Abstract
Specific interactions of the classical swine fever virus internal ribosomal entry site (IRES) with 40S ribosomal subunits and eukaryotic translation initiation factor (eIF)3 enable 43S preinitiation complexes containing eIF3 and eIF2–GTP–Met-tRNAMeti to bind directly to the initiation codon, yielding 48S initiation complexes. We report that eIF5B or eIF5B/eIF3 also promote Met-tRNAMeti binding to IRES–40S complexes, forming 48S complexes that can assemble elongation-competent ribosomes. Although 48S complexes assembled both by eIF2/eIF3- and eIF5B/eIF3-mediated Met-tRNAMeti recruitment were destabilized by eIF1, dissociation of 48S complexes formed with eIF2 could be out-competed by efficient subunit joining. Deletion of IRES domain II, which is responsible for conformational changes induced in 40S subunits by IRES binding, eliminated the sensitivity of 48S complexes assembled by eIF2/eIF3- and eIF5B/eIF3-mediated mechanisms to eIF1-induced destabilization. However, 48S complexes formed by the eIF5B/eIF3-mediated mechanism on the truncated IRES could not undergo efficient subunit joining, as reported previously for analogous complexes assembled with eIF2, indicating that domain II is essential for general conformational changes in 48S complexes, irrespective of how they were assembled, that are required for eIF5-induced hydrolysis of eIF2-bound GTP and/or subunit joining.
Keywords: classical swine fever virus, eIF1, eIF2, eIF5B, IRES
Introduction
The canonical 5′ end-dependent mechanism of translation initiation requires >10 eukaryotic translation initiation factors (eIFs) and occurs in two stages—formation of the 48S initiation complex and subsequent joining of a 60S subunit (Pestova et al, 2007). The first step is recruitment of initiator tRNA (Met-tRNAMeti) to a 40S subunit by eIF2–GTP, binding of which is stabilized by eIF1, eIF1A and eIF3, resulting in formation of a 43S preinitiation complex. eIF4F, eIF4A and eIF4B cooperatively unwind the cap-proximal region of mRNA to allow attachment of the 43S complex. After attachment, 43S complexes scan downstream to the initiation codon, where they stop and form 48S initiation complexes with established codon–anticodon base pairing in the peptidyl (P)-site of the 40S subunit: eIF1 has a key role in discriminating against initiation at near-cognate or poor-context triplets. Codon–anticodon recognition triggers eIF5-mediated hydrolysis of eIF2-bound GTP and release of inorganic phosphate (Pi) that commit the scanning ribosome to an initiation codon. Finally, eIF5B mediates displacement of factors from the 40S subunit and its joining with a 60S subunit to form an elongation-competent 80S ribosome. eIF2–GDP is recycled to eIF2–GTP by eIF2B. Elongation, during which the ribosome catalyses peptide synthesis, involves cycles of delivery of aminoacyl-tRNA by elongation factor eEF1A to the ribosomal aminoacyl (A)-site, peptide bond formation and translocation of peptidyl-tRNA from A- to P-sites by eEF2.
A few classes of viral mRNAs are translated in a cap-independent mode as a result of internal ribosomal entry that requires fewer eIFs than canonical initiation. Hepatitis C virus (HCV, a hepacivirus) and classical swine fever virus (CSFV, a pestivirus) exemplify one class of internal ribosomal entry sites (IRES). Initiation on the approximately 300-nt-long hepacivirus and pestivirus (HP) IRESs is determined by their ability to bind independently to 40S subunits and eIF3 (Pestova et al, 1998b, 2005). These interactions enable 43S complexes to attach directly to the initiation codon without scanning. Initiation on these IRESs therefore does not require eIFs 4F/4A/4B/1/1A. As in canonical initiation, eIF5 and eIF5B mediate subsequent subunit joining (Locker et al, 2007). HP IRESs consist of two principal domains, II and III (Figure 4A). Domain II is a long hairpin with internal loops, and domain III comprises a basal pseudoknot, several branched hairpins and an apical four-way helical junction. eIF3 binds to the apical region of domain III, whereas 40S subunits bind to the IRES at four sites: domain II binds ribosomal protein (rp) S5 near the E-site (see below), the pseudoknot binds near the mRNA exit channel and the platform, domain IIId likely binds to rRNA expansion segment ES7 and the four-way junction binds to the body near ES6 (Sizova et al, 1998; Kolupaeva et al, 2000a, 2000b; Lukavsky et al, 2000; Kieft et al, 2001; Spahn et al, 2001; Boehringer et al, 2005). These interactions result in the IRES-proximal coding sequence entering the mRNA-binding channel of the 40S subunit so that the initiation codon is placed in the P-site where it base pairs with Met-tRNAMeti (Pestova et al, 1998b; Ji et al, 2004; Otto and Puglisi, 2004). Domain II contains important conserved sequence motifs, including the apical hairpin, and has an overall bent conformation introduced by an asymmetric internal loop (Odreman-Macchioli et al, 2001; Lukavsky et al, 2003; Locker et al, 2007). Deletion of domain II from HP IRESs leads to reduced translational activity, unstable binding of IRES mRNA in the ribosomal mRNA-binding channel and impaired eIF5-mediated hydrolysis of eIF2-bound GTP and subunit joining (Kolupaeva et al, 2000a, 2000b; Fletcher and Jackson, 2002; Ji et al, 2004; Otto and Puglisi, 2004; Locker et al, 2007). Although domain II does not determine the affinity of binding of the HCV IRES to the 40S subunit (Kieft et al, 2001), it does interact with the head and platform near the E-site, inducing rotation of the head and conformational changes in the mRNA-binding channel (Spahn et al, 2001). HP-like IRESs have recently been identified in several picornaviruses, including simian picornavirus type 9 (SPV9), which have an HP-like domain III that binds independently to eIF3 and the 40S subunit, but mostly have an unconventional domain II (Pisarev et al, 2005; Hellen and de Breyne, 2007; de Breyne et al, 2008).
Figure 4.
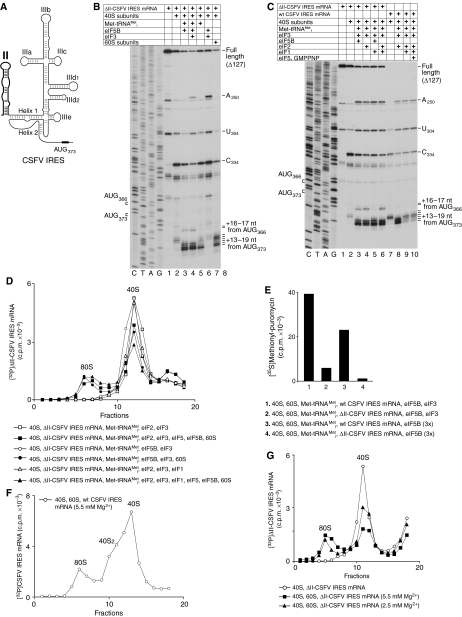
Influence of domain II on eIF2/eIF3- and eIF5B/eIF3-dependent initiation on the CSFV IRES. (A) Schematic representation of the CSFV IRES secondary structure showing domain II in bold. (B, C) Toeprinting analysis of 40S/IRES and 80S/IRES binary complexes, and 48S initiation complexes assembled on wt and ΔII-CSFV IRESs with of Met-tRNAMeti and eIFs as indicated. The positions of ribosomal complexes are shown relative to the AUG codons located in the P-site. Toeprints at A250 and U304 are characteristic of bound eIF3, and the toeprint at C334 reflects binding of a 40S subunit (Pestova et al, 1998b). Lanes C, T, A and G show the cDNA sequence corresponding to ΔII-CSFV IRES mRNA. The positions of the initiation codons AUG373 and AUG366 are indicated on the left. (D, F, G) Ribosomal association of 32P-labelled ΔII-CSFV-IRES mRNA (D, G) and 32P-labelled wt-CSFV IRES mRNA (F) after incubation with translation components, as indicated, assayed after sucrose gradient centrifugation by Cerenkov counting. Upper fractions have been omitted for clarity. Sedimentation was from right to left. The positions of 40S subunits and 80S ribosomes are indicated. (E) Methionyl-puromycin synthesis by 80S ribosomes assembled on wt and ΔII-CSFV IRESs with 35S-labelled Met-tRNAMeti and eIFs as indicated. Data obtained after subtraction of background (reaction mixtures containing 80S ribosomes and 35S-labelled Met-tRNAMeti) are presented as the mean of three independent experiments.
Mammalian cells respond to viral infection by phosphorylation of eIF2α by protein kinases PKR (double-stranded RNA-dependent protein kinase) and PERK (PKR-like ER kinase) (Dever et al, 2007). Phosphorylation converts eIF2–GDP, from a substrate of eIF2B, to its inhibitor, thereby lowering the level of eIF2–GTP–Met-tRNAMeti (TC) in cells and reducing translation globally. However, recent reports indicate that HCV IRES-mediated translation is refractory to reduced TC availability and less sensitive to PKR activation and phosphorylation of eIF2α than cap-dependent initiation (Rivas-Estilla et al, 2002; Vyas et al, 2003; Robert et al, 2006). The mechanism by which HCV IRES-mediated initiation escapes the consequence of eIF2α phosphorylation is not known.
We have addressed this question using the CSFV IRES, and report here that IRES/40S complexes can bind to Met-tRNAMeti in the presence of eIF5B and eIF3, at a level comparable to that of conventional eIF2-mediated binding, yielding 48S initiation complexes that can undergo subunit joining to form elongation-competent 80S ribosomes. However, such initiation complexes assembled in the absence of eIF2 were completely dissociated by eIF1. The IRES lacking domain II was also able to form 48S complexes by the eIF5B/eIF3-mediated mechanism that, in contrast to similar complexes assembled on the wt IRES, were resistant to eIF1-induced disassembly. However, these initiation complexes, like 48S complexes assembled by the eIF2-dependent mechanism on similarly truncated CSFV/HCV IRESs (Locker et al, 2007) were unable to undergo efficient subunit joining, which suggests that initiation complexes assembled by eIF2- or eIF5B/eIF3-mediated mechanisms on the CSFV IRES lacking domain II have an impaired ability to undergo some general conformational changes required for both efficient hydrolysis of eIF2-bound GTP and subsequent subunit joining. Elongation-competent 80S ribosomes could also assemble on the wt CSFV IRES at the physiological Mg2+ concentration in the absence of initiation factors, but the efficiency of such factor-independent initiation was only approximately 10% of initiation in the presence of eIF2. The possible contributions of eIF5B/eIF3-mediated and factor-independent initiation mechanisms to the resistance of CSFV IRES-mediated initiation to a reduction in TC levels are discussed.
Results
Resistance of translation mediated by CSFV and HCV IRESs to inhibition by eIF2α phosphorylation
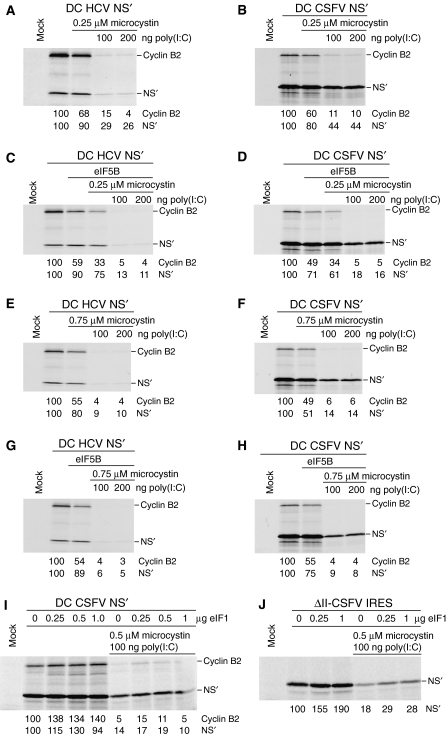
The sensitivity of end-mediated initiation and initiation on HCV and CSFV IRESs to inhibition by eIF2α phosphorylation was compared by translating dicistronic mRNAs in rabbit reticulocyte lysate (RRL) that had been pretreated with poly(I:C) and microcystin (a protein phosphatase inhibitor) to activate PKR and phosphorylate eIF2α (Price et al, 1991). Under these conditions, eIF2α phosphorylation was nearly complete (data not shown). Dicistronic mRNAs contained IRESs and approximately 50 nt of homologous coding sequence inserted between the cyclin B2 cistron and a downstream cistron encoding a truncated influenza NS protein (NS′) (Reynolds et al, 1995; Pestova et al, 1998b). Repression of end- and IRES-mediated translation by poly(I:C) was greater when microcystin was also present, but HCV and CSFV IRESs retained two- to fourfold more activity than end-mediated translation in all conditions (Figure 1A and B; data not shown), consistent with reports that the HCV IRES is partially resistant to reductions in eIF2–GTP–Met-tRNAMeti levels in vivo and in cell-free translation reactions (Rivas-Estilla et al, 2002; Robert et al, 2006). The CSFV IRES consistently remained slightly more active than the HCV IRES when TC levels were reduced and was therefore chosen as a model to characterize the basis for the resistance of HP IRESs to inhibition by eIF2α phosphorylation.
Figure 1.
Resistance of translation mediated by CSFV and HCV IRESs to inhibition by PKR-mediated phosphorylation of eIF2. Translation of (A, C, E, G) DC HCV mRNA, (B, D, F, H, I) DC CSFV mRNA and (J) ΔII-CSFV IRES mRNA in RRL preincubated with (A, B, E, F) microcystin and poly (I:C), (C, D, G, H) microcystin, poly(I:C) and eIF5B and (I, J) microcystin, poly(I:C) and eIF1. Translation products were quantified relative to translation in the absence of poly(I:C), microcystin, and exogenous eIF1 and eIF5B, which was defined as 100%.
eIF2-independent initiation on the CSFV IRES
Two prior observations suggested mechanisms that might enable the CSFV IRES to retain partial activity when TC levels are low. Thus, it has been reported that 80S ribosomes assembled on the HCV IRES without initiation factors can undergo elongation factor-mediated translation at an elevated (5 mM) Mg2+ concentration (Lancaster et al, 2006). Second, studies with the SPV9 IRES revealed that it promotes eIF2-independent binding of Met-tRNAMeti to 40S subunits that was stimulated by eIF3 (de Breyne et al, 2008). These findings prompted us to assay elongation by 80S ribosomes assembled on the CSFV IRES independently of initiation factors, and to investigate whether combinations of eIFs other than eIF2 promote binding of Met-tRNAMeti to CSFV IRES/40S complexes.
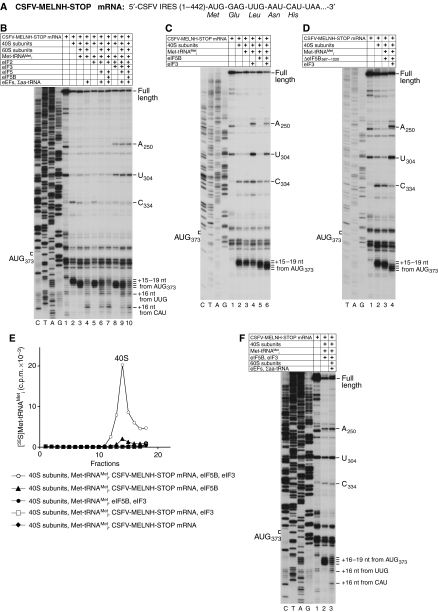
The possibility that 80S ribosomes assembled on the CSFV IRES independently of initiation factors might be elongation-competent, as on the HCV IRES, was first assayed by toeprinting analysis using CSFV-MELNH-STOP mRNA in which the sixth codon downstream of the IRES had been replaced by a UAA termination codon (Figure 2A). This mRNA retained the downstream sequence required for optimal IRES function (Fletcher et al, 2002). Consistent with previous reports (Pestova et al, 1998b), binary 40S/CSFV-MELNH-STOP mRNA complexes yielded toeprints +15–+17 nt downstream of the initiation codon AUG373, with the strongest stop at +16 nt (Figure 2B, lane 2). Addition of 60S subunits and Met-tRNAMeti together, but not individually to reaction mixtures containing 40S subunits and this mRNA, caused the ribosome toeprint to shift to positions +16–+19 nt from AUG373, with the most prominent bands at +17–+18 nt (Figure 2B, lane 3; data not shown). The appearance of toeprints at these positions has previously been reported to occur as a consequence of eIF2-mediated binding of Met-tRNAMeti to AUG373 in the P-site of CSFV IRES/40S subunit complexes in the presence and in the absence of eIF3 (Pestova et al, 1998b), which can be seen in Figure 2B, lanes 5, 8. This similarity suggested that cognate (initiator) tRNA had bound to AUG373 in the P-site of 80S ribosomes assembled on this IRES and that it had induced a conformational change in IRES/80S ribosome complexes similar to that induced in 40S/IRES complexes. Inclusion of eEF1H, eEF2 and total cytoplasmic tRNA aminoacylated with Glu, Leu, Asn and His (Σaa-tRNA) in reaction mixtures strongly reduced the +16–+19 nt toeprints and caused a prominent new toeprint to appear 16 nt downstream of the CAU triplet that occupies the P-site of elongating ribosomes arrested at the stop codon (Figure 2B, lane 4). Met-tRNAMeti-containing 80S ribosomes assembled on CSFV-MELNH-STOP mRNA without eIFs were therefore elongation-competent. The appearance of a low-intensity toeprint 16 nt downstream of the third codon (UUG: leucine) in all elongation reactions (Figures 2B, F and 3D) indicated that elongating 80S ribosomes failed to quantitatively reach the stop codon, likely due to the relatively low abundance in Σaa-tRNA preparations of Asn-tRNAAsn (which is required to decode the fourth codon), as noted previously for other aa-tRNAs (Pestova and Hellen, 2003; Alkalaeva et al, 2006). This deficiency could not be overcome by adding more Σaa-tRNA to reaction mixtures, because total tRNA preparations contain contaminating RNAs (Pestova and Hellen, 2001) that interfere nonspecifically with in vitro translation reconstitution (Pestova and Kolupaeva, 2002; Pisarev et al, 2007a). Although translational components were initially incubated at a physiological (2.5 mM) Mg2+ concentration in all experiments described above, the Mg2+ concentration was increased by 8 mM during reverse transcription. Thus, the observed elongation by 80S ribosomes assembled on CSFV-MELNH-STOP mRNA could potentially have occurred not at 2.5 mM Mg2+ during initial incubation, but instead at 10.5 mM Mg2+ during subsequent toeprinting. The ability of the CSFV-MELNH-STOP mRNA to assemble elongation-competent 80S ribosomes without eIFs at 2.5 mM Mg2+ was therefore verified in experiments that included sucrose density gradient centrifugation (see below).
Figure 2.
eIF2-dependent and eIF2-independent formation of elongation-competent 80S ribosomes on CSFV IRES mRNA. (A) Schematic representation of CSFV-MELNH-STOP mRNA. (B–D, F) Toeprinting analysis of binary 40S/IRES, 48S initiation, 80S initiation and 80S elongation complexes assembled on CSFV-MELNH-STOP mRNA with translation components as indicated. The positions of ribosomal complexes are shown relative to the mRNA codon in the ribosomal P-site. Toeprints at A250 and U304 are characteristic of bound eIF3, and the toeprint at C334 reflects binding of a 40S subunit (Pestova et al, 1998b). Lanes C, T, A and G show the cDNA sequence corresponding to CSFV-MELNH-STOP mRNA. The position of the initiation codon AUG373 is indicated on the left. (E) Binding of 35S-labelled Met-tRNAMeti to 40S subunits in the presence of CSFV-MELNH-STOP mRNA and eIFs as indicated. Ribosomal complexes were fractionated by sucrose density gradient centrifugation and analysed by scintillation counting of 35S-labelled Met-tRNAMeti. Sedimentation was from right to left. Upper fractions have been omitted for clarity. The position of 40S subunits is indicated.
Consistent with a previous report (Locker et al, 2007), efficient conversion of conventional 48S complexes assembled with eIF2 on CSFV-MELNH-STOP mRNA into elongation-competent 80S ribosomes required eIF5 and eIF5B (Figure 2B, lanes 7, 10). Elongation in the presence of eIF5 alone was very inefficient even for 48S complexes assembled without eIF3 (Figure 2B, lane 6), consistent with observations that eIF2 predominantly remains bound to 40S subunits after eIF5-induced hydrolysis of eIF2-bound GTP (Pisarev et al, 2006) and therefore continues to block subunit association.
Experiments to characterize eIF2-independent binding of Met-tRNAMeti to CSFV IRES/40S complexes revealed that, in contrast to the SPV9 IRES, the CSFV IRES did not promote factor-independent binding of Met-tRNAMeti to IRES/40S complexes alone or with only eIF3, but promoted binding of Met-tRNAMeti to IRES/40S complexes in the presence of eIF5B and, more efficiently, with eIF5B and eIF3 (Figure 2C and D). Thus, no shift in the toeprints corresponding to IRES/40S complexes was observed upon addition of Met-tRNAMeti alone or with eIF3 (Figure 2C, lanes 3, 4), but a partial shift occurred on inclusion of Met-tRNAMeti and native full-length eIF5B or recombinant N-terminally truncated ΔeIF5B587–1220 (Figure 2C, lane 5; Figure 2D, lane 3). A complete shift occurred in the presence of Met-tRNAMeti, eIF5B/ΔeIF5B587–1220 and eIF3 (Figure 2C, lane 6; Figure 2D, lane 4) that was identical to the shift reported for eIF2-mediated binding of Met-tRNAMeti to CSFV IRES/40S complexes (Pestova et al, 1998b). eIF5B-dependent binding of Met-tRNAMeti to IRES/40S complexes was confirmed in sucrose density gradient centrifugation experiments (Figure 2E). Ten-fold less Met-tRNAMeti bound to CSFV IRES/40S complexes in the presence of eIF5B alone (Figure 2E, filled triangles) than with eIF5B and eIF3 (Figure 2E, open circles). However, the difference in binding of Met-tRNAMeti in the presence and absence of eIF3 detected in this assay could have been exacerbated by the stringency of sucrose density gradient centrifugation. Ribosomal attachment of Met-tRNAMeti did not occur without the IRES (Figure 2E, filled circles) or eIF5B (Figure 2E, open squares and filled diamonds). 40S ribosomal complexes assembled on CSFV-MELNH-STOP mRNA with Met-tRNAMeti, eIF5B and eIF3 joined with 60S subunits to form elongation-competent 80S ribosomes (Figure 2F).
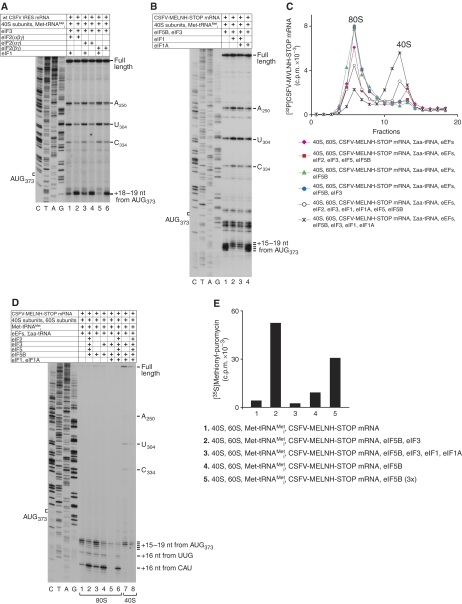
eIF1 substantially destabilizes 48S complexes assembled on CSFV and SPV9 IRESs in the presence of eIF2, and abolishes eIF2-independent binding of Met-tRNAMeti to SPV9 IRES/40S complexes (Pestova et al, 1998a; de Breyne et al, 2008). In the absence of eIF1, α-subunit-deficient eIF2 (Anthony et al, 1990), β-subunit-deficient eIF2 (Pisarev et al, 2006) and complete eIF2 showed very similar activity in 48S complex formation on the CSFV IRES (Figure 3A, lanes 2, 4, 6). eIF1 strongly reduced 48S complex formation on this IRES even in the presence of complete eIF2, as reported previously, and had an even greater effect on 48S complexes assembled with eIF2α-deficient eIF2 (Figure 3A, lanes 1, 3, 5). The stabilizing effect of eIF2α on 48S complexes assembled on this IRES was similar to that reported for 48S complexes assembled during canonical 5′-end dependent initiation (Pisarev et al, 2006). Initiation complexes assembled on the CSFV IRES with eIF5B and eIF3 were more prone to eIF1-induced destabilization than complexes assembled with eIF2. Thus, in contrast to initiation complexes assembled with eIF2/eIF3, toeprints at positions +18–19 nt downstream of the AUG triplet (which are characteristic of initiation complexes containing P-site Met-tRNAMeti) were not observed even at a low level in reaction mixtures containing eIF5B/eIF3 in the presence of either eIF1 alone or together with eIF1A (Figure 3B). This indicates that, similar to SPV9 IRES/40S/Met-tRNAMeti complexes (de Breyne et al, 2008), initiation complexes formed on the CSFV IRES with eIF5B/eIF3 were completely dissociated by eIF1. Although in all these experiments eIF1 was added to reaction mixtures simultaneously with other translational components, delayed addition of eIF1 to initiation complexes preformed on the CSFV IRES with eIF2/eIF3 or eIF5B/eIF3 in its absence resulted in the same level of dissociation (data not shown). Therefore, as in the case of initiation complexes formed on near-cognate initiation codons and initiation codons in bad nucleotide context (Pestova and Kolupaeva, 2002), eIF1 could actively dissociate initiation complexes preformed on the CSFV IRES.
Figure 3.
Influence of eIF1 on eIF2-dependent and eIF2-independent attachment of Met-tRNAMeti to 40S/CSFV IRES complexes. (A, B) eIF1-mediated dissociation of 48S complexes assembled on wt CSFV IRES (A) and CSFV-MELNH-STOP (B) mRNAs in the presence of eIF2/eIF3 (A) and eIF5B/eIF3 (B) assayed by toeprinting. The positions of ribosomal complexes are shown relative to the mRNA codon in the ribosomal P-site. Toeprints at A250 and U304 are characteristic of bound eIF3, and the toeprint at C334 reflects binding of a 40S subunit (Pestova et al, 1998b). Lanes C, T, A and G show cDNA sequences corresponding to wt CSFV IRES and CSFV-MELNH-STOP mRNAs, as appropriate. The position of the initiation codon AUG373 is indicated on the left. (C) Ribosomal association of 32P-labelled CSFV-MELNH-STOP mRNA after incubation with translation components, as indicated, assayed after sucrose gradient centrifugation by Cerenkov counting. Upper fractions have been omitted for clarity. Sedimentation was from right to left. The positions of 40S subunits and 80S ribosomes are indicated. (D) Toeprinting analysis of 80S- and 40S-containing fractions shown in (C). The positions of ribosomal complexes are shown relative to the mRNA codon in the ribosomal P-site. Lanes C, T, A and G show the cDNA sequence corresponding to CSFV-MELNH-STOP mRNA. (E) Methionyl-puromycin synthesis by 80S ribosomes assembled on CSFV-MELNH-STOP mRNA with 35S-labelled Met-tRNAMeti and different eIFs as indicated. Data obtained after subtraction of background (reaction mixtures containing 80S ribosomes and 35S-labelled Met-tRNAMeti) are presented as the mean of three independent experiments.
As discussed above, during toeprint analysis, the observed initiation/elongation events might have occurred not at 2.5 mM Mg2+ during initial incubation, but instead at the elevated Mg2+ concentration used during the subsequent primer extension step. To assay whether factor-independent assembly of elongation-competent 80S ribosomes and eIF5B/eIF3-dependent formation of 48S complexes on the CSFV IRES can occur at a physiological (2.5 mM) Mg2+ concentration, ribosomal elongation complexes were assembled on [32P]CSFV-MELNH-STOP mRNA with different combinations of eIFs at 2.5 mM Mg2+ and separated from reaction components (factors and aa-tRNAs) by centrifugation through sucrose density gradients prepared in buffer also containing 2.5 mM Mg2+; the positions of 40S subunits and 80S ribosomes on this mRNA in 40S- and 80S-containing fractions were subsequently determined by toeprinting. After incubation with 40S/eIF2/eIF3/eIF5/eIF5B/Σaa-tRNA/60S/eEFs or 40S/eIF3/eIF5B/Σaa-tRNA/60S/eEFs, almost all CSFV-MELNH-STOP mRNA was associated with 80S ribosomes (Figure 3C, red squares and blue circles), and toeprinting analysis of 80S ribosome-containing peak fractions yielded a predominant stop 16 nt downstream of the CAU triplet (Figure 3D, lanes 2 and 4), indicating that they mostly comprised elongating ribosomes arrested at the stop codon. Thus, in the absence of eIF1/eIF1A, the eIF2/eIF3- and eIF5B/eIF3-dependent modes of initiation were equally efficient in forming elongation-competent 80S ribosomes on the IRES. The amount of elongation-competent 80S ribosomes formed by initiation in the presence of eIF5B alone constituted approximately 25% of that formed with both eIF5B and eIF3 (Figure 3D, compare lanes 3 and 4), although the overall amount of 80S ribosomes assembled on the CSFV IRES in both cases was very similar (Figure 3C, green triangles and blue circles). Inclusion of eIF1/eIF1A into reaction mixtures containing eIF2/eIF3 reduced formation of elongation-competent 80S ribosomes to approximately 60% (Figure 3C, open circles; Figure 3D, lane 6). Considering that eIF1 dissociated approximately 90% of 48S initiation complexes assembled on the CSFV IRES (Figure 3A; Figure 4C, lane 9), such a relatively high yield of elongation-competent ribosomes (∼60%) formed in the presence of eIF1/1A must reflect successful competition of eIF5/5B-promoted subunit joining with eIF1-induced dissociation of 48S complexes, as in the absence of 60S subunits, eIF5 alone or with eIF5B did not protect 48S complexes from eIF1-induced dissociation even in the presence of GMPPNP (Figure 4C, lane 10; data not shown). Similar competition between eIF1-induced dissociation and ribosomal subunit joining was reported previously for 48S complexes formed on the AUG located 1 nt from the 5′ end of mRNA (Pestova and Kolupaeva, 2002). On the other hand, assembly of 80S ribosomes on the IRES was almost completely abolished by inclusion of eIF1/eIF1A in reaction mixtures with Met-tRNAMeti and eIF5B/eIF3 (Figure 3C, crosses; Figure 3D, lane 5). Consistently, the efficient methionylpuromycin (MP) synthesis observed in reaction mixtures containing 40S/eIF3/eIF5B/Met-tRNAMeti/60S and the CSFV IRES was almost completely inhibited by inclusion of eIF1/eIF1A (Figure 3E, lanes 2 and 3). MP synthesis in reaction mixtures containing eIF5B alone constituted approximately 15–20% of that observed in the presence of eIF5B/eIF3, but could be increased by raising eIF5B's concentration (Figure 3E, lanes 4, 5). In control reactions, MP synthesis was not stimulated by eIF3/eIF5B in the absence of the CSFV IRES as compared with the background (reaction mixtures containing 40S/Met-tRNAMeti/60S) (data not shown). After incubation with 40S/60S/Σaa-tRNA/eEFs, approximately 20% of CSFV-MELNH-STOP mRNA was associated with 40S subunits, and the rest was bound to 80S ribosomes (Figure 3C, magenta diamonds). However, toeprinting analysis of 80S ribosome-containing fractions indicated that only approximately 15% of 80S ribosomes underwent elongation leading to the appearance of the toeprint 16 nt downstream of the CAU triplet (Figure 3D, lane 1). Thus, in contrast to the HCV IRES (Lancaster et al, 2006), elongation-competent 80S ribosomes could assemble on the CSFV IRES without initiation factors at 2.5 mM Mg2+, but with only approximately 10% of the efficiency observed for eIF2- or eIF5B/eIF3-mediated initiation in the absence of eIF1/eIF1A.
Although eIF5B/eIF3-mediated initiation on the CSFV IRES in the absence of eIF1 was considerably more efficient in the in vitro-reconstituted system than in the factor-independent initiation, this difference could be minimized in the cellular environment containing eIF1, and factor-independent initiation could even contribute more to eIF2-independent translation on this IRES, unless cells possess an as-yet undiscovered mechanism that protects initiation complexes assembled on the IRES with eIF5B/eIF3 from eIF1-induced dissociation. Supplementation with exogenous eIF5B of RRL treated with poly(I:C) and microcystin did not increase the resistance of HCV or CSFV IRESs to inhibition by eIF2α phosphorylation, but even slightly inhibited translation (Figure 1, panels C and D versus panels A and B, and panels G and H versus panels E and F), indicating either that eIF5B is not limiting in RRL, or that in the presence of eIF1, the contribution of eIF5B/eIF3-mediated initiation to the overall level of initiation on the IRES in these conditions is low. Some RRL preparations contain reduced levels of eIF1 (our unpublished observations), but the relatively high resistance of HP IRESs to inhibition by eIF2α phosphorylation was not due to the RRL preparations used in the experiments described here containing so little eIF1 that eIF5B/eIF3-mediated initiation is allowed, because translation on the CSFV IRES in treated or untreated RRL was not inhibited by addition of eIF1 (Figure 1I).
Influence of domain II on eIF2-dependent and eIF2-independent initiation on the CSFV IRES
Domain II of the HCV IRES is responsible for the substantial conformational changes in 40S subunits induced by IRES binding (Spahn et al, 2001) and for efficient eIF5-induced hydrolysis of eIF2-bound GTP in 48S complexes assembled on HCV/CSFV IRESs (Locker et al, 2007). We therefore first investigated the requirement for domain II of the CSFV IRES for its ability to recruit Met-tRNAMeti to 40S subunits in the absence of eIF2, using a mutant (ΔII-CSFV IRES) lacking nt 1–127 of the 5′ UTR, including all of domain II. As reported (Kolupaeva et al, 2000b), the ΔII-CSFV IRES efficiently formed binary complexes with 40S subunits that were characterized by a strong toeprint at C334 in Helix 1 of the pseudoknot and toeprints +13–+15 nt from AUG373 that were much weaker than on the wt IRES (Figure 4B, lane 2; Figure 4C, lanes 2, 7), suggesting a defect in accommodation in the ribosomal mRNA-binding cleft of ΔII-CSFV IRES mRNA. As with the wt CSFV IRES, eIF5B and eIF3 together promoted very efficient binding of Met-tRNAMeti to 40S/ΔII-CSFV IRES complexes, causing new toeprints to appear +16–+19 nt from AUG373, most prominently at +18–+19 nt (Figure 4B, lane 4; Figure 4C, lane 3). The efficiency of attachment of Met-tRNAMeti to 40S/IRES complexes in the presence of eIF5B/eIF3 and of eIF2/eIF3 was similar for both ΔII-CSFV IRES and wt CSFV IRES mRNAs, at least at the concentration of translation components used in this study (Figure 4C, lanes 4, 8). Attachment of Met-tRNAMeti to 40S/ΔII-CSFV IRES complexes formed with eIF5B alone was only marginally lower than in the presence of eIF5B and eIF3 (Figure 4B, lanes 3, 4). In all cases, eIF5B-dependent binding of Met-tRNAMeti to 40S/ΔII-CSFV IRES complexes was confirmed by sucrose density gradient centrifugation (data not shown). Importantly, in contrast to the wt CSFV IRES mRNA, substantial binding of Met-tRNAMeti to 40S/ΔII-CSFV IRES complexes occurred in the absence of all factors (Figure 4B, lane 5 versus Figure 2C, lane 3). Moreover, eIF1 could not dissociate initiation complexes assembled on ΔII-CSFV IRES mRNA with either eIF5B/eIF3 or eIF2/eIF3 (Figure 4C, lanes 5, 6 versus lanes 3, 4). Thus, the conformational changes in 40S subunits that are induced by domain II were not required for eIF5B- or eIF5B/eIF3-dependent binding of Met-tRNAMeti to 40S/CSFV IRES complexes. Moreover, in the absence of such changes, 40S/CSFV IRES complexes were able to bind to Met-tRNAMeti in the absence of all initiation factors. However, ribosomal conformational changes induced by domain II were responsible for eIF1-mediated dissociation of initiation complexes assembled in the presence of either eIF2/eIF3 or eIF5B/eIF3. Interestingly, in the case of ΔII-CSFV IRES mRNA, a small quantity of initiation complexes formed on AUG366, 7 nt upstream of the initiation codon AUG373 (Figure 4B, lanes 3, 4; Figure 4C, lanes 3, 4, 6).
Deletion of domain II impairs eIF5-induced GTP hydrolysis and release of eIF2/GDP from 48S complexes assembled on the mutant CSFV IRES and consequently also inhibits ribosomal subunit joining (Locker et al, 2007). We therefore investigated whether 48S complexes assembled on ΔII-CSFV IRES mRNA with eIF5B/eIF3 could undergo more efficient subunit joining than complexes assembled with eIF2. As in the case of 48S complexes assembled with eIF2/eIF3 (Figure 4D, filled squares), only 20–25% of initiation complexes assembled with eIF5B/eIF3 underwent subunit joining (Figure 4D, filled circles). Consistently, the level of MP synthesis in the reaction mixtures containing 40S/eIF5B/eIF3/Met-tRNAMeti/60S and the ΔII-CSFV IRES was approximately sevenfold lower than with the wt IRES (Figure 4E, lanes 1, 2). To investigate whether inefficient subunit joining and, as a result, low levels of MP synthesis were caused by impaired dissociation of eIF3 from 48S complexes assembled on the ΔII-CSFV IRES in the presence of eIF5B/eIF3, MP synthesis was investigated for 48S complexes assembled with eIF5B alone on wt and ΔII-CSFV IRESs. MP synthesis with the wt IRES in reaction mixtures containing eIF5B without eIF3 constituted approximately 60% of that observed in the presence of eIF5B/eIF3, whereas almost no MP synthesis occurred in reaction mixtures containing the ΔII-CSFV IRES (Figure 4E, lanes 3, 4). This indicated that 48S complexes assembled on the ΔII-CSFV IRES in the presence of eIF5B have a defect other than impaired dissociation of eIF3. To investigate whether binding of ΔII-CSFV IRES RNA to 40S subunits directly impairs their ability to join 60S subunits, association of 60S subunits with IRES/40S complexes at different Mg2+ concentrations was compared for the wt and ΔII-CSFV IRESs. Association of 60S subunits with 40S/wt CFSV IRES complexes was low and even at an elevated (5.5 mM) Mg2+ concentration, joining was only approximately 10–15% complete (Figure 4F, data not shown), whereas binary 40S/ΔII-CSFV IRES complexes had a greater ability to associate with 60S subunits, and at 5.5 mM Mg2+, approximately 45% formed 80S ribosomes (Figure 4G). We also noted that irrespective of the Mg2+ concentration, the wt CSFV IRES tended to form complexes containing 40S subunit dimers. Interestingly, toeprinting analysis of 80S ribosomes, formed on the ΔII-CSFV IRES without factors, showed that the intensity of stops +13–+15 nt from AUG373 increased relative to binary 40S/ΔII-CSFV IRES complexes, with the most prominent band shifting from position +14 to +15, and that stops appeared at positions +18–+19, which are, in fact, a general characteristic of 48S complexes containing Met-tRNAMeti in the P-site (Figure 4B, lane 7). Thus, deletion of domain II increased, rather than impaired, the ability of binary 40S/CSFV IRES complexes to associate with 60S subunits. We therefore conclude that irrespective of the mode of assembly (through eIF2 or eIF5B), initiation complexes formed on the ΔII-CSFV IRES likely have an impaired ability to undergo some general conformational changes required for efficient eIF5-induced hydrolysis of eIF2-bound GTP as well as for subsequent subunit joining; these changes could occur upon codon recognition and might, at least in part, involve adjustment of the position of initiator tRNA in the P-site.
The resistance of translation mediated by the ΔII-CSFV and wt CSFV IRESs to inhibition by eIF2α phosphorylation were similar (Figure 1I, lane 6; Figure 1J, lane 5). However, in contrast to the wt IRES, addition of eIF1 to treated or untreated RRL increased translation of ΔII-CSFV IRES mRNA up to twofold (Figure 1I and J). The reason for this increase is not immediately obvious, but we note that eIF1 slightly enhanced joining of 60S subunits to 48S complexes assembled on the ΔII-CSFV IRES with eIF2/eIF3 (Figure 4D, filled triangles versus filled squares).
Discussion
Translation in RRL promoted by HCV and CSFV IRESs was more resistant to inhibition by phosphorylation of eIF2α than canonical end-dependent initiation, consistent with previous reports (Rivas-Estilla et al, 2002; Robert et al, 2006). We investigated mechanism(s) that might underlie the ability of HP IRESs to function when TC levels are reduced and found that 40S/wt-CSFV IRES complexes can bind to initiator tRNA in the presence of eIF5B alone, and that binding in the presence of eIF5B and eIF3 occurs at a level similar to that of conventional eIF2-mediated binding. eIF5B and its prokaryotic homologue, initiation factor IF2, which promotes binding of initiator tRNA to the prokaryotic 30S ribosomal subunit, have similar ribosomal binding sites (Allen et al, 2005; Unbehaun et al, 2007). Although in contrast to IF2, eIF5B does not normally promote attachment of initiator tRNA to the 40S subunit, data reported here indicate that in some circumstances, eIF5B can stabilize ribosomal binding of initiator tRNA. The fact that eIF3 can stimulate binding of initiator tRNA to the 40S subunit, mediated by eIF2 (in canonical initiation), by eIF5B (on the CSFV IRES) or even without other initiation factors (on the SPV9 IRES), indicates that eIF3 has a general role in this process, perhaps inducing common conformational changes in the 40S subunit that increases its affinity to initiator tRNA.
The unconventional 48S complexes formed with eIF5B/eIF3 were competent to form active 80S ribosomes, but they completely lacked resistance to eIF1-induced dissociation. Importantly, conventional 48S complexes assembled on the wt CSFV IRES with eIF2 are also very susceptible to eIF1-induced dissociation (Pestova et al, 1998a). However, in reaction mixtures allowing subunit joining, approximately 60% of initiation complexes assembled with eIF2/eIF3 formed 80S ribosomes in the presence of eIF1, whereas initiation complexes assembled with eIF5B/eIF3 did not undergo subunit joining in identical conditions. In the absence of 60S subunits, neither eIF5 nor eIF5B protected 48S complexes assembled with eIF2/eIF3 against eIF1-induced destabilization, and it is, therefore, likely that the conversion (∼60%) of initiation complexes into active 80S ribosomes was because ribosomal subunit joining out-competed eIF1-induced dissociation of 48S complexes. It is currently unclear whether translation of wt CSFV mRNA in vivo relies solely on subunit joining predominating over eIF1-induced dissociation of 48S complexes assembled with eIF2/eIF3, or if there is an as-yet unidentified cellular mechanism that protects 48S complexes from destabilization by eIF1. In this context, it may be relevant that 40S subunits bound to the HCV IRES in translation extracts are preferentially associated with RACK1 and nucleolin (Yu et al, 2005), and that the RNA-binding protein La enhances HCV translation (Costa-Mattioli et al, 2004). However, if there is no such mechanism to protect 48S complexes assembled with eIF5B/eIF3 from destabilization, it is difficult to imagine how eIF5B/eIF3-mediated initiation could contribute substantially to the resistance of HP IRESs to inhibition by reduction in TC levels.
Cryo-EM studies revealed that substantial conformational changes occur in the 40S subunit upon binding of the HCV IRES that are dependent on domain II (Spahn et al, 2001). Although domain III binds to the 40S subunit at several sites, these interactions do not substantially alter the 40S subunit's conformation, at least at the current level of resolution (∼20 Å), whereas interaction of domain II with the head and platform near the E-site induces rotation of the head relative to the body and conformational changes in the platform that induce interaction of h16 and rpS3, apparently closing the rpS14-rpS3 latch at the exit side of the mRNA-binding channel and opening the h18-h34 entry latch.
To investigate whether conformational changes induced by the IRES constitute the basis for the particular sensitivity of 48S complexes assembled on the wt CSFV IRES to eIF1-mediated destabilization, we assayed 48S complexes formed on a mutant IRES lacking domain II (‘ΔII-IRES'). Although accommodation of the coding region into the ribosomal mRNA-binding cleft was impaired in binary 40S/ΔII-IRES complexes, as reported by Kolupaeva et al (2000b), these complexes, similar to binary complexes formed on the wt IRES, efficiently recruited initiator tRNA in the presence of eIF2/eIF3, eIF5B/eIF3 or eIF5B alone (at least at the concentrations of translation components employed in the present study). However, in contrast to 48S complexes formed on the wt IRES, complexes formed on the ΔII-IRES were completely resistant to eIF1-induced destabilization. It has been reported that 48S complexes assembled with eIF2 on HCV and CSFV IRESs lacking domain II do not support efficient eIF5-mediated hydrolysis of eIF2-bound GTP and that ribosomal subunit joining is consequently impaired (Ji et al, 2004; Otto and Puglisi, 2004; Locker et al, 2007). We now found that 48S complexes assembled on the ΔII-IRES in an eIF5B- or eIF5B/eIF3-dependent manner were similarly unable to undergo efficient subunit joining, even though eIF2 was not involved in the initiation process. It is, therefore, likely that irrespective of the mode of assembly, initiation complexes formed on the ΔII-IRES have an impaired ability to undergo general conformational changes that are required for efficient hydrolysis of eIF2-bound GTP and subsequent subunit joining. Such conformational changes could result from establishment of codon–anticodon interaction in the P-site and might, at least in part, involve adjustment of the position of initiator tRNA in the P-site. It is likely that binding of CSFV/HCV IRESs lacking domain II to 40S subunits locks them in a relatively rigid conformation, so that they cannot respond properly to establishment of codon–anticodon base pairing. It is not known whether conformational changes in the 40S subunit caused by binding of the wt HCV IRES (Spahn et al, 2001) directly mimic or substitute for those that occur upon initiation codon recognition in canonical 48S initiation complexes assembled by the cap-dependent mechanism, or if the presence of domain II simply endows ribosomal complexes with a higher degree of flexibility permitting them to respond to binding of initiator tRNA and establishment of codon–anticodon base pairing, in which case the conformation of fully formed 48S complexes would likely differ from that of binary 40S/IRES complexes. Determination of the structure of a 48S complex assembled on an HP IRES would resolve this issue.
Interestingly, binary 40S subunit complexes assembled on the CSFV ΔII-IRES bound to initiator tRNA in a factor-independent manner, in contrast to analogous complexes assembled on the wt IRES. This implies that conformational changes caused by domain II destabilize P-site tRNA binding. These changes may be further amplified by the conformational changes that eIF1 likely induces in the 40S subunit (Lomakin et al, 2003; Passmore et al, 2007), thereby accounting for the enhanced sensitivity of 48S complexes assembled on the wt IRES to eIF1-induced destabilization. As in contrast to prokaryotic 30S subunits, eukaryotic 40S subunits do not bind to initiator tRNA directly, it is likely that association of 40S subunits with the ΔII-IRES induces some conformational changes, promoting recruitment of initiator tRNA. However, it cannot be excluded that binding of initiator tRNA is mostly stabilized by interaction with the AUG codon already correctly positioned in the P-site. Similar factor-independent binding of initiator tRNA was recently reported for binary 40S/wt SPV9 IRES complexes (de Breyne et al, 2008). It may be significant that the predicted structure of SPV9 IRES domain II is quite different from that of domain II of HCV and CSFV IRESs, and that it lacks the conserved apical loop which is characteristic of domain II of HP IRESs and is critical for their function in promoting hydrolysis of eIF2-bound GTP and subunit joining (Locker et al, 2007). The conformational changes in 40S subunits caused by binding of the SPV9 IRES might differ, at least in magnitude, from those induced by the HCV IRES. It is likely relevant that 48S complexes assembled on the SPV9 IRES were much more resistant to eIF1-induced destabilization than complexes assembled on the wt CSFV IRES (de Breyne et al, 2008; this report).
In conclusion, the presence of domain II allows initiation complexes assembled on the wt CSFV IRES to adopt a conformation that permits hydrolysis of eIF2-bound GTP and/or ribosomal subunit joining, but at the same time renders these complexes very sensitive to eIF1-induced dissociation, which can, at least in part, be overcome by the efficient ribosomal subunit joining process in addition to any hypothetical cellular mechanism that might more directly protect 48S complexes from destabilization.
In contrast to the HCV IRES (Lancaster et al, 2006), binary 40S/wt-CSFV IRES complexes did not associate efficiently with 60S subunits even at elevated (5.5 mM) Mg2+, conditions in which only approximately 15% of binary 40S/IRES complexes were able to form 80S ribosomes (Figure 4F). Deletion of domain II only modestly increased the direct association of binary 40S/IRES complexes with 60S subunits, indicating that even in the absence of conformational changes caused by domain II, binding of the IRES to 40S subunits impairs their ability to form 80S ribosomes. However, in the presence of tRNA, approximately 80% of binary 40S/wt-CSFV IRES complexes joined with 60S subunits even at 2.5 mM Mg2+ (Figure 3C). Although we could not detect direct binding of initiator tRNA to 40S/wt-CSFV IRES complexes by toeprinting, such an interaction might be too weak to detect in this way, but could nevertheless be sufficient to stimulate subunit joining. However, only a small proportion of 80S complexes assembled in the absence of initiation factors was elongation-competent, and the overall efficiency of productive factor-independent initiation on the CSFV IRES was only approximately 10% of that observed with eIF2 (Figure 3D, compare lanes 1, 2).
Taken together, our data indicate that although eIF5B/eIF3-mediated initiation on the CSFV IRES was considerably more efficient than factor-independent initiation in the in vitro-reconstituted system in the absence of eIF1, this difference is likely minimized in the cellular environment containing eIF1, unless cells possess a mechanism that protects initiation complexes formed with eIF5B/eIF3 from dissociation. In that case, both mechanisms could contribute to initiation on the CSFV IRES when the TC level in virus-infected cells is reduced by eIF2α phosphorylation. However, these considerations should not overshadow the likelihood that TCs may simply have a higher affinity for 40S/IRES/eIF3 complexes than for 40S subunits that are not associated with the IRES, which would allow HP IRESs to compete efficiently with mRNAs translated by cap-dependent initiation for limiting amounts of TCs.
Materials and methods
Plasmids and primers
Expression vectors for His6-tagged eIF1, eIF1A, eIF5, N-terminally truncated ΔeIF5B587–1220 (Pestova et al, 1998a, 2000) and Escherichia coli methionyl-tRNA synthetase (Lomakin et al, 2006) and transcription vectors pXL.HCV(40–373).NS′ (Reynolds et al, 1995), pCSFV(1-442).NS′, pXL.CSFV(1–442).NS′ (Pestova et al, 1998b) and pCSFV(128–442).NS′ (Kolupaeva et al, 2000b) have been described. The vector pCSFV(1–442)-MELNH-STOP, in which the sixth and seventh codons (UUU-GAA) were substituted by the nucleotides UAA-AAA, was generated by polymerase chain reaction mutagenesis of pCSFV(1-442)NS′ and insertion of the corresponding PCR fragment between EcoRI and BamHI sites of pUC19. pXL.HCV(40–373).NS′ (for DC HCV mRNA), pCSFV(1–442).NS′ (for wt CSFV IRES mRNA), pXL.CSFV(1–442).NS′ (for DC CSFV mRNA) and pCSFV(128–442).NS′ (for ΔII-CSFV IRES mRNA) were linearized with EcoRI, and pCSFV(1–442)-MELNH-STOP (for CSFV-MELNH-STOP mRNA) was linearized with HindIII. mRNAs were transcribed with T7 polymerase. For experiments that involved sucrose density gradient centrifugation, 32P-labelled CSFV-MELNH-STOP, wt CSFV IRES and ΔII-CSFV IRES mRNAs (3 × 105 c.p.m./μg) were transcribed in the presence of [α32P]CTP.
Purification of initiation factors, ribosomal subunits and aminoacylation of tRNA
Ribosomal 40S and 60S subunits, native eIF2, eIF3, β subunit-deficient eIF2αγ, eEF1H and eEF2, and rabbit aminoacyl-tRNA synthetases were purified from RRL (Green Hectares, Oregon, WI), and recombinant eIF1, eIF1A, eIF5, ΔeIF5B587–1220 and E. coli methionyl-tRNA synthetase were expressed in E. coli BL21(DE3) and purified as described previously (Pestova and Hellen, 2003; Lomakin et al, 2006; Kolupaeva et al, 2007; Pisarev et al, 2007b). Native total rabbit tRNA (Novagen) was aminoacylated with Glu, Leu, Asn and His as described previously (Pestova and Hellen, 2003). Native tRNAMeti was partially purified from total tRNA by gel-filtration on Superdex 75 and reverse phase chromatography on a Waters 3.9 × 300 mm Delta Pak C4 column (Unbehaun et al, 2004) and aminoacylated with E. coli methionyl-tRNA synthetase. For sucrose density gradient centrifugation and methionyl-puromycin synthesis experiments, tRNAMeti was aminoacylated using 35S-labelled Met (5 × 105 c.p.m./pmol).
Assembly of ribosomal complexes
Binary 40S/IRES and 48S initiation complexes were assembled by incubating 1 pmol CSFV-MHVL-STOP, wt CSFV IRES or ΔII-CSFV IRES mRNAs with 3 pmol 40S subunits and combinations of 8 pmol Met-tRNAMeti, 10 pmol eIF2, 10 pmol eIF2αγ, 10 pmol eIF2βγ, 10 pmol eIF3, 8 pmol native eIF5B, 8 pmol ΔeIF5B587–1220, 15 pmol eIF1 and 15 pmol eIF1A (as indicated in the text) for 10 min at 37°C in 40 μl reaction mixtures containing buffer A (20 mM Tris, pH 7.5, 100 mM KAc, 2.5 mM MgCl2, 2 mM dithiothreitol, 40 U RNasin, 0.25 mM spermidine and 0.4 mM GTP). To obtain 80S initiation complexes, assembled 48S initiation complexes were supplemented with 4 pmol 60S subunits and different combinations of 10 pmol eIF5 and 10 pmol eIF5B, and incubated for another 10 min at 37°C. For elongation, reaction mixtures containing 80S initiation complexes were supplemented with 15 μg Σaa-tRNA, 10 pmol eEF1H and 10 pmol eEF2 and incubated at 37°C for an additional 10 min. For experiments that involved sucrose density gradient centrifugation, ribosomal complexes were assembled in 120 μl reaction mixtures with either 32P-labelled CSFV-MHVL-STOP, wt CSFV IRES and ΔII-CSFV IRES mRNAs or with 35S-labelled Met-tRNAMeti, as indicated in the text.
Toeprinting analysis
Assembled 48S initiation and 80S elongation complexes were analysed by primer extension using AMV reverse transcriptase and a primer end-labelled with γ32P-labelled ATP and complementary to the NS′-coding sequence, essentially as described previously (Pestova et al, 1998b).
Sucrose density gradient centrifugation
Ribosomal complexes assembled with 32P-labelled CSFV-MHVL-STOP, wt CSFV IRES and ΔII-CSFV IRES mRNAs or with 35S-labelled Met-tRNAMeti were subjected to centrifugation through 10–30% sucrose density gradients prepared in buffer A (without GTP) in a Beckman SW55 rotor at 53 000 r.p.m. for 75 min. Ribosomal association of [32P]mRNA was measured by Cerenkov counting of an aliquot of each fraction. Fractions that corresponded to ribosomal complexes were analysed by toeprinting as described above. Ribosomal association of 35S-labelled Met-tRNAMeti was measured by scintillation counting of an aliquot of each fraction.
Methionyl-puromycin synthesis
Methionyl-puromycin synthesis was assayed essentially as described previously (Pestova et al, 2000). One pmol of CSFV-MHVL-STOP, wt CSFV IRES or ΔII-CSFV IRES mRNA, 3 pmol 40S subunits, 8 pmol 35S-labelled Met-tRNAMeti, 10 pmol eIF3, 8 pmol native eIF5B, 15 pmol eIF1, 15 pmol eIF1A and 4 pmol 60S subunits were incubated in different combinations (as indicated in the text) in 40 μl reaction mixtures containing buffer A for 10 min at 37°C. After assembly, ribosomal complexes were incubated with 1 mM puromycin and extracted with ethyl acetate.
In vitro translation assays
DC HCV, DC CSFV and ΔII-CSFV IRES mRNAs (20 μg/ml) were translated in RRL (Promega, Madison, WI) supplemented with 0.5 mCi/ml 35S-labelled Met (43.5 TBq/mmol) for 60 min at 30°C, following pre-incubation of RRL for 5 min at 30°C with poly(I:C) with or without microcystin, eIF5B or eIF1, as indicated in the text. Translation products were analysed by 15% SDS–polyacrylamide gel electrophoresis. Gels were quantified by Molecular Dynamics PhosphoImager analysis.
Acknowledgments
We are indebted to WC Merrick for the generous gift of α-less eIF2. This work was supported by NIH Grant GM59660 to TVP and NIH Grant AI-51340 to CUTH.
References
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV (2006) In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 [DOI] [PubMed] [Google Scholar]
- Anthony DD Jr, Kinzy TG, Merrick WC (1990) Affinity labeling of eukaryotic initiation factor 2 and elongation factor 1 alpha beta gamma with GTP analogs. Arch Biochem Biophys 281: 157–162 [DOI] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H (2005) Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13: 1695–1706 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Svitkin Y, Sonenberg N (2004) La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol 24: 6861–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Breyne S, Yu Y, Pestova TV, Hellen CUT (2008) Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA 14: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Dhar AC, Sicheri F (2007) The eIF2α Kinases. Translational Control in Biology and Medicine, pp 319–344. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Fletcher SP, Ali IK, Kaminski A, Digard P, Jackson RJ (2002) The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA 8: 1558–1571 [PMC free article] [PubMed] [Google Scholar]
- Fletcher SP, Jackson RJ (2002) Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J Virol 76: 5024–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT, de Breyne S (2007) A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol 81: 5850–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA (2004) Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosomal entry site RNA. Proc Natl Acad Sci USA 101: 16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA (2001) Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7: 194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, de Breyne S, Pestova TV, Hellen CUT (2007) In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol 430: 409–439 [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CUT (2000a) An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol 74: 6242–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CUT (2000b) Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA 6: 1791–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AM, Jan E, Sarnow P (2006) Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12: 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ (2007) HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV (2003) Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CUT, Pestova TV (2006) The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD (2003) Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 10: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD (2000) Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol 7: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Odreman-Macchioli F, Baralle FE, Buratti E (2001) Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J Biol Chem 276: 41648–41655 [DOI] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD (2004) The pathway of HCV IRES-mediated translation initiation. Cell 119: 369–380 [DOI] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT (1998a) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT (2001) Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA 7: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT (2003) Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev 17: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT (2007) The Mechanism of Translation Initiation in Eukaryotes. Translational Control in Biology and Medicine, pp 87–128. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT (1998b) A prokaryotic like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CUT, Pestova TV (2007a) Recycling of eukaryotic post-termination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CUT, Pestova TV (2006) Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20: 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Shirokikh NE, Hellen CUT (2005) Translation initiation by factor independent binding of eukaryotic ribosomes to internal ribosomal entry sites. CR Biol 328: 589–605 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CUT, Pestova TV (2007b) Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Price NT, Welsh GI, Proud CG (1991) Prosphorylation of only serine-51 in protein synthesis initiation factor-2 is associated with inhibition of peptide-chain initiation in reticulocyte lysates. Biochem Biophys Res Commun 176: 993–999 [DOI] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ (1995) Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J 14: 6010–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Estilla AM, Svitkin Y, Lopez Lastra M, Hatzoglou M, Sherker A, Koromilas AE (2002) PKR-dependent mechanisms of gene expression from a subgenomic hepatitis C virus clone. J Virol 76: 10637–10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR, Pelletier J (2006) Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell 17: 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CUT (1998) Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol 72: 4775–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J (2001) Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV (2004) Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Marintchev A, Lomakin IB, Didenko T, Wagner G, Hellen CUT, Pestova TV (2007) Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J 26: 3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas J, Elia A, Clemens MJ (2003) Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. RNA 9: 858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Ji H, Doudna JA, Leary JA (2005) Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci 14: 1438–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]