Abstract
The mechanism of function of the bacterial flagellar switch, which determines the direction of flagellar rotation and is essential for chemotaxis, has remained an enigma for many years. Here we show that the switch complex associates with the membrane-bound respiratory protein fumarate reductase (FRD). We provide evidence that FRD binds to preparations of isolated switch complexes, forms a 1:1 complex with the switch protein FliG, and that this interaction is required for both flagellar assembly and switching the direction of flagellar rotation. We further show that fumarate, known to be a clockwise/switch factor, affects the direction of flagellar rotation through FRD. These results not only uncover a new component important for switching and flagellar assembly, but they also reveal that FRD, an enzyme known to be primarily expressed and functional under anaerobic conditions in Escherichia coli, nonetheless, has important, unexpected functions under aerobic conditions.
Keywords: bacterial chemotaxis, bacterial motility, flagellar assembly, FliG, fumarate reductase
Introduction
Bacterial chemotaxis is considered by many as the signalling system for which the level of understanding is most advanced (see, for a review, Eisenbach, 2004). The object of chemotactic signalling in bacteria is to change the direction of flagellar rotation in response to stimulation, which results in a change in swimming behaviour and direction (Larsen et al, 1974). The structural element that is responsible for switching the direction of flagellar rotation is the switch—a ring-like complex (termed the C-ring) at the base of the flagellar motor, which is one of nature's most intriguing molecular machines (see, for reviews, Macnab, 1995; Eisenbach and Caplan, 1998). The switch complex is built from multiple units of the proteins FliG, FliM, and FliN. It is striking how little is known about switch function despite (a) the centrality of switching for chemotaxis, (b) the many known aspects of switch structure, (c) the availability of functional switching mutants, and (d) the extensive effort invested in studying the switch (see, for reviews, Eisenbach and Caplan, 1998; Berg, 2003; Kojima and Blair, 2004). It is known that the default direction of rotation (i.e., the direction in the absence of intracellular signalling) is counterclockwise (see, for a review, Eisenbach, 1996), and that phosphorylation-controlled binding of CheY to the switch protein, FliM, enables the switch to shift to clockwise rotation (see, for a review, Eisenbach, 2004). However, the post-binding processes that occur at the switch are still a mystery.
Over a decade ago, Marwan et al (1990) found that fumarate is a factor needed for the archeal Halobacterium salinarium to swim back and forth. Subsequently, this dicarboxylate was found to be a clockwise switching factor in Escherichia coli. In cytoplasm-free but CheY-containing cell envelopes, it enabled the flagella to switch their direction of rotation (Barak and Eisenbach, 1992; Barak et al, 1996). By employing mutants having exceptionally low or high intracellular levels of fumarate due to the absence of one of the enzymes that act on fumarate, that is, succinate dehydrogenase (SDH) or fumarase, Prasad et al (1998) and Montrone et al (1998) found that, in intact E. coli cells, fumarate increases both the fraction of time spent in clockwise rotation and switching frequency. These effects were due, in part, to reduction of the standard free energy difference between the clockwise and counterclockwise states of the switch (Prasad et al, 1998). Fumarase, a cytosolic protein, was also shown to be involved in a phosphorylation-independent response to some repellents (Montrone et al, 1996).
The effects of fumarate in intact cells of E. coli were independent of the presence of CheY in the cell, indicating that fumarate exerted its action on the switch rather than on CheY (Prasad et al, 1998). In the present communication, we discovered that the enzyme fumarate reductase (FRD) is the target of fumarate at the switch.
FRD and its closely related homologue SDH are four-subunit membrane-bound enzymes (see Supplementary Figure s1 and Supplementary data), usually thought to be involved in electron transport reactions (see, for review, Cecchini et al, 2002). It is well documented that FRD functions in anaerobic respiration of E. coli and that SDH is involved in aerobic respiration. We provide evidence for a reversible interaction between FRD (hitherto unknown to be associated with motility or flagella) and FliG of the flagellar switch, and we demonstrate that mutants lacking frd are defective in flagellar assembly and switching and are not responsive to fumarate.
Results
Fumarate does not bind to any of the known switch proteins
We initiated this work by trying to determine whether fumarate binds to the switch complex. We isolated the intact switch complex of E. coli flagella (see Materials and methods and Supplementary Figure s2), incubated it with [14C]fumarate, and separated it from the medium by centrifugation. We detected no binding of [14C]fumarate (assayed in the range of 5–50 μM [14C]fumarate) to the isolated switch complex. We also measured the binding of [14C]fumarate to each of the three purified switch proteins. We used both equilibrium dialysis and centrifugal ultrafiltration, described in Materials and methods and Supplementary data, to measure binding of [14C]fumarate in the range 0.5–10 000 μM to each of the three switch proteins (10–200 μM). No binding was detected.
Potential targets of fumarate binding to the flagellar switch
The absence of detectable direct binding to any switch protein suggested that another protein may transmit the fumarate effect to the switch. This protein is expected to be membrane-bound because earlier it was shown that fumarate enhances switching even in envelopes devoid of cytoplasm (Barak and Eisenbach, 1992; Barak et al, 1996). Potential candidates for transmitting the fumarate effect to the switch are SDH and FRD—enzymes similar in amino-acid composition, 3D structure (Supplementary Figure s1), cofactors, and mechanisms of function (Cecchini et al, 2002). Both interact with fumarate and are membrane-bound. Thus, we tested the interaction of SDH and FRD with the switch.
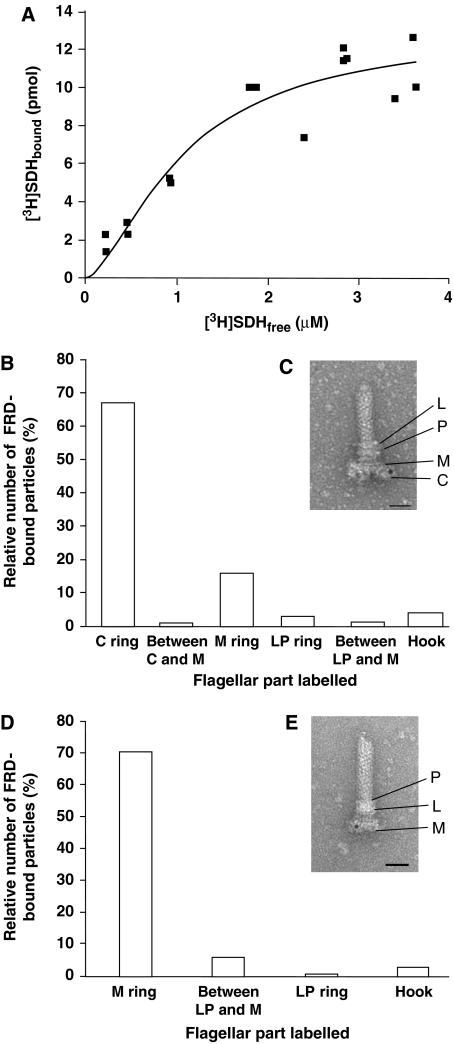
Evidence consistent with SDH binding to the switch complex was observed following incubation of [3H]SDH with the isolated intact switch complex and separation by centrifugation (Figure 1A; Kd=1.1±0.3 μM; mean±s.e.m.). To reduce nonspecific SDH binding, the experiment (detailed in Materials and methods) was carried out in the presence of at least 40-fold excess BSA, high salt concentration, and detergent. Insufficient amounts of the switch complex made direct binding measurements with an irrelevant protein impractical.
Figure 1.
SDH and FRD binding to the switch complex. (A) Concentration-dependent SDH binding to the isolated switch complex. See Materials and methods for details. The free SDH concentration was calculated by subtracting the concentration of bound SDH from the added SDH concentration. The data shown are normalized and derived from three different switch batches. The concentration of the switch was 0.1 mg protein/ml. The connecting line between the experimental points is a theoretical fit, performed by Origin computing program according to the Hill model (R2=0.90). (B) Protein A-gold particles bound to FRD in isolated extended flagellar basal bodies. The preparation of extended basal bodies was preincubated sequentially with FRD, anti-FRD antibody, and Protein A-gold. The number of gold particles counted was 1985. A total of 1265 basal bodies were counted; 715 were labelled at C-ring, M-ring, or between these rings, 87 were nonspecifically labelled in other parts of the basal body, and 463 were not labelled. The average number of gold particles associated with a basal body was 2.5 (the most common being 2 or 3 particles/basal body; range=1–10 particles/basal body). For the negative control with boiled FRD (not shown in the figure), a total of 382 basal bodies were counted: 3 (0.8%) were labelled at C- or M-ring, 4 (1%) were nonspecifically labelled in other parts of the basal body, and 375 (98%) were not labelled. Similar numbers were observed with BSA substituting for boiled FRD. (C) Image of an isolated extended flagellar basal body, treated as in (B) (bar=10 nm). (D) Protein A-gold particles bound to FRD in isolated basal bodies lacking the structure of the C-ring. Gold particles counted were 1619. Of the 1679 basal bodies counted, 1156 were labelled at the M-ring, 155 were nonspecifically labelled in other parts of the basal body, and 368 were not labelled. The average number of gold particles associated with such a basal body was 1.3 (the most common being 1 particle/body; range=1–6 particles/basal body). (E) Image of an isolated basal body lacking the structure of the C-ring, treated as in (B) (bar=10 nm).
To examine FRD binding to the switch, we employed immuno-electron microscopy, which requires much less material and does not involve chemical modification of the enzyme. We isolated flagellar hook basal bodies (termed ‘extended basal bodies'; Francis et al, 1994) and allowed them to bind FRD. (The basal body is the rotor of the flagellar motor, connected to the flagellar filament by means of a hook. It consists of a rod surrounded by coaxial rings: the M ring, a doublet ring embedded in the cytoplasmic membrane; the P ring, embedded in the peptidoglycan layer; and the L ring, embedded in the outer membrane (Figure 1E). Extended basal bodies also contain the C-ring (Figure 1C).) BSA was used as a control both for nonspecific binding/adsorption of the antibodies to the basal bodies and detection of protein A-Gold trapping by the basal body particles in the labelling protocol. Denatured FRD was used as a negative control. After binding, all the samples were incubated sequentially with anti-FRD antibody and protein A-Gold. FRD was localized to the C-ring (i.e., the switch complex) and, to a much lesser extent, to the M-ring (Figure 1B and C). Neither BSA nor denatured FRD was similarly localized. Thus, both SDH and FRD appear to bind to the switch complex.
To substantiate FRD binding to the M-ring, we assessed by immuno-electron microscopy its binding to a preparation of basal bodies that lack the C-ring structure and do not contain the switch proteins FliM and FliN (Francis et al, 1994). However, they do contain FliG, as ∼80% of the basal bodies are labelled following anti-FliG immunolabelling (Supplementary Figure s3) and as previously demonstrated (Francis et al, 1992, 1994). FRD binding to this preparation was evident (Figure 1D and E). The observation that both the C-ring and M-ring bound FRD raised the possibility that the site of FRD and, probably, SDH binding was FliG. This is because FliG is the only switch protein that was present in both basal-body preparations and because it appears to be associated with both the C-ring and M-ring (Oosawa et al, 1994; Thomas et al, 2001).
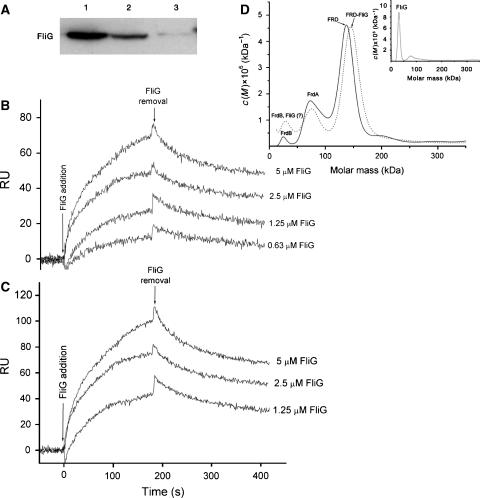
To explore this possibility, we employed three independent experimental approaches. (a) Following incubation with His-tagged FliG, biotinylated SDH and FRD were brought down by streptavidin–agarose beads and probed for the presence of FliG by western blotting with anti-His-tagged FliG antibody. His-tagged FliG colocalized with either SDH or FRD (Figure 2A). (b) Real-time surface plasmon resonance analysis showed that both purified SDH (Figure 2B) and FRD (Figure 2C), when individually immobilized onto the sensor surface, bound His-tagged FliG in a concentration-dependent manner. The dissociation rate constants, obtained on the basis of the Langmuir 1:1 bimolecular kinetic model, were similar for SDH and FRD (2 × 10−3 s−1), as were the association rate constants (3000 and 5000 M−1 s−1 for SDH and FRD, respectively) and the resulting dissociation constants (Kd=0.6±0.1 and 0.4±0.3 μM; mean±s.d. for SDH and FRD, respectively). Earlier tests showed that immobilized SDH bound neither FliM nor FliN (Prasad, 2001). The similarity between the Kd value obtained in this experiment for SDH binding to FliG and the Kd value for SDH binding to the isolated switch complex in Figure 1A strongly supports the argument that the SDH–switch binding in Figure 1A was specific. (c) Analytical ultracentrifugation was used to assess complex formation between His-tagged FliG and FRD. The shift in the location of the major protein peak from the molar mass of FRD to that of a 1:1 FliG–FRD complex when His-tagged FliG was present (Figure 2D) confirmed the formation of a complex and identified its stoichiometry.
Figure 2.
Binding of purified His-tagged FliG to purified FRD or SDH. (A) Pull-down assay. FliG was incubated with biotinylated FRD (lane 1), biotinylated SDH (lane 2), or, as a negative control, with neither FRD nor SDH (lane 3), precipitated with streptavidin–agarose beads, and probed with an anti-His-tagged FliG antibody. (B, C) Real-time interaction of His-tagged FliG with biotinylated SDH (B) or biotinylated FRD (C), measured by surface plasmon resonance. RU, resonance units. The values shown are after subtraction of the value of the empty channel. The kinetic and equilibrium constants, shown in the text, were obtained by a global fit, using the Langmuir 1:1 bimolecular kinetic model (A+B⇔AB; fit quality: χ2=5 and 11 for the SDH and FRD curves, respectively). (D) Analytical ultracentrifugation of the FliG-FRD complex. The c(M) distribution is a variant of the distribution of Lamm equation solutions using Sedfit (see Supplementary data). Three samples were run: FRD alone (2 μM; solid line), His-tagged FliG alone (16 μM; inset), and both together (2 μM each; dotted line). The major peak observed in the His-tagged FliG and FRD mixture (dotted line) was at 150 kDa (corresponding to a 1:1 FliG:FRD complex; 158 kDa theoretical value, calculated from the amino-acid composition). The changes of the sedimentation rates of the lesser peaks of FRD were minor in the presence of His-tagged FliG. The major peak of His-tagged FliG (inset) was at 32 kDa (37 kDa theoretical); the low peak at 75 kDa probably reflects a FliG dimer. The peaks of FRD (solid line) correspond, in the order of decreasing abundance, to FRD (134 kDa, consisting of all four subunits FrdA, FrdB, FrdC, and FrdD; 121 kDa theoretical), FrdA (71 kDa; 66 kDa theoretical), and FrdB (23 kDa; 27 kDa theoretical). FrdC and FrdD are not observed in the figure, as expected, due to their extreme hydrophobicity (Cecchini et al, 2002), which precludes them from the solution (either sedimenting to the bottom or adhering to the wall of the tube).
The seminal issues, however, are whether this binding has physiological significance, and whether either or both SDH and FRD are responsible for the action of fumarate on the switch.
FRD but not SDH affects the functions of FliG
FliG is central to the function of the flagellar motor. It is known to be directly involved in motor assembly, torque generation, and switching (see, for a review, Kojima and Blair, 2004). Consequently, defects in this protein may lead to loss of flagella, paralysis, or biased direction of flagellar rotation. Assuming that the interaction of FliG with FRD or SDH is required for its function, absence of FRD or SDH may lead to any one of these phenotypes. Therefore, we prepared three deletion mutants: a Δfrd mutant, deleted for the genes encoding all the subunits of FRD; a Δsdh mutant in which two of the four genes encoding SDH were deleted, resulting in complete absence of SDH (Montrone, 1996; Prasad et al, 1998); and a double Δfrd Δsdh mutant.
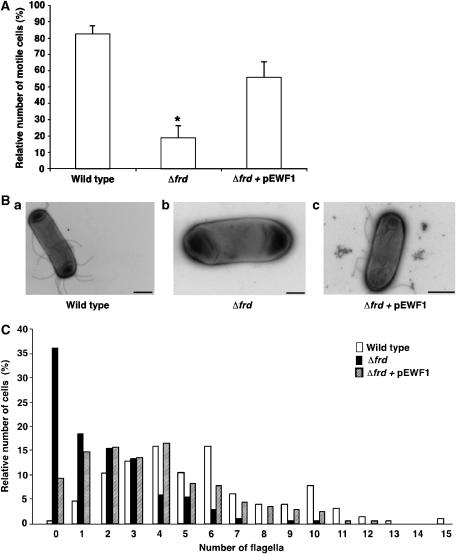
The Δsdh mutant did not differ from its wild-type parent with respect to motility (data not shown), whereas, strikingly, the Δfrd mutant and the double mutant were barely motile. As shown for the Δfrd mutant (Figure 3A), many cells did not swim at all, others swam more slowly than usual and, in most of these latter cases, the movement was wobbly. This behaviour resulted from a decrease in the number of flagella (Figure 3B and C). The wild-type parent had a median of 5 flagella/cell, but the Δfrd mutant had a median of only 1 flagellum/cell, with many cells having no flagella at all. Similar data (not shown) were obtained for the double mutant. To verify that the observed phenotype was due to the absence of FRD, we complemented the frd deletion with a plasmid, producing a single copy of FRD under its native promoter (pEWF1). The plasmid restored, at least partially, the number of flagella (median of 3 flagella/cell; Figure 3B and C) and increased the fraction of motile cells (Figure 3A). As the frd deletion did not affect the expression level of FliG, as evident from western blots with anti-FliG antibody (Supplementary Figure s4), the results suggest that FRD is required for normal flagellar assembly.
Figure 3.
Effects of frd and sdh deletions on swimming, assembly of flagella, and switching the direction of flagellar rotation. (A) Percentage of motile cells. Swimming cells were video-recorded and the fraction of motile cells was determined blindly. Values shown are the mean±s.e.m. of three experiments, 100–200 cells for each strain in each experiment. The asterisk indicates a statistically significant difference from the other columns (P<0.01 and P<0.05 for wild-type and Δfrd+pEWF1 columns, respectively; one-way ANOVA plus Tukey–Kramer tests). (B) Typical micrographs of strains RP437 (a; bar=1 μm), RP437Δfrd (b; 0.5 μm), and RP437Δfrd containing pEWF1 (c; 1 μm). (C) Distribution of the number of flagella per cell. Cells were negatively stained with uranyl acetate and photographed using a transmission electron microscope. The number of flagella of ∼200 cells of each strain were counted without prior knowledge of the strain being counted. The difference in the number of flagella between the strains was very significant (P<0.001; Krusal–Wallis test). Open columns, strain RP437; solid columns, RP437Δfrd; hatched columns, RP437Δfrd containing pEWF1.
FRD deletion could potentially reduce the energy level and elevate the fumarate level in the cell, contributing to the observed phenotypes of the Δfrd mutant. No evidence for these scenarios was found. We measured the intracellular ATP concentration and found it to be similar in all the strains used, with a variation of ∼10% from the value of 2.5±0.1 mM measured for the wild-type strain. (It should be mentioned, however, that although removal of FRD did not have a significant effect on cellular ATP levels, its absence might have other, unexplored effects on cellular metabolism.) The possible contribution of an elevated fumarate level was ruled out earlier by showing that, in cells deleted of the genes encoding fumarase, elevated cellular fumarate has no effect on the motility level (Montrone et al, 1998; Prasad et al, 1998).
To examine the effect of frd deletion on flagellar rotation of those cells that had residual flagella, we tethered them through a flagellum to a glass slide and monitored the rotation of the cells (Silverman and Simon, 1974). The tethered Δfrd cells rarely switched their direction of rotation, making a reversal once every 2.5 min on average (Table I, line 2) and rotating counterclockwise 99.9% of the time (line 1). The addition of the strong repellent benzoate increased the probability of clockwise rotation in both the Δfrd strain and its wild-type parent (line 3 versus line 1). However, in the Δfrd strain, the fraction of responsive cells was smaller (line 5), and the level of clockwise rotation achieved after this stimulation was significantly lower than that in the wild type (line 3). In contrast, the switching frequency was higher than that in the wild type (line 4), possibly reflecting futile attempts to switch to clockwise rotation. These results are consistent with the clockwise-promoting effect of fumarate in the intact cells of E. coli (Montrone et al, 1998; Prasad et al, 1998) and with the possibility, investigated below, that FRD is the protein that transmits the fumarate effect to the switch.
Table 1.
Comparison between wild-type and Δfrd strains with respect to the parameters of flagellar rotation
| Parameter testeda | Wild type | Δfrd | P-value for the difference between the strainsb |
|---|---|---|---|
| Before stimulation | |||
| 1. Time fraction spent in clockwise rotation±s.e.m. (%) | 2.0±0.9 | 0.1±0.1 | 0.25 |
| 2. Switching frequency±s.e.m. (min−1) | 2.1±0.5 | 0.4±0.2 | <0.009 |
| After stimulation | |||
| 3. Time fraction spent in clockwise rotation±s.e.m. (%) | 70±4 | 13±4 | <0.0001 |
| 4. Switching frequency±s.e.m. (min−1) | 0.7±0.2 | 16±2 | <0.0001 |
| 5. Fraction of responsive cells (%) | 96 | 75 | 0.001 |
| aThe rotation of 46–59 tethered cells was monitored for periods of 40 s before and subsequent to stimulation with the repellent benzoate (50 mM). | |||
| bCalculated by the Mann–Whitney test, except for the fraction of responsive cells, which was calculated by the Fisher's exact test. | |||
FRD transmits the effect of fumarate to the flagellar switch
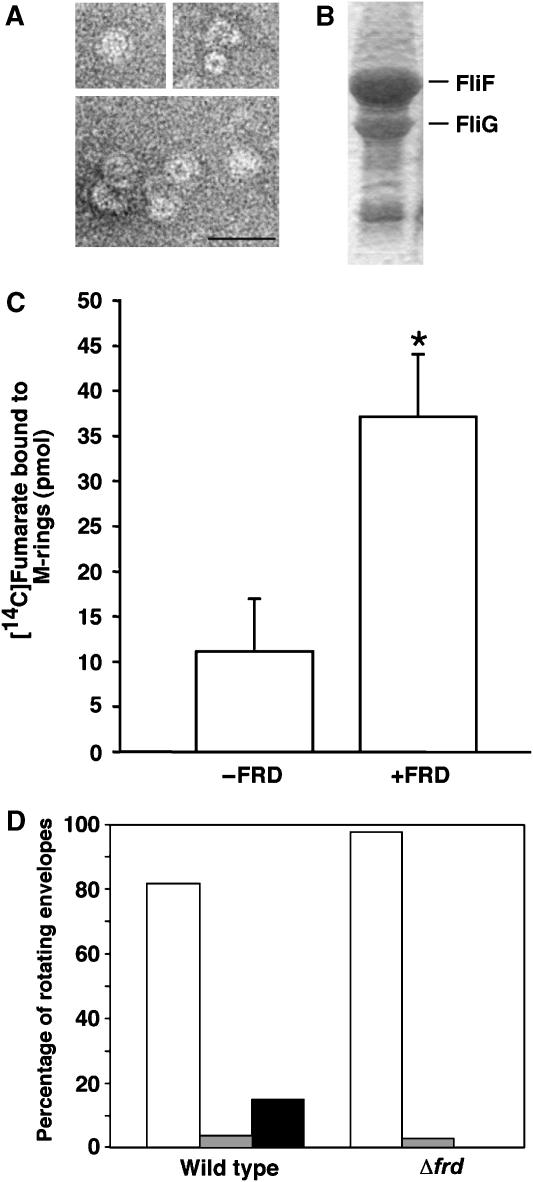
To examine this possibility, we employed two approaches. First, we studied the binding of [14C]fumarate to a preparation of M-rings associated with FliG (Figure 4A and B)—the minimal structure shown to bind FRD (Figure 1D and E)—in the presence and absence of FRD. The presence of FRD significantly increased the binding of fumarate to this preparation (Figure 4C). Next, we measured the effect of fumarate on flagellar rotation using tethered, cytoplasm-free envelopes containing His-tagged CheY. In intact bacteria, fumarate mainly increases the fraction of time spent in clockwise rotation (Montrone et al, 1998; Prasad et al, 1998). In envelopes, fumarate enables switching: tethered envelopes require this dicarboxylate to switch from one direction of rotation to the other (Barak and Eisenbach, 1992; Barak et al, 1996). If fumarate binding to the switch complex depends on FRD, envelopes prepared from the Δfrd mutant should be incapable of switching either in the presence or absence of fumarate. The results confirmed this prediction. Although fumarate was present, none of the Δfrd envelopes switched directions, rotating exclusively in one direction, mostly counterclockwise. In contrast, wild-type envelopes showed switching (Figure 4D). These results (Figure 4C and D), taken together with the observation that Δsdh mutant envelopes continue to switch when fumarate is added (Barak et al, 1996), suggest that, FRD, but not SDH, transmits the fumarate effect to the switch and that FRD may be a functional component of the switching mechanism of E. coli.
Figure 4.
FRD transmits the effect of fumarate to FliG at the switch–motor complex. (A) Electron micrographs of purified M-rings. Bar=50 nm. (B) SDS gel of the M-rings (composed of the protein FliF), demonstrating the co-occurrence of FliG. (C) Binding of fumarate to the M-ring in the presence and absence of FRD. Fumarate binding was measured with 50 μM [14C]fumarate and 20 μM FRD. The concentration of the M-ring proteins was 10 μM. The data shown are the means (±s.e.m.) of 3–4 repetitions. The asterisk indicates a statistically significant difference (P<0.04, unpaired t-test). (D) Effect of frd deletion on the rotation of tethered, CheY-containing, cytoplasm-free envelopes in the presence of fumarate at room temperature. The figure shows the distribution of envelopes according to their direction of rotation. Open columns, counterclockwise-rotating cells; hatched columns, clockwise-rotating cells; solid column, switching cells. The numbers of monitored envelopes were 115 and 39 for the wild-type strain and the Δfrd mutant, respectively.
Discussion
In this study, we have made three important discoveries, none of which were predictable on the basis of pre-existing knowledge. First, we demonstrated that FRD is important for the function of the switch and appears to form a dynamic complex with it. This conclusion is based on the observations that (a) absence of FRD results in a sharp decrease in the number of flagella per cell and, consequently, in cell motility (Figure 3), without affecting the level of FliG in the cell (Supplementary Figure s4); (b) the ability of existing flagella to switch rotation to the clockwise direction is much reduced in the absence of FRD (Table I); and (c) FRD forms a 1:1 complex with the switch protein FliG (Figures 1 and 2). These observations thus demonstrate that FRD is involved in both flagellar assembly and switching to clockwise rotation.
Second, we identified FRD as the target binding site of fumarate at the switch, enabling fumarate to exert its stimulating effect on clockwise switching (Figure 4).
Third, we have demonstrated that FRD has an important role under aerobic conditions in E. coli, unassociated with ATP production. All the results shown were obtained under aerobic conditions, where FRD is minimally expressed (∼4% of the expression level compared to anaerobic conditions; Jones and Gunsalus, 1985) and, according to the current understanding, it has no essential function (Cecchini et al, 2002). Furthermore, although FRD, similar to SDH, is known to be involved in the energy-conversion system, it has not been associated until now with flagellar function or with the motility system. Our observations raise significant new questions about the relationship between FRD and the switch.
Complex formation between the switch and FRD
The formation of a complex between FRD/SDH and FliG at the switch was demonstrated by five different lines of evidence (Figures 1 and 2). One of them, surface plasmon resonance analysis, indicated that the complex formed is not permanent, slowly dissociating when FliG is removed (Figure 2B and C). These observations suggest that the complex formed is in a dynamic equilibrium with non-bound FRD. Further studies are needed to determine which subunits of FRD and domain of FliG are involved in complex formation. The similarity between the phenotypes of the Δfrd mutant (Figure 3) and those of fliG mutants with tryptophan replacements at residue 163 or 165 in the middle domain of FliG (Brown et al, 2007) raises the possibility that the FliG region(s) affected by these tryptophan replacements are involved in the interaction with FRD.
Comparative roles of FRD and SDH
An intriguing question is why the phenotypes are specific for FRD and not for SDH, even though both homologues bind to FliG with similar affinities (Figure 2). Two alternative explanations seem plausible. (a) The similarity of the affinities suggests that either or both of the cytoplasmic localized A and B subunits, known to be highly similar in both enzymes (Supplementary Figure s1), participate in this binding. Thus, they are reasonable candidates for interaction with the switch complex, which resides in the cytoplasm and is tethered to the membrane (Khan et al, 1992; Francis et al, 1994; Macnab, 1995). This suggests that the subtle differences between SdhAB and FrdAB (e.g., the more exposed C-terminus of FrdB) are sufficient to make only the latter active in switching and flagellar assembly. (b) The part of the molecule that is responsible for the observed activities of FRD may be the membrane domain, which is the region of these proteins with significant differences between FRD and SDH (Supplementary Figure s1).
Functional differences between FRD and SDH also may account for their different effects on switching. FRD is a more versatile enzyme, functional over a much wider range of physiological conditions than SDH (Ackrell et al, 1993; Léger et al, 2001). For example, FRD may be able to compensate for the changes in the metabolic conditions better than SDH, and it functions with a variety of diverse quinones, whereas SDH appears to function only with higher potential quinones (Maklashina et al, 2006). One could speculate that these differences in activity could affect the conformations of FRD and SDH and thus the interaction with FliG at the switch complex.
It is interesting to note that the flagellar master regulon induces the expression of all FRD subunits and represses those of SDH (Prüss et al, 2003). This correlation between the expression of flagellar genes and frd genes, in contrast to the sdh genes, is consistent with the present study showing an association between flagellar number and FRD. It is further supported by the finding of Landini and Zehnder (2002) that E. coli grown under anaerobic conditions has increased flagellation.
Flagellum-related activities of FRD and its involvement in electron transport
In E. coli, although FRD is considered to be exclusively involved in anaerobic respiration and electron transport, this versatile enzyme can physiologically substitute for SDH in aerobic respiration (Guest, 1981). The question is whether the observations made herein are related to the electron transport properties of FRD or to some hitherto unrecognized protein–protein interactions. It is clear that E. coli FRD does not generate a proton-motive force during electron and proton transfer (Cecchini et al, 2002; Lancaster, 2002). Therefore, the observations reported in this article are not directly related to enhanced energization. This conclusion is in accordance with the previous observation of Barak et al (1996) that the promoting effect of fumarate on switching in cytoplasm-free envelopes of E. coli is not related to oxidation–reduction processes because other dicarboxylates, which can bind FRD and SDH but cannot accept electrons, also cause switching although less efficiently. Thus, the electron transfer from menaquinol to fumarate is unlikely to be involved in fumarate's effect on switching. However, this does not exclude the possibility that FRD is just one part of a larger energy-conversion complex associated with the switch–motor complex.
Potential mode by which FRD may affect the switch
The results of this study do not answer the question how fumarate binding to FRD enables switching or how FRD enhances flagellar assembly. It is well established that binding of a dicarboxylate or full reduction of the iron–sulphur clusters results in long-range conformational changes in FRD (Iverson et al, 2000; Hudson et al, 2005). It is plausible that the binding of fumarate or one of its analogues induces a conformational change in FRD that may modify either the interaction with FliG or the physiological response. These possibilities are clearly speculative and more data are required to resolve this issue.
FRD in other bacterial species
At present, it is not clear whether the observations reported here are restricted to E. coli and similar species. It is known from available databases of bacterial genomes that there are many motile bacterial species that do not possess FRD. FRD is considered an evolutionary precursor of SDH (Ackrell et al, 1992), and may have evolved under anaerobic conditions. Involvement of FRD with the flagellar switch–motor complex may have arisen to support under anaerobic conditions the high energetic requirements for the synthesis and function of the flagella. Facultative anaerobic bacteria such as E. coli often contain both FRD and SDH. In these cases, FRD may maintain its association with the flagella. In contrast, in strictly aerobic bacteria lacking FRD, perhaps SDH or another membrane protein may substitute for FRD. Such possibilities remain to be explored.
Materials and methods
Chemicals
2,6-Dichloroindophenol, phenazine methosulphate, succinate (sodium salt), fumarate (sodium salt), polyoxyethylene(9)dodecylether, and [14C]fumarate were obtained from Sigma (St Louis, MO, USA); Ni-NTA matrices were from QIAGEN (Hilden, Germany); sodium boro[3H]hydride, [14C]leucine, and [3H]leucine were from Amersham (Buckinghamshire, UK). All the compounds were of the highest purity grade available. (See Supplementary data for the complete list.)
Bacterial strains
The strains and plasmids used in this study are listed in Table II. Strains RP437Δfrd and RP437ΔsdhΔfrd were constructed by P1 transduction as described (Silhavy et al, 1984). A P1 phage lysate prepared on DW35 (ΔfrdABCD, zjd∷Tn10) was used to infect RP437 and RP437Δsdh. Positive colonies were selected for tetracycline resistance and for their inability to use fumarate for growth on fumarate/glycerol minimal medium under anaerobic conditions as described (Westenberg et al, 1993).
Table 2.
List of strains and plasmids used in this study
| Strain/plasmid | Relevant genotype | Reference and/or source |
|---|---|---|
| Strains | ||
| BL21(λDE3) | HsdS gal ompT rB− mB− [ΔC1857(Ts)indI Sam7 min5 lacUV5-T7 gene 1] | Studier et al (1990), Novagen |
| BL21(λDE3)pLysS | HsdS gal ompT rB− mB− [ΔC1857(Ts)indI Sam7 min5 lacUV5-T7 gene 1] | Studier et al (1990), Novagen |
| RP437 | Wild type for chemotaxis; his thr leu metE thi eda rpsL | Parkinson (1978) |
| DW35 | zjd∷Tn10Δ(frdABCD)18sdhC∷kan | Westenberg et al (1993) |
| RP437Δsdh | Wild type for chemotaxis; his thr leu metE thi eda rpsL; Δsdh | Montrone (1996) |
| RP437Δfrd | Wild type for chemotaxis; his thr leu metE thi eda rpsL; Δfrd | This study |
| RP437ΔsdhΔfrd | Wild type for chemotaxis; his thr leu metE thi eda rpsL; ΔsdhΔfrd | This study |
| DFB225ΔfliG | RP437 parent containing an fliG in-frame deletion | Received from David Blair (Lloyd et al, 1996) |
| M15 [pREP4] | LacZ StrRKanR | Zamenhof and Villarejo (1972), QIAGEN |
| Plasmids | ||
| pQE12-CheY-His tag | 6 × His–CheY overproducing | Received from A Wolfe |
| pKOT107 | FliG and FliF overproducing | Oosawa et al (1994) |
| pFAS | SDH overproducing | Maklashina et al (1998) |
| pKLR2 | FliM and FliN overproducing | Oosawa et al (1994) |
| pEWG1 | 10 × His–FliG overproducing | This study |
| pEWG2 | 10 × His–FliG | This study |
| pBAD24 | Expression vector containing the PBad promoter; Apr | Guzman et al (1995) |
| pH3 | FRD overproducing | Blaut et al (1989) |
| pEWF1 | FRD single-copy producing | This study |
Constructs
For the construction of the plasmid pEWG1 (a His-tagged FliG expression vector), the gene fliG was amplified from the E. coli chromosome by PCR, with the primer sequences 5′-GGAATTCCATATGAGTAACCTGACAGGCACC-3′ and 5′-CGGGATCCTCAGACAGGTATCCTCG-3′. The amplified PCR fragments were cloned into the pET19b vector (Novagen, Darmstadt, Germany) with NdeI/BamHI sites. The construct was confirmed by DNA sequencing. To verify that the His-tagged FliG is functional, we constructed the plasmid pEWG2, which expresses His-tagged FliG under the arabinose promoter. Strain DFB225 deleted of fliG was transformed with the pEWG2 plasmid (a His-tagged FliG expression vector), described below, and the functionality of the His-tagged FliG was confirmed by the formation of expanding rings on tryptone broth semi-solid agar plates. The plasmid pEWG2 was constructed in two ligation steps. First, the His-tagged fliG fragment was excised from pEWG1 by digestion with NcoI and BamHI, followed by ligation with pBAD24 predigested with NcoI and SmaI. Next, the BamHI site was filled and ligated with the SmaI blunt end to form pEWG2.
To construct the plasmid pEWF1 (FRD expression vector), plasmid pH3 was digested with HindIII to release the complete frd coding sequence including its natural promoter and ligated into a similarly digested pZS24-lac/ara-MCS-1 plasmid (containing the pSC-101 origin of replication). The functional status of FRD expressed from pEWF1 was verified by the ability of the RP437Δfrd mutant, transformed with this plasmid, to grow under anaerobic conditions in a minimal medium of glycerol/fumarate.
Preparations
Switch complex and M-ring. To prepare the switch complex, we expressed both the plasmids pKOT107 (overproducing FliG and FliF of Salmonella typhimurium) and pKLR2 (overproducing FliM and FliN of S. typhimurium) in E. coli strain BL21(λDE3). To prepare the M-ring, we expressed only pKOT107. The overproduced switch complex (having the form of a ring) and M-ring were purified by differential centrifugation as described by Sagi et al (2003). The preparation of isolated switch complexes was tested for functionality by measuring, as described below, phosphorylation-dependent binding of [14C]CheY to them (Sagi et al, 2003).
Flagellar hook basal bodies. Extended basal bodies for electron microscopic studies were isolated as described by Francis et al (1994) from S. typhimurium cells of strain SJW134 (a wild-type strain except for the lack of expressed flagellin gene; Thomas et al, 2001) grown to late-log phase. The cells were cooled on ice, pelleted, lysed, and the outer membrane was dissociated with KOH, pH 11. To prepare specimens of basal bodies lacking the C-ring, extended basal bodies were spun down and the pellet was incubated in glycine (50 mM, pH 3.5) for 1 h at 0°C. The sample was again pelleted and resuspended in Tris–HCl (10 mM, pH 8), EDTA (5 mM), and Triton X-100 (0.1% v/v). The samples were kept at 0°C during all the steps.
Flagellated, CheY-containing, cytoplasm-free envelopes. Flagellated cell envelopes (Eisenbach and Adler, 1981) were prepared by penicillin treatment followed by osmotic lysis as described (Ravid and Eisenbach, 1984). The lysis medium contained purified, His-tagged CheY (36 μM, for inclusion in the formed envelopes; Ravid et al, 1986), Tris–HCl (50 mM, pH 7.9), Tetren (0.1 mM), and DL-lactate (2 mM).
Antibodies. Three polyclonal anti-FliG antibodies were used: anti-His-tagged FliG antibody for the pull-down experiments (Figure 2A); anti-FliG antibody received from K Oosawa for the immunolabelling of basal bodies (Supplementary Figure s3); and anti-FliG antibody obtained from M Macnab for western blots of bacterial extracts (Supplementary Figure s4). The polyclonal antibodies against His-tagged FliG (a band cut from gel) and those against FRD (purified native protein) were raised in rabbits as described (Harlow and Lane, 1988). To purify the antibodies, we used Immunolink beads (Pierce, Rockford, IL, USA). A lysate of a ΔfliG or Δfrd strain was used to remove all reactive material according to the manufacturer's protocol. The non-retained antibody was checked for purity and specificity by western blotting. The resulting antibodies were highly specific (Supplementary Figure s5).
Protein purification
His-tagged FliG and His-tagged CheY. E. coli 10 × His–FliG (for use in binding assays with SDH and FRD) was expressed from the plasmid pEWG1, which had been transformed into E. coli strain BL21(λDE3)pLysS (Table II). E. coli 6 × His–CheY, demonstrated earlier to be functional (Sagi et al, 2003), was produced from the plasmid pQE12, pre-transformed into E. coli strain M15 (Table II). Both proteins were purified by Ni-NTA affinity chromatography (QIAGEN) according to the manufacturer's instructions. Proteins were concentrated by ultrafiltration (Ultracel Amicon) and dialysed against the buffer used in the assay. The purities of the isolated proteins were high (Supplementary Figure s2).
SDH and FRD. E. coli SDH and FRD were purified to a high degree (Supplementary Figure s2) as described by Tornroth et al (2002) and Luna-Chavez et al (2000), respectively. Purified SDH and FRD had specific activities of 17 and 20 μmol/min/mg protein, as determined by the succinate:phenazine methosulphate/2,6-dichloroindophenol reaction at 37°C (Ackrell et al, 1978). The isolation procedure retains quinone and lipids in isolated SDH and FRD (Luna-Chavez et al, 2000; Yankovskaya et al, 2003) and yields a 1:1:1:1 stoichiometry of subunits in both enzymes (Supplementary Figure s2).
Switch proteins. His-tagged FliM was purified to >95% purity from E. coli strain BL21(λDE3)pLysS, carrying the plasmid pEWM1 that overexpresses S. typhimurium's His-tagged FliM, as described by Bren and Eisenbach (1998). S. typhimurium FliG (for use in binding assays with [14C]fumarate) and FliN were overexpressed in E. coli strain BL21(λDE3)pLysS and partially purified as described by Welch et al (1993).
Protein modifications
Radiolabelling of His-tagged CheY. Labelling of 6 × His–CheY was performed in vivo by metabolic incorporation of [14C]leucine during growth, as described by Sagi et al (2003).
Radiolabelling of SDH. SDH was radiolabelled by methylating its free amino groups with formaldehyde and NaB[3H]4, as described previously (Blat and Eisenbach, 1996). This radiolabelling approach had the disadvantage that it reduced the enzymatic activity of SDH by about half.
Biotinylation of SDH and FRD. The proteins in KPi (20 mM, pH 7.5) were added to 5- to 10-fold molar excess of biotin-3-sulpho-N-hydroxy succinimide ester (sodium salt), incubated overnight at 4°C, and dialysed against the binding assay buffer. Biotinylation was verified with streptavidin–agarose beads (Pierce) according to the manufacturer's protocol.
Binding of [14C]CheY, [3H]SDH, and [14C]fumarate to the switch complex and to the M-ring
To assess binding of radiolabelled compounds to the switch complex or to the M-rings, samples were incubated at room temperature (25°C) with the radiolabelled protein or radiolabelled fumarate for 5 min (with the switch complex) or 1 h (with M-rings). For [14C]CheY or [14C]fumarate binding, 1.6 mg/ml switch complex was used. For [3H]SDH binding, 0.1 mg/ml switch complex was used. The switch complex was incubated with [14C]CheY (3.6 μM, 13–17 d.p.m./pmol), [3H]SDH (0.25–4 μM, 2600–7400 d.p.m./pmol), or [14C]fumarate (10–500 μM, 14 d.p.m./pmol) in a solution of Tris–HCl (50 mM, pH 7.9), MgSO4 (5 mM), and BSA (10 mg/ml or, in the case of fumarate binding, 1 mg/ml). In addition, the solution contained [3H]leucine or [14C]leucine (600 d.p.m./pmol), as appropriate, to correct for the entrapped volume as described by Sagi et al (2003). For [3H]SDH binding, the solution also contained NaCl (150 mM) and Thesit (0.05%; w/v). For phosphorylation conditions in the case of [14C]CheY binding, AcP (25 mM) was added to the reaction mixture. Following the incubation, the switch complex was centrifuged at 200 000 g for 10 min at 4°C. The pellet was resuspended in 100 μl of Tris–HCl (50 mM, pH 7.9). Aliquots of the reaction mixture before centrifugation and aliquots of the supernatant and pellet subsequent to centrifugation were counted for 14C and 3H with a liquid scintillation analyser.
Binding of [14C]fumarate to isolated switch proteins
The binding of [14C]fumarate to the switch proteins (distinct from its binding to the switch complex, described above) was carried out by equilibrium dialysis (a classical approach for studying the binding of a small molecule to a protein) and centrifugal ultrafiltration (a technique employed even for low-affinity binding) as detailed in Supplementary data.
Binding of FRD or SDH to FliG
Pull-down assay. Biotinylated FRD or SDH (4 μM) was mixed with His-tagged FliG (8 μM) in solution A containing KPi (20 mM, pH 7.5), NaCl (150 mM), and Thesit (0.05%), followed by overnight incubation with shaking at 4°C. Subsequently, the reaction mixture was incubated with streptavidin–agarose beads and BSA (1.6 mg/ml) for 1 h at 4°C, washed with solution A, boiled for 5 min with SDS sample buffer, applied to 12% SDS–PAGE, and the gels probed with anti-His-tagged FliG antibody.
Measurement of binding by surface plasmon resonance. Measurement of FliG-FRD/SDH binding by surface plasmon resonance was carried out with a ProteOn™ XPR36 Protein Interaction Array System (Bio-Rad). Biotinylated FRD and SDH were individually immobilized onto five NeutrAvidin-coated channels of a Proteon NLC (Bio-Rad) sensor chip, leaving one channel blank as reference. Immobilization was achieved by injecting biotinylated FRD (180 μl) in a solution of KPi (50 mM, pH 7.4), MgSO4 (5 mM), NaCl (150 mM), and the surfactant Tween 20 (0.005%) at a rate of 30 μl/min. About 750–900 RU of biotinylated FRD and SDH was typically immobilized under these conditions. His-tagged FliG was passed continuously over the immobilized proteins in each channel at a rate of 100 μl/min. Regeneration of the surface was carried out with short pulses of NaOH (10 mM) for 30 s at a flow rate of 100 μl/min. As a negative reference, we used interferon α2. The dissociation and rate constants were calculated using the ProteOn Manager v.2 (Bio-Rad).
Sedimentation velocity measurement. Sedimentation velocity measurements were performed in an analytical ultracentrifuge as detailed in Supplementary data.
Electron microscopy assays
FRD binding to basal bodies. Basal bodies were incubated at 0°C for ∼12–14 h with activated FRD (1 mg/ml). Anti-FRD antibody was added and the solution was incubated at 0°C for 15 min. Protein A-gold was added to the complex and incubated for 10 min at 0°C. The samples were centrifuged in a Beckman TLA rotor at 50k r.p.m. for 5 min. The pellet was washed 3 × , applied to carbon-coated grids, and stained with 2% uranyl acetate. The samples were imaged with an FEI Morgani 268 electron microscope equipped with an AMT CCD camera, 1k × 1k (FEI, The Netherlands) or Philips CM12 TEM (Phillips Electronic Instruments Co., Mahwah, NJ).
Negative staining for counting flagella. Cells were grown on tryptone broth to mid-logarithmic phase and washed with KPi (pH 7.4, 20 mM). The cell suspension (4 μl) was deposited on a nitrocellulose-coated electron microscopy grid. The sample was blotted with a filter paper and washed twice with droplets of 1% uranyl acetate.
Determination of the direction of flagellar rotation by tethering assay
The direction of flagellar rotation of intact cells and cell envelopes was determined by the tethering assay (Silverman and Simon, 1974), detailed in Supplementary data.
Measurement of intracellular ATP
Bacteria were grown on tryptone broth to a mid-exponential phase, frozen in aliquots of 1 × 107 cells, and diluted 1:6 in nucleotide-releasing buffer for bacterial cells (Lumac bv, The Netherlands). The ATP level in the cells was measured by the luciferin–luciferase method (Stanley and Williams, 1969), using a Lumac/3M Biocounter, model 2010A. Each measurement was preformed in duplicate or triplicate. The calculations were based on an internal volume of 1.2 × 10−15 l/cell and 2.2 μl/mg protein (Welch et al, 1995).
Statistical analysis
All statistical analyses were carried out by Instat software (GraphPad Software, San Diego, CA).
Supplementary Material
Supplementary Information
Acknowledgments
We are indebted to G Schreiber for professional advice and continuous assistance in the protein–protein binding experiments, to M Dines and S Cohen (Bio-Rad, Haifa, Israel) for their invaluable help in the Proteon experiments, to R Barak for assistance and advice in the studies with cytoplasm-free envelopes, to A Gakamsky for statistical analysis, and to P Reuven for the title. We thank DF Blair for providing strain DFB225ΔfliG, and M Macnab and K Oosawa for the anti-FliG antibodies. ME is an incumbent of the Jack and Simon Djanogly Professorial Chair in Biochemistry. This study was supported by a US–Israel Binational Science Foundation research grant 2005199 to ME and GC.
References
- Ackrell BA, Armstrong FA, Cochran B, Sucheta A, Yu T (1993) Classification of fumarate reductases and succinate dehydrogenases based upon their contrasting behaviour in the reduced benzylviologen/fumarate assay. FEBS Lett 326: 92–94 [DOI] [PubMed] [Google Scholar]
- Ackrell BA, Kearney EB, Singer TP (1978) Mammalian succinate dehydrogenase. Methods Enzymol 53: 466–483 [DOI] [PubMed] [Google Scholar]
- Ackrell BAC, Johnson MK, Gunsalus RP, Cecchini G (1992) Structure and function of succinate dehydrogenase and fumarate reductase. In Chemistry and Biochemistry of Flavoenzymes, Müller F (ed), Vol. III, pp 229–297. Boca Raton, Fl: CRC Press [Google Scholar]
- Barak R, Eisenbach M (1992) Fumarate or a fumarate metabolite restores switching ability to rotating flagella of bacterial envelopes. J Bacteriol 174: 643–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R, Giebel I, Eisenbach M (1996) The specificity of fumarate as a switching factor of the bacterial flagellar motor. Mol Microbiol 19: 139–144 [DOI] [PubMed] [Google Scholar]
- Berg HC (2003) The rotary motor of bacterial flagella. Annu Rev Biochem 72: 19–54 [DOI] [PubMed] [Google Scholar]
- Blat Y, Eisenbach M (1996) Mutants with defective phosphatase activity show no phosphorylation-dependent oligomerization of CheZ, the phosphatase of bacterial chemotaxis. J Biol Chem 271: 1232–1236 [DOI] [PubMed] [Google Scholar]
- Blaut M, Whittaker K, Valdovinos A, Ackrell BA, Gunsalus RP, Cecchini G (1989) Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J Biol Chem 264: 13599–13604 [PubMed] [Google Scholar]
- Bren A, Eisenbach M (1998) The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol 278: 507–514 [DOI] [PubMed] [Google Scholar]
- Brown PN, Terrazas M, Paul K, Blair DF (2007) Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol 189: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini G, Schroder I, Gunsalus RP, Maklashina E (2002) Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim Biophys Acta 1553: 140–157 [DOI] [PubMed] [Google Scholar]
- Eisenbach M (1996) Control of bacterial chemotaxis. Mol Microbiol 20: 903–910 [DOI] [PubMed] [Google Scholar]
- Eisenbach M (2004) Chemotaxis. London: Imperial College Press [Google Scholar]
- Eisenbach M, Adler J (1981) Bacterial cell envelopes with functional flagella. J Biol Chem 256: 8807–8814 [PubMed] [Google Scholar]
- Eisenbach M, Caplan SR (1998) Bacterial chemotaxis: unsolved mystery of the flagellar switch. Curr Biol 8: R444–R446 [DOI] [PubMed] [Google Scholar]
- Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM (1992) Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-Ring face of the basal body. Proc Natl Acad Sci USA 89: 6304–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NR, Sosinsky GE, Thomas D, DeRosier DJ (1994) Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Guest JR (1981) Partial replacement of succinate dehydrogenase function by phage- and plasmid-specified fumarate reductase in Escherichia coli. J Gen Microbiol 122: 171–179 [DOI] [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies: A Laboratory Manual, pp 298–299. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Hudson JM, Heffron K, Kotlyar V, Sher Y, Maklashina E, Cecchini G, Armstrong FA (2005) Electron transfer and catalytic control by the iron–sulfur clusters in a respiratory enzyme, E. coli fumarate reductase. J Am Chem Soc 127: 6977–6989 [DOI] [PubMed] [Google Scholar]
- Iverson TM, Luna-Chavez C, Schroder I, Cecchini G, Rees DC (2000) Analyzing your complexes: structure of the quinol-fumarate reductase respiratory complex. Curr Opin Struct Biol 10: 448–455 [DOI] [PubMed] [Google Scholar]
- Jones HM, Gunsalus RP (1985) Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate. J Bacteriol 164: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IH, Reese TS, Khan S (1992) The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA 89: 5956–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Blair DF (2004) The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol 233: 93–134 [DOI] [PubMed] [Google Scholar]
- Lancaster CR (2002) Succinate:quinone oxidoreductases: an overview. Biochim Biophys Acta 1553: 1–6 [DOI] [PubMed] [Google Scholar]
- Landini P, Zehnder AJB (2002) The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol 184: 1522–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SH, Reader RW, Kort EN, Tso W-W, Adler J (1974) Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature 249: 74–77 [DOI] [PubMed] [Google Scholar]
- Léger C, Heffron K, Pershad HR, Maklashina E, Luna-Chavez C, Cecchini G, Ackrell BA, Armstrong FA (2001) Enzyme electrokinetics: energetics of succinate oxidation by fumarate reductase and succinate dehydrogenase. Biochemistry 40: 11234–11245 [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Tang H, Wang X, Billings S, Blair DF (1996) Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol 178: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Chavez C, Iverson TM, Rees DC, Cecchini G (2000) Overexpression, purification, and crystallization of the membrane-bound fumarate reductase from Escherichia coli. Protein Expr Purif 19: 188–196 [DOI] [PubMed] [Google Scholar]
- Macnab RM (1995) Flagellar switch. In Two-Component Signal Transduction, Hoch JA, Silhavy TJ (eds), pp 181–199. Washington, DC: American Society for Microbiology [Google Scholar]
- Maklashina E, Berthold DA, Cecchini G (1998) Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J Bacteriol 180: 5989–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklashina E, Hellwig P, Rothery RA, Kotlyar V, Sher Y, Weiner JH, Cecchini G (2006) Differences in protonation of ubiquinone and menaquinone in fumarate reductase from Escherichia coli. J Biol Chem 281: 26655–26664 [DOI] [PubMed] [Google Scholar]
- Marwan W, Schafer W, Oesterhelt D (1990) Signal transduction in Halobacterium depends on fumarate. EMBO J 9: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrone M (1996) Untersuchungen zur Funktion von Fumarat als Schaltfaktor in der photophobischen Reaktion von Halobacterium salinarium und der chemophobischen Reaktion bei Escherichia coli. PhD thesis, University of Munich, Munich
- Montrone M, Eisenbach M, Oesterhelt D, Marwan W (1998) Regulation of switching frequency and bias of the bacterial flagellar motor by CheY and fumarate. J Bacteriol 180: 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrone M, Oesterhelt D, Marwan W (1996) Phosphorylation-independent bacterial chemoresponses correlate with changes in the cytoplasmic level of fumarate. J Bacteriol 178: 6882–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa K, Ueno T, Aizawa S (1994) Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol 176: 3683–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS (1978) Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol 135: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K (2001) Function of fumarate in bacterial chemotaxis. PhD thesis, Department of Biological Chemistry, The Weizmann Institute of Science, Rehovot, Israel
- Prasad K, Caplan SR, Eisenbach M (1998) Fumarate modulates bacterial flagellar rotation by lowering the free energy difference between the clockwise and counterclockwise states of the motor. J Mol Biol 280: 821–828 [DOI] [PubMed] [Google Scholar]
- Prüss BM, Campbell JW, VanDyk TK, Zhu C, Kogan Y, Matsumura P (2003) FlhD/FlhC is a regulator of anaerobic respiration and the Entner–Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J Bacteriol 185: 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid S, Eisenbach M (1984) Direction of flagellar rotation in bacterial cell envelopes. J Bacteriol 158: 222–230 [Erratum: 159 (1984) 1433] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid S, Matsumura P, Eisenbach M (1986) Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc Natl Acad Sci USA 83: 7157–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi Y, Khan S, Eisenbach M (2003) Binding of the chemotaxis response regulator CheY to the isolated, intact switch complex of the bacterial flagellar motor: lack of cooperativity. J Biol Chem 278: 25867–25871 [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW (1984) Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Silverman M, Simon M (1974) Flagellar rotation and the mechanism of bacterial motility. Nature 249: 73–74 [DOI] [PubMed] [Google Scholar]
- Stanley PE, Williams SG (1969) Use of liquid scintillation spectrometer for determining adenosine triphosphate by luciferase enzyme. Anal Biochem 29: 381–392 [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185: 60–89 [DOI] [PubMed] [Google Scholar]
- Thomas D, Morgan DG, DeRosier DJ (2001) Structures of bacterial flagellar motors from two FliF–FliG gene fusion mutants. J Bacteriol 183: 6404–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornroth S, Yankovskaya V, Cecchini G, Iwata S (2002) Purification, crystallisation and preliminary crystallographic studies of succinate:ubiquinone oxidoreductase from Escherichia coli. Biochim Biophys Acta 1553: 171–176 [DOI] [PubMed] [Google Scholar]
- Welch M, Margolin Y, Caplan SR, Eisenbach M (1995) Rotational asymmetry of Escherichia coli flagellar motor in the presence of arsenate. Biochim Biophys Acta 1268: 81–87 [DOI] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S-I, Eisenbach M (1993) Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA 90: 8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenberg DJ, Gunsalus RP, Ackrell BA, Sices H, Cecchini G (1993) Escherichia coli fumarate reductase frdC and frdD mutants. Identification of amino acid residues involved in catalytic activity with quinones. J Biol Chem 268: 815–822 [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299: 700–704 [DOI] [PubMed] [Google Scholar]
- Zamenhof PJ, Villarejo M (1972) Construction and properties of Escherichia coli strains exhibiting complementation of galactosidase fragments in vivo. J Bacteriol 110: 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information