), or in the presence of FOA (panels +FOA). For each strain, several independent abp1Δ isolates were analysed and they all showed very similar results. (C) Similar results as in (B) but performed with strain SPG27, where the entire cenH element was replaced by ura4+. In this case, the results obtained with four independent abp1Δ isolates are presented. (D) Swi6 deposition at the silent domain of the mating-type region was determined in wild-type (lanes wt) and abp1Δ cells (lanes abp1Δ) by ChIP analysis using αSwi6 antibodies. Immunoprecipitated material was analysed as in Figure 3 with primers to amplify specific bands of the IR-R and IR-L regions (bands IRr and IR), the cenH element (bands cenH/c1) and a region just left of mat3 (bands mat3/r2) (see Supplementary Table SII for a description of the primers used). Lanes swi6Δ correspond to immunoprecipitation experiments performed with αSwi6 antibodies in a swi6Δ strain, missing Swi6. Lanes – correspond to mock immunoprecipitation experiments in which no antibodies were added. Numbers below each lane correspond to the ratio of the corresponding mating-type-specific band with respect to the control act band.
These observations point out the possibility that the switching defects observed in abp1Δ cells could reflect a contribution of Abp1 to heterochromatin assembly at the mating-type locus. Actually, Abp1 was reported to have a modest contribution to heterochromatin assembly at centromeres, as centromeric silencing was found to be slightly reduced in abp1Δ cells (Nakagawa et al, 2002). To test this hypothesis, we analysed the effects of an abp1Δ deletion on heterochromatin-mediated silencing at the mating-type locus. In these experiments, we used several reporter strains carrying an ura4+ gene inserted at different locations across the silent heterochromatic domain of the mating-type locus (Figure 5B, top). In all these strains, the reporter ura4+ gene is strongly silenced (Grewal and Klar, 1997; Thon et al, 2002), so that they grow poorly in the absence of uracil and strongly in the presence of fluoroorotic acid monohydrate (FOA) (Figure 5B, rows wt). In all these cases, silencing is relieved in swi6Δ or crl4Δ mutants (Figure 5B, rows swi6Δ and crl4Δ), which show extremely poor growth in the presence of FOA. Deletion of abp1, however, does not result in any significant relief of silencing, as indicated by the robust growth observed in the presence of FOA (Figure 5B, rows abp1Δ). Moreover, in the absence of uracil, both wild-type and abp1Δ cells show similar growth. The effect of deleting abp1 was also analysed in strain SPG27, where the entire cenH element was replaced by ura4+ (Figure 5C). In this strain, silencing is less strong as it relies mainly on the Atf1–Pcr1 pathway (Grewal and Klar, 1996). As a consequence, SPG27 cells show significant growth both in the absence of uracil and in the presence of FOA (Figure 5C, row wt), being more sensitive to slight changes in the levels of heterochromatin assembly factors. As shown in Figure 5C, in this strain, silencing is slightly reinforced in abp1Δ cells, as shown by the reduced growth observed in the absence of uracil (Figure 5C, rows abp1Δ). Concomitantly, growth in the presence of FOA increases. In these experiments, several independent abp1Δ isolates were analysed for each reporter strain and they all showed very similar effects.
ChIP analysis confirmed these results (Figure 5D). In these experiments, Swi6 distribution at the silent chromatin domain of the mating-type locus was determined in wild-type (wt) and abp1Δ cells. Swi6 deposition was analysed at the IR repeats, as well as at a region of the cenH element (c1) and at a region just left from mat3 (r2) (Figure 5D, bands IRr, IR, cenH/c1 and mat3/r2). Both in wild-type and abp1Δ cells, immunoprecipitation with αSwi6 antibodies results in a similar enrichment of the four mating-type-specific bands indicated above (Figure 5D, lanes wt and abp1Δ), when compared to similar immunoprecipitation experiments performed in a swi6Δ strain, missing Swi6 (Figure 5D, lanes swi6Δ), or when no antibodies were added (Figure 5D, lanes –). These results show that, in abp1Δ cells, Swi6 deposition at the mating-type region is not decreased to any significant extent.
Altogether, these observations indicate that deletion of abp1 does not disrupt heterochromatin organisation at the mating-type region. In contrast, in the SPG27 strain, silencing appears to increase in abp1Δ cells, suggesting that deletion of abp1 actually reinforces heterochromatin-dependent silencing at the mating-type locus.
The switching defect of abp1Δ cells is suppressed in swi2Δ and swi5Δ mutants
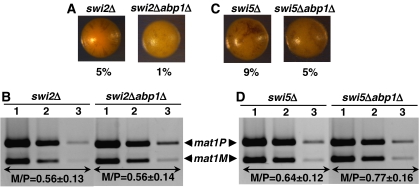
In abp1Δ cells, mat3 is the preferred donor locus during mating-type switching (Figure 1). It is known that donor selection depends on the functionality of the Swi2–Swi5 complex (Jia et al, 2004b). Swi5 is a general DNA recombination/repair factor that forms a complex with Rhp51/Rad51, a DNA recombination factor that is also required for mating-type switching (Akamatsu et al, 2003; Grishchuk et al, 2004). Localisation of Swi5 to the mating-type locus depends on Swi2 that binds to the SRE located adjacent to mat3 and, only in mat1M cells, spreads to mat2 (Jia et al, 2004b). Therefore, these two factors cooperate to promote recombination and to regulate directionality of mating-type switching.
To analyse whether the switching defect of abp1Δ cells depends on Swi2–Swi5, we determined the effects of deleting abp1 in swi2Δ and swi5Δ mutants. As reported earlier (Jia et al, 2004b), mat2 is the preferred donor locus both in swi2Δ (M/P=0.56±0.13) and swi5Δ cells (M/P=0.64±0.12) (Figure 6B and D). In these mutant backgrounds, deletion of abp1 does not alter donor selection as a similar preference for mat2 is observed in swi2Δabp1Δ (M/P=0.56±0.14) (Figure 6B) and swi5Δabp1Δ double mutants (M/P=0.77±0.16) (Figure 6D), which also show low sporulation frequencies (Figure 6A and C). These results indicate that preferred utilisation of mat3 in abp1Δ cells depends on the functionality of the Swi2–Swi5 complex.
Figure 6.
Analysis of the effects on mating-type switching of deleting abp1 in swi2Δ (A, B) and swi5Δ (C, D) cells. (A, C) Iodine staining of the indicated strains. Numbers below each panel correspond to the frequencies of sporulation. (B, D) Quantitative multiplex PCR determination of the predominant mating type adopted by each of the indicated strains. PCR reactions were performed as in Figure 1D. The M/P ratios (±s.d.) are indicated below the corresponding lanes.
Deletion of abp1 abolishes spreading of Swi2 across heterochromatin
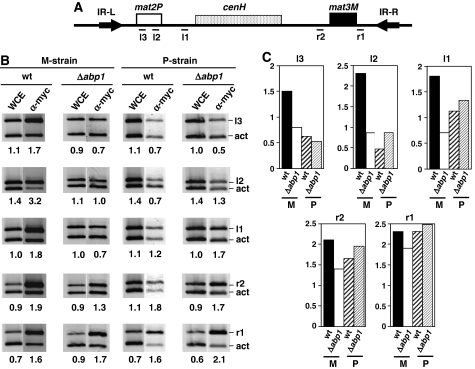
Efficient utilisation of mat2 as a donor depends on spreading of the Swi2–Swi5 complex across the entire silent heterochromatic domain of the mating-type locus so as to reach mat2 (Jia et al, 2004b). Therefore, it is possible that deletion of abp1 affects spreading of Swi2–Swi5 across heterochromatin. To address this question, we performed ChIP experiments to determine the effects of deleting abp1 on the distribution of Swi2 across the silent domain of the mating-type region (Figure 7). For this purpose, Swi2 was myc tagged in both a stable M-strain (mat1smto) (Engelke et al, 1987) and a stable P-strain (mat1PΔ17) (Arcangioli and Klar, 1991). It was reported earlier (Jia et al, 2004b) that, in a P-strain, Swi2 distribution is constrained to mat3, whereas in an M-strain, it spreads to mat2. In these experiments, we determined the presence of Swi2 at two different sites located within mat2 (l2 and l3), as well as at two locations surrounding mat3 (r1 and r2) and at the region corresponding to the major Abp1-binding site (l1), located between mat2 and cenH (Figure 7A). Consistent with previously reported results by others (Jia et al, 2004b), in the M-strain, Swi2 associates with all these five locations (Figure 7B, lanes wt in M-strain, and Figure 7C), whereas in the P-strain, it is found enriched only at the mat3 region, especially at region r1 that spans the SRE (Figure 7B, lanes wt in P-strain, and Figure 7C). In the M-strain, deletion of abp1 strongly alters the distribution of Swi2, so that, in abp1Δ cells, Swi2 is detected at mat3 locations (r1 and r2), but not at mat2 (l3 and l2) or at region l1 (Figure 7B, lanes abp1Δ in M-strain, and Figure 7C). In contrast, in the P-strain, deletion of abp1 does not significantly alter the distribution of Swi2, so that, in this case, it is detected only at mat3 locations, both in wild-type and abp1Δ cells (Figure 7B, lanes wt and abp1Δ in P-strain, and Figure 7C). Swi5 distribution is also likely to be affected in abp1Δ cells, as binding of Swi5 to the mating-type region, as well as its spreading through heterochromatin, is known to depend on the presence of Swi2 (Jia et al, 2004b). Altogether, these results indicate that, in the mating-type locus, Abp1 is required for efficient spreading of the Swi2–Swi5 complex across heterochromatin.
Figure 7.
Deletion of abp1 impairs spreading of Swi2–Swi5 to mat2. The effects of deleting abp1 on the distribution of Swi2–myc along the silent domain of the mating-type locus were determined in both a stable M-strain (mat1smto) and a stable P-strain (mat1PΔ17) by ChIP analysis using α-myc antibodies. (A) Regions of the silent domain of the mating-type locus where the presence of Swi2–myc was analysed (see Supplementary Table SII for a description of the primers used). (B) ChIP analysis corresponding to wild-type (wt) and abp1Δ cells derived from either a stable M-strain (panel M-strain) or a stable P-strain (panel P-strain). Material obtained after immunoprecipitation with α-myc antibodies was analysed as described in Figure 3 using primers specific for the indicated regions (bands r1, r2, l1, l2 and l3). Lanes WCE correspond to PCR products obtained from the input material before immunoprecipitation. Lanes α-myc correspond to the products obtained from the immunoprecipitated material. Numbers below each lane correspond to the ratio of the corresponding mating-type-specific band with respect to the control act band. (C) Quantitative analysis of the results shown in (B). For each of the mating-type-specific regions analysed, the relative fold of enrichment of the corresponding band in the immunoprecipitated material with respect to the input material (WCE) is presented for both wild type (wt) and abp1Δ cells in a stable M or P background.
Discussion
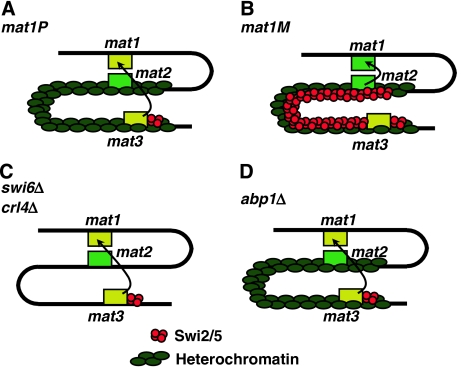
Here, we have shown that, in the fission yeast S. pombe, Abp1 contributes to the regulation of directionality of mating-type switching. A model to account for the results reported here is summarised in Figure 8. Directionality of switching depends on the cell-type-specific distribution of the Swi2–Swi5 complex that binds to the SRE located adjacent to mat3 and, only in mat1M cells, spreads to mat2 (Jia et al, 2004b). As a consequence, mat1P cells preferentially use mat3 as a donor (Figure 8A) and, vice versa, mat1M cells use mat2 (Figure 8B), thus ensuring efficient switching. Here, we have shown that deletion of abp1 abolishes spreading of Swi2–Swi5 to mat2 and, therefore, abp1Δ cells are unable to use mat2 as a donor locus during switching. It was shown that spreading of Swi2–Swi5 to mat2 is mediated by the heterochromatin organisation of the mating-type locus (Jia et al, 2004b), so that mutants of heterochromatin assembly factors (i.e. swi6Δ and crl4Δ) are also unable to use mat2 as a donor (Figure 8C). However, we have shown here that heterochromatin organisation at the mating-type locus is preserved in abp1Δ cells. Moreover, our results also show that, in abp1Δ cells, utilisation of mat3 as a donor depends on swi2 and swi5, indicating that deletion of abp1 does not affect the functionality of the Swi2–Swi5 complex. Altogether, these results indicate that Abp1 is actually required for efficient spreading of Swi2–Swi5 across heterochromatin at the mating-type locus (Figure 8D).
Figure 8.
Model to account for the contribution of Abp1 to the regulation of directionality of mating-type switching. In mat1P cells, the Swi2–Swi5 complex localises to mat3 (A) and, only in mat1M cells, spreads to mat2 (B), allowing its use as a donor during switching. Spreading of Swi2–Swi5 to mat2 is mediated by heterochromatin being abolished by mutations in heterochromatin assembly factors (i.e. swi6Δ and crl4Δ), which prevent use of mat2 as a donor (C). In abp1Δ cells, heterochromatin organisation of the mating-type locus is preserved but spreading of Swi2–Swi5 to mat2 is impaired and, as a consequence, mat2 is not efficiently used as a donor during switching (D).
Further work is required to determine the precise molecular mechanism(s) underlying the contribution of Abp1 to heterochromatin-mediated spreading of the Swi2–Swi5 complex. However, our results and those recently reported by others (Cam et al, 2007) indicate that Abp1 binds to the mating-type locus, where a major Abp1-binding site maps close to the cenH element. In addition to the mating-type locus, Abp1 also localises to multiple other sites, including Tf2 retrotransposons, centromeric dh repeats and some autonomously replicating sequences (ARS) (Cam et al, 2007). Binding at the mating-type locus strongly suggests a direct contribution to spreading of Swi2–Swi5. In this context, Abp1, which is a sequence-specific DNA-binding protein that recognises AT-rich DNA (Murakami et al, 1996; Halverson et al, 1997; Kikuchi et al, 2002), is likely to be involved in recruitment of factors that facilitate such spreading. Actually, in the mating-type locus, Abp1 was shown to contribute to recruitment of the HDAC Crl3 (Cam et al, 2007), a component of SHREC (Sugiyama et al, 2007), that is required for heterochromatin assembly at the mating-type locus (Ekwall and Ruusala, 1994; Yamada et al, 2005). However, Abp1–Crl3 interaction does not appear to have a major contribution to heterochromatin assembly at the mating-type locus as, on the one hand, our results indicate that deletion of abp1 does not impair silencing and/or Swi6 deposition at the mating-type locus and, moreover, in abp1Δ cells, high levels of Crl3 are detected at the silent domain of the mating-type region (Cam et al, 2007). Whether recruitment of Crl3 accounts to any extent for the contribution of Abp1 to the regulation of mating-type switching remains, however, to be determined. On the other hand, the possibility that Abp1 would directly mediate binding of the Swi2–Swi5 complex appears unlikely, as no specific enrichment of Swi2 is detected at the major Abp1-binding site of the mating-type locus. We cannot, however, exclude the possibility that Abp1 might bind additional sites within the mating-type locus, as both our binding studies and those performed by others (Cam et al, 2007) do not cover the entire locus. Actually, several potential Abp1-binding sites are found within the mating-type locus (Murakami et al, 1996; Halverson et al, 1997; Thon et al, 1999; Baum and Clarke, 2000).
Abp1 is known to participate in the regulation of multiple processes. In particular, Abp1 was shown to contribute to silencing of Tf2 retrotransposons through the recruitment of Crl3 and a second HDAC, Crl6 (Cam et al, 2007). Interestingly, silencing of Tf2 retrotransposons does not depend on Swi6, nor it does require other heterochromatin factors such as Crl4, Rik1 and Chp1 (Greenall et al, 2006), indicating that, also in this case, Abp1 is not involved in heterochromatin assembly.
At centromeres, it was proposed that Abp1 contributes to heterochromatin assembly, as it binds to several sites located within centromeric heterochromatin (Baum and Clarke, 2000; Cam et al, 2007), and deletion of abp1 results in a modest relief of centromeric silencing, which is enhanced in abp1Δcbh1Δ cells (Nakagawa et al, 2002). These observations are in contrast with our results showing that, at the mating-type locus, Abp1 is not required for heterochromatin assembly and silencing. In contrast, silencing at the mating-type locus appears to be slightly reinforced in abp1Δ cells. Most likely, this apparent contradiction is the consequence of disruption of centromeric heterochromatin, which could increase intracellular levels of silencing factors favouring their deposition at other heterochromatic sites, such as the mating-type locus. Similar effects were reported in Saccharomyces cerevisiae for sir4 mutants, where telomeric silencing is reduced and, concomitantly, silencing at the rDNA locus increases (Smith et al, 1998). The molecular basis of the contribution of Abp1, as well as of Cbh1 and Cbh2, to heterochromatin assembly at centromeres is, however, not fully understood.
Finally, Abp1 is also likely to participate in DNA replication, as it was found to interact physically with Cdc23 (MCM10), a protein implicated in the initiation of DNA replication (Locovei et al, 2006). Consistent with a role in DNA replication, abp1Δ cells grow slowly, due to a delay in S-phase, and abp1 interacts genetically with a number of genes involved in DNA replication, including cdc23, orc1 and orc2. Actually, Abp1 was first identified on the basis of its specific binding to ARS (Murakami et al, 1996) and it associates with some ARS in vivo (Cam et al, 2007). All these observations strongly support a contribution of Abp1 to DNA replication. Therefore, it is possible that the contribution of Abp1 to the regulation of mating-type switching is linked to DNA replication. At the mating-type locus, DNA replication has an essential function in the establishment of the SSB/DSB imprint that triggers recombination and gene conversion. After switching, the pattern of localisation of Swi2–Swi5 must be modified accordingly to the new mating information located at mat1. It is tempting to speculate that relocalisation of Swi2–Swi5 might take place during the next round of DNA replication, where Abp1 could exert an effect as a licensing factor that facilitates spreading of the complex in mat1M cells but not in mat1P cells.
Abp1, as well as the closely related Cbh1 and Cbh2 proteins, show significant homology to human CENP-B (Murakami et al, 1996; Halverson et al, 1997; Lee et al, 1997; Ireland et al, 2001). Homology is high at the N-terminal half, which contains the DNA-binding domain and mediates sequence-specific DNA recognition, and the DDE domain, which has putative endonuclease activity being similar to the catalytic domain of pogo transposases (Ireland et al, 2001). However, the C-terminal domain, which mediates protein–protein interactions, is less well conserved. Abp1, Cbh1 and Cbh2 have redundant functions in the regulation of several processes (Halverson et al, 1997; Baum and Clarke, 2000; Ireland et al, 2001; Nakagawa et al, 2002; Cam et al, 2007). In most cases, however, single mutants show only weak phenotypes that are strongly enhanced in double mutants. In contrast, the contribution of Abp1 to the regulation of directionality of mating-type switching is strong and specific, indicating that it constitutes a major function of Abp1.
Materials and methods
Media, genetic procedures and cytological procedures
S. pombe cells were grown in complete medium (YES) or Edinburgh minimal medium (EMM), according to standard procedures.
To estimate the efficiency of switching, individual colonies grown on sporulation medium were exposed to iodine vapours according to Thon and Klar (1993). The frequency of sporulation was determined from a 3-day-old culture by phase-contrast microscopy visualisation.
The effect of deleting abp1 on silencing at the mating-type was determined by plating 10 μl of serial 10-fold dilutions of fresh growing cultures in EMM plates lacking uracil (−URA) or supplemented with 1 μg/μl of 5-FOA.
Strains and plasmids
Strains used in these experiments are summarised in Supplementary Table SI. abp1Δ∷KAN, abp1Δ∷NAT, abp1Δ∷HPH, cbh1Δ∷NAT, cbh2Δ∷KAN, swi2Δ∷KAN and swi5Δ∷KAN deletions were obtained by replacing the entire ORF of the corresponding gene with PCR products carrying KanMX6, NatMX6 or HphMX6 markers obtained from pFA6a plasmids (Bähler et al, 1998; Hentges et al, 2005; Van Driessche et al, 2005), using appropriate primers containing a region of approximately 80 nt of homology to either side of the insertion site. Abp1 was tagged with 3HA and Swi2 with 13myc using plasmids pFA6a–3HA and pFA6a–13myc, respectively (Bähler et al, 1998; Van Driessche et al, 2005). To express Abp1 in abp1Δ cells, plasmid pREP81 was used (Basi et al, 1993).
Southern analysis
For Southern analysis, 20 μg of genomic DNA was digested with HindIII, run on a 0.6% agarose-TBE gel and blotted onto a nylon membrane (Hybond-N+; Amersham Biosciences). A 9-kb SalI–SphI mat1M fragment, obtained from plasmid pON177 (Styrkársdóttir et al, 1993), was 32P-labelled with Ready-To-Go™ DNA Labeling Beads (Amersham Biosciences) and used as a probe. This fragment spans the region corresponding to the mat1 locus and contains mat3M information at mat1, so that, in addition to the 10.4-kb HindIII fragment of the mat1 locus, it also recognises a 4.2-kb fragment corresponding to the mat3M locus.
Quantitative multiplex PCR analysis
Switching efficiency was analysed by quantitative multiplex PCR analysis of genomic DNA to determine the genetic content at the mat1 locus (Jia et al, 2004b). Primers used in these experiments were MT1 (common to mat1P and mat1M) 5′-AGAAGAGAGAGTAGTTGAAG-3′; MP (mat1P specific) 5′-ACGGTAGTCATCGGTCTTCC-3′ and MM (mat1M specific) 5′-TACGTTCAGTAGACGTAGTG-3′. Serial dilutions (10-fold) of the products obtained after PCR amplification were run on a 1.5% agarose-TBE gel, stained with 0.5 μg/ml ethidium bromide and captured using a Syngene GeneGenius System (Syngene, Cambridge, UK) equipped with GeneSnap version 6 gel documentation software. GeneTools version 3 software (Syngene) was used for quantitative analysis.
ChIP experiments
ChIP experiments were performed as described elsewhere (Pidoux et al, 2004). When the distribution of Swi6 was determined, IP was carried out with rabbit polyclonal αSwi6 antibodies (Abcam no. 14898) and ProteinA-Sepharose™ CL-4B beads (GE Healthcare no. 17-0780-01). In the case of Abp1–HA, rabbit polyclonal αHA antibodies (Abcam no. 9110) and ProteinA-agarose beads (Upstate no. 16–157) were used and, for Swi2, we used goat polyclonal α-myc antibodies (Abcam no. 9132) and ProteinG-PLUS-Agarose beads (Santa Cruz Biotechnology; SC-2002). PCR products were run on 2% agarose-TBE gels, visualised with 0.5 μg/ml ethidium bromide and captured using a Syngene GeneGenius System (Syngene) equipped with GeneSnap version 6 gel documentation software. GeneTools version 3 software (Syngene) was used for quantitative analysis. Primers used are summarised in Supplementary Table SII.
Supplementary Material
Supplementary Table SI
Supplementary Table SII
Acknowledgments
We are thankful to Drs R Allshire, F Antequera, B Arcangoli, J Ayté, R Egel, O Nielsen, SIS Grewal and G Thon for materials. We are also thankful to T Cavazza for help with some of the experiments. LA-A acknowledges receipt of a doctoral fellowship from MEC. F-XM, acknowledges receipt of an I3P postdoctoral contract from CSIC. This study was supported by grants from the MEC (BMC2003-243, BMC2006-1627 and CSD2006-00049), the CIRIT (2001SGR00344) and the EU (LSHB-CT-2004-511965). This study was carried out within the framework of the CeRBa of the Generalitat de Catalunya.
References
- Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H (2003) Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci USA 100: 15770–15775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B (1998) A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J 10: 4503–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B, Klar AJS (1991) A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J 10: 3025–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B, Thon G (2004) Mating-type cassettes: structure, switching and silencing. In The Molecular Biology of Schizosaccharomyces pombe, Egel R (ed), pp 129–144. Berlin, Heidelberg, New York: Springer-Verlag [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K (1993) TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 15: 131–136 [DOI] [PubMed] [Google Scholar]
- Baum M, Clarke L (2000) Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol 20: 2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresch C, Müller G, Egel R (1968) Genes involved in meiosis and sporulation of a yeast. Mol Gen Genet 102: 301–306 [DOI] [PubMed] [Google Scholar]
- Cam HP, Noma K-i, Ebina H, Levin HL, Grewal SIS (2007) Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451: 431–436 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Ruusala T (1994) Mutations in Rik1, Clr2, Clr3 and Clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke U, Grabowski L, Gutz H, Heim L, Schmidt H (1987) Molecular characterisation of h-mutants of Shizosaccharomyces pombe. Curr Genet 12: 535–542 [Google Scholar]
- Greenall A, Williams ES, Martin KA, Palmer JM, Gray J, Liu C, Whitehall SK (2006) Hip3 interacts with the HIRA proteins Hip1 and Sml9 and is required for transcriptional silencing and accurate chromosome segregation. J Biol Chem 281: 8732–8739 [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Klar AJS (1996) Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86: 95–101 [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Klar AJS (1997) A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146: 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk AL, Kraenhenbuehl R, Molnar F, Fleck O, Kohli J (2004) Genetic and cytological characterization of the RecA-homologous proteins Rad51 and Dmc1 of Schizosaccharomyces pombe. Curr Genet 44: 317–328 [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K-i, Ayoub N, Cohen A, Grewald SIS (2002) Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- Halverson D, Baum M, Stryker J, Carbon J, Clarke L (1997) A centromere DNA-binding protein from fission yeast affects chromosome segregation and has homology to human CENP-B. J Cell Biol 136: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM (2005) Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Ireland JT, Gutkin GI, Clarke L (2001) Functional redundancies, distinct localizations and interactions among three fission yeast homologs of centromere protein-B. Genetics 157: 1191–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJS (1998) The chromo and SET domains of the Crl4 protein are essential for silencing in fission yeast. Nat Genet 19: 192–195 [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SIS (2004a) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Jia S, Yamada T, Grewal SIS (2004b) Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119: 469–480 [DOI] [PubMed] [Google Scholar]
- Kikuchi J, Iwahara J, Kigawa T, Murakami Y, Okazaki T, Yokoyama S (2002) Solution structure determination of the two DNA-binding domains in the Shizosaccharomyces pombe Abp1 protein by a combination of dipolar coupling and diffusion anisotropy restraints. J Biomol NMR 22: 333–347 [DOI] [PubMed] [Google Scholar]
- Lee J-K, Huberman JA, Hurwitz J (1997) Purification and characterization of a CENP-B homologue protein that binds the centromeric K-type repeat DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci USA 94: 8427–8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locovei AM, Spiga M-G, Tanaka K, Murakami Y, D'Urso G (2006) The CENP-B homolog, Abp1, interacts with the initiation protein Cdc23(MCM10) and is required for efficient DNA replication in fission yeast. Cell Div 1: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Huberman JA, Hurwitz J (1996) Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA 93: 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Lee J-K, Hurwitz J, Allshire RC, Nakayama J-i, Grewal SIS, Tanaka K, Murakami Y (2002) Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev 16: 1766–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SIS (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewald SIS (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293: 1150–1155 [DOI] [PubMed] [Google Scholar]
- Pidoux A, Mellone B, Allshire R (2004) Analysis of chromatin in fission yeast. Methods 33: 252–259 [DOI] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SIS (2003) Sir2 regulates histone lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13: 1240–1246 [DOI] [PubMed] [Google Scholar]
- Smith J, Brachmann C, Pillus L, Boeke J (1998) Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149: 1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkársdóttir U, Egel R, Nielsen O (1993) The smt-0 mutation which abolishes mating-type switching in fission yeast is a deletion. Curr Genet 23: 184–186 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K-i, Zofall M, Kobayashi R, Grewal SIS (2007) SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Thon G, Bjerling P, Bünner CM, Verhein-Hansen J (2002) Expression-state boundaries in the mating-type region of fission yeast. Genetics 161: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Bjerling P, Nielsen IS (1999) Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics 151: 945–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Klar AJS (1993) Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics 134: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Borgstrom B, Weilguny D, Egel R, Cooper JP, Nielsen O (2004) The fission yeast protein Rik1 is required for telomere clustering during meiosis. J Cell Biol 165: 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J (2005) Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast 22: 1061–1068 [DOI] [PubMed] [Google Scholar]
- Vengrova S, Dalgaard JZ (2004) RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev 18: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S, Dalgaard JZ (2006) The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep 7: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI (2005) The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table SI
Supplementary Table SII