Abstract
Glutaredoxins (Grxs) are small oxidoreductases that reduce disulphide bonds or protein-glutathione mixed disulphides. More than 30 distinct grx genes are expressed in higher plants, but little is currently known concerning their functional diversity. This study presents biochemical and spectroscopic evidence for incorporation of a [2Fe–2S] cluster in two heterologously expressed chloroplastic Grxs, GrxS14 and GrxS16, and in vitro cysteine desulphurase-mediated assembly of an identical [2Fe–2S] cluster in apo-GrxS14. These Grxs possess the same monothiol CGFS active site as yeast Grx5 and both were able to complement a yeast grx5 mutant defective in Fe–S cluster assembly. In vitro kinetic studies monitored by CD spectroscopy indicate that [2Fe–2S] clusters on GrxS14 are rapidly and quantitatively transferred to apo chloroplast ferredoxin. These data demonstrate that chloroplast CGFS Grxs have the potential to function as scaffold proteins for the assembly of [2Fe–2S] clusters that can be transferred intact to physiologically relevant acceptor proteins. Alternatively, they may function in the storage and/or delivery of preformed Fe–S clusters or in the regulation of the chloroplastic Fe–S cluster assembly machinery.
Keywords: chloroplast, glutaredoxin, iron–sulphur protein, plant
Introduction
Iron–sulphur (Fe–S) proteins are intimately involved in numerous essential biological processes, such as photosynthesis, respiration and the metabolism of carbon, oxygen, hydrogen, nitrogen and sulphur (Johnson and Smith, 2005). However, little is known concerning the mechanism of Fe–S cluster biogenesis in plants. Much of the current understanding of Fe–S cluster biogenesis stems from investigation of components of the bacterial isc (iron–sulphur cluster assembly), suf (sulfur mobilization) and nif (nitrogen fixation) operons (Johnson et al, 2005) and identification and characterization of homologous ISC-type proteins in yeast and mammalian mitochondria (Lill and Mühlenhoff, 2005). The ISC, SUF and NIF Fe–S cluster assembly machineries share a common basic mechanism involving cysteine desulphurase (IscS, SufS and NifS)-mediated assembly of [2Fe–2S] or [4Fe–4S] clusters on U-type (IscU, SufU and N-terminal domain of NifU), A-type (IscA, SufA and NifIscA) and Nfu-type (corresponding to the C-terminal domain of NifU) scaffold proteins, and subsequent intact cluster transfer into acceptor apo-proteins. In the case of the ISC machinery, [2Fe–2S] cluster transfer from IscU is facilitated by specific molecular co-chaperones (HscA and HscB) in an ATP-dependent reaction (Chandramouli and Johnson, 2006) and [4Fe–4S] cluster assembly on dimeric IscU occurs at the subunit interface via reductive coupling of two [2Fe–2S] clusters (Chandramouli et al, 2007). However, little is currently known concerning the detailed mechanism of Fe–S cluster assembly and transfer involving scaffold proteins. In plants, Fe–S cluster biosynthesis primarily occurs in mitochondria, using the ISC machinery with Isu, IscA and Nfu as potential scaffold proteins, and in chloroplasts using the SUF machinery with SufA, SufB and Nfu proteins as potential scaffold proteins (Balk and Lobreaux, 2005; Ye et al, 2006b; Layer et al, 2007).
Glutaredoxins (Grxs) are small proteins that normally function in the reduction of disulphide bridges or glutathionylated proteins. However, recent studies in Saccharomyces cerevisiae and Escherichia coli have indicated specific roles for some Grxs in facilitating Fe–S cluster biosynthesis (Rodríguez-Manzaneque et al, 2002; Mühlenhoff et al, 2003; Achebach et al, 2004). Yeast cells deleted for the GRX5 gene were found to be more sensitive to oxidative stress, to accumulate free iron and to have impaired mitochondrial Fe–S cluster biogenesis and respiratory growth (Rodríguez-Manzaneque et al, 1999, 2002). Other prokaryotic and eukaryotic CGFS Grxs have been shown to be efficient functional substitutes for yeast Grx5 (Cheng et al, 2006; Molina-Navarro et al, 2006). Although the specific function of yeast Grx5 in Fe–S cluster biogenesis remains to be elucidated, 55Fe radiolabelling studies of knockout mutants implicate a role in mediating transfer of clusters preassembled on the Iscu1p scaffold protein into acceptor proteins (Mühlenhoff et al, 2003). The discovery that Grx5 is also required for vertebrate haem synthesis raises the possibility that Grx5 is a key determinant for channelling Fe into haem and Fe–S cluster biosynthesis in mammals (Wingert et al, 2005).
The most obvious role for Grxs in Fe–S cluster biogenesis lies in facilitating Fe–S cluster assembly or transfer by reducing disulphides on scaffold or apo forms of Fe–S proteins. However, the discovery that some Grxs can assemble Fe–S clusters suggests the possibility of alternative roles in Fe–S cluster assembly or transfer. Poplar GrxC1 (CGYC active site) and human Grx2 (CSYS active site) are homodimers with a subunit-bridging [2Fe–2S] cluster ligated by one active site cysteine of each monomer and the cysteines of two external glutathione (GSH) molecules (Feng et al, 2006; Johansson et al, 2007; Rouhier et al, 2007). The cluster-containing dimeric form of human Grx2 was proposed to function as a redox sensor for the activation of Grx2 in case of oxidative stress (Lillig et al, 2005). Although this is a viable hypothesis, mutagenesis studies on poplar GrxC1 indicate that incorporation of a [2Fe–2S] cluster is likely to be a general feature of plant Grxs possessing a glycine adjacent to the catalytic cysteine (Rouhier et al, 2007). Hence CGFS Grxs, such as yeast Grx5, might have the capacity to incorporate a Fe–S cluster. Moreover, the requirement of GSH for the export of a Fe–S cluster (or a precursor thereof) from mitochondria to facilitate the assembly of cytosolic Fe–S proteins in S. cerevisiae (Lill and Mühlenhoff, 2005) provides further circumstantial evidence in support of a role for GSH- and Grx-ligated [2Fe–2S] clusters in Fe–S biogenesis.
In higher plants, around 30 different Grx isoforms can be classified into three distinct subgroups depending on their active site sequences (Rouhier et al, 2004). The first class, which contains Grxs with C[P/G/S][Y/F][C/S] motifs other than CGFS, is homologous to the classical dithiol Grxs such as E. coli Grx1 and 3, yeast Grx1 and 2 and mammalian Grx1 and 2. The second class has a strictly conserved CGFS active site sequence and includes Grxs homologous to yeast Grx3, 4 and 5 or E. coli Grx4. Plants have generally four members in this group (GrxS14 to S17). The properties of proteins of the third class, which is specific to higher plants and involves a CC[M/L][C/S] active site, are largely unknown.
This study presents biochemical, spectroscopic and analytical evidence for the incorporation of [2Fe–2S] clusters in two plant chloroplast CGFS Grxs, GrxS14 and S16, and both in vivo and in vitro evidence for their involvement in the maturation of Fe–S proteins. The results demonstrate that monothiol Grxs have the potential to function as scaffold proteins for de novo synthesis and efficient delivery of [2Fe–2S] clusters, as Fe–S cluster delivery or storage proteins for mediating the transfer of Fe–S clusters from ISC or SUF scaffold proteins to acceptor proteins, or as sensors of the cellular Fe–S cluster status in Fe homoeostasis.
Results
The plant CGFS Grx subgroup
In silico analysis of Grxs from different kingdoms reveals that four or five Grxs with CGFS active site are generally present in higher plants and in Chlamydomonas reinhardtii, whereas only three are present in S. cerevisiae, two in most other fungi and in mammals, one in Synechocystis and in E. coli (Rouhier et al, 2004). In Populus trichocarpa, GrxS14 and S15 are small proteins (171 and 172 amino acids, respectively, including the transit peptide sequence) with a single repeat of the Grx module. GrxS16 is larger (296 amino acids including the transit peptide sequence) with an N-terminal extension linked to the Grx module. GrxS17 is larger (492 amino acids) and displays an N-terminal Trx-like domain with a WCDAS active site followed by three successive Grx modules. A careful examination of amino-acid sequence alignments (Supplementary Figure 1) indicates that although not present in all CGFS Grxs, a second cysteine is found in 60% of the 250 CGFS Grxs found in GenBank at a conserved position closer to the C terminus. It is conserved in GrxS14 and S16 (Cys 87 in GrxS14 and Cys 221 in GrxS16, numbering based on the recombinant mature protein sequences), in the third module of GrxS17 and in GrxC1, whereas it is absent in GrxS15, in the second Grx domain of GrxS17 and in GrxC4 and only partly conserved in the first Grx domain of plant GrxS17. In ScGrx5, these two cysteines are able to form a disulphide bridge in the presence of oxidized GSH (Tamarit et al, 2003).
Subcellular localization of the CGFS Grxs
We have determined the localization of all the CGFS Grxs that possess an N-terminal transit sequence. GrxS17, predicted to be cytosolic, does not seem to possess such an extension and its localization has not been characterized further. The full-length sequences of the three other Grxs devoid of the stop codon were introduced in frame before the GFP sequence and the construction was used to bombard tobacco leaves. As shown in Figure 1, the fluorescence of GrxS14 and S16 strictly coincides with the one of the chlorophyll, whereas the fluorescence of GrxS15 superimposed well with one of the mitochondrial marker. Therefore, GrxS14 and S16 are chloroplastic and GrxS15 is mitochondrial.
Figure 1.
Subcellular localization of CGFS Grxs by GFP fusion. (A) GrxS14, (B) GrxS16 and (C) GrxS15. From left to right: visible light, autofluorescence of chlorophyll (red) or mitochondrial marker (white); fluorescence of the constructions and merged images. Only one of the guard cells shows chloroplast-localized GFP, because a small numbers of cells were transfected. As the mitochondrial marker (DsRed) is co-transfected with the GFP construction, it is only visible in the cell that expresses GFP.
Some poplar monothiol but not dithiol Grxs rescue the defects of a yeast mutant lacking Grx5
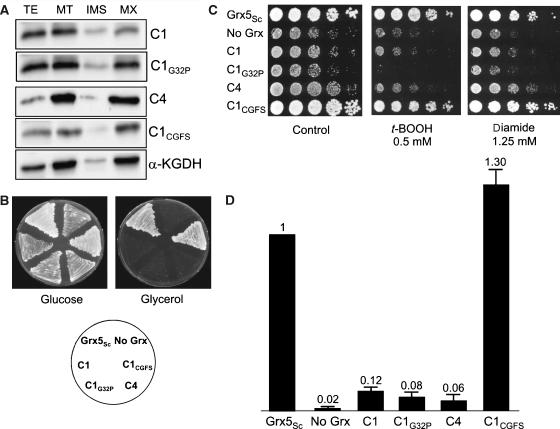
To determine whether the four poplar monothiol Grxs rescue the defects of a yeast Δgrx5 mutant, we targeted the proteins at the mitochondrial matrix (Molina et al, 2004). All the proteins were adequately compartmentalized in the mitochondrial matrix (Figure 2A). Of the two poplar Grxs with a single Grx domain, GrxS14 and S15, only the first one rescued the grx5 mutant defects in respiratory growth (Figure 2B) and its sensitivity to oxidants (Figure 2C). These defects were also efficiently rescued by GrxS16 and S17. To test whether a single Grx domain is sufficient for the function of GrxS17, we fused only the most C-terminal domain of S17 (from amino acid 398 to 492) to the mitochondrial targeting sequence of Grx5 (S17398−492). This protein was also compartmentalized at the mitochondrial matrix (Figure 2A), although a double band appeared. The band with lower mobility probably corresponds to unprocessed precursor still compartmentalizing at yeast mitochondria. S17398−492 suppressed partially the growth phenotypes of the grx5 mutant, in particular growth in respiratory conditions (Figure 2B). The ratio of activities of the mitochondrial enzymes aconitase (containing Fe–S clusters) and malate dehydrogenase (without Fe–S clusters) was used as a measure of the efficiency of the Fe–S cluster assembly in mitochondrial proteins (Molina et al, 2004). This ratio was measured in strains carrying all these constructions in a chromosomal grx5 background (Figure 2D). Ratios were comparable with wild type in strains expressing the mitochondrial forms of S14, S16 and S17, in accordance with the growth phenotypes. In contrast, both S15 and the truncated form of S17 exhibited much lower enzyme ratios (Figure 2D). For all the strains tested, absolute malate dehydrogenase levels were basically similar, and doxycycline addition to growth media lowered aconitase activity to basal levels (data not shown). For the monothiol Grxs analysed, there appears to be a correlation between efficiency to express active aconitase and growth phenotypes, except that poplar mitochondrial S15 did not rescue at all the growth defects of the yeast mutant, although still being able to synthesize low levels of mature aconitase. The anomalous results with S15 and S17398−492 are puzzling and may indicate functional diversity within the general class of monothiol Grxs (see below) or that aconitase maturation does not provide a good measure of the efficiency of general Fe–S cluster biosynthesis in mitochondria. The latter is supported by a very recent study of the requirements for mitochondrial aconitase Fe–S cluster maturation in S. cerevisiae, which indicated a specific requirement for Isa1p, Isa2p and the Iba57, proteins that are not required for general Fe–S cluster biogenesis in mitochondria (Gelling et al, 2008).
Figure 2.
Rescue of the S. cerevisiae grx5 mutant defects by poplar monothiol glutaredoxins. (A) Compartmentalization of GrxS14, S15, S16, S17 and S17398−492 in the mitochondrial matrix of S. cerevisiae cells. Cultures were grown exponentially in YPLactate medium at 30°C to about 3 × 107 cells ml−1, before mitochondrial isolation and subfractionation. TE, total cell extract; MT, mitochondrial fraction; IMS, intermembrane space; MX, matrix. Proteins (20 μg) were loaded in the TE lanes, and 5 μg was loaded in the other lanes. Anti-HA anti-lipoic acid antibodies were used in the western blot to detect the HA-tagged proteins, and the matrix marker α-ketoglutarate dehydrogenase (α-KGDH). (B) Growth on glucose (YPD plates) or glycerol (YPGly plates), after 3 days at 30°C. (C) Sensitivity to t-BOOH or diamide of the strains after 3 days at 30°C on YPD plates. (D) Ratio between aconitase and malate dehydrogenase activities in exponential cultures at 30°C in YPGalactose medium.
To determine whether the capacity to bind a Fe–S cluster is sufficient for grx5 complementation, we then used poplar GrxC1 (CGYC), which incorporates a [2Fe–2S] centre and GrxC1 G32P (CPYC) and GrxC4 (CPYC) which do not (Rouhier et al, 2007). Although all these Grxs were targeted to the matrix (Figure 3A), none of these proteins rescued (i) the inability of a grx5 mutant for respiratory growth, (ii) the sensitivity to oxidants and (iii) the capacity to assembly a Fe–S cluster in aconitase (Figure 3B–D). None of the three dithiol Grxs, even the one binding a Fe–S cluster, is functional in yeast mitochondria for the maturation of Fe–S proteins. To determine whether this is caused by structural incompatibility of the dithiol Grxs with the Fe–S cluster biosynthetic machinery or more specifically by the different active site sequences with either dithiol or monothiol motifs, we modified the CGYC and CPYC active sites of GrxC1 and GrxC4 into CGFS to mimic the active site sequence of Grx5. The resulting GrxC1 CGFS fully substituted for yeast Grx5 with respect to all phenotypes analysed (Figure 3), whereas GrxC4 CGFS did not (data not shown). The GrxC1 CGFS rescuing effects did not occur when its expression from the tet promoter was switched off by doxycycline addition to the growth medium (data not shown). We therefore conclude that the requirement for a monothiol Grx active site could preclude poplar dithiol Grxs from functionally rescuing a grx5 mutant, but in some cases, exemplified by the GrxC4 CGFS derivatives, other sequence or structural requirements are needed.
Figure 3.
Rescue of the S. cerevisiae grx5 mutant defects by poplar dithiol glutaredoxins. (A) Compartmentalization of GrxC1, C1G32P, C4 and C1CGFS in the mitochondrial matrix of S. cerevisiae cells. Growth conditions and western blot analyses are similar to those described in Figure 2. TE, total cell extract; MT, mitochondrial fraction; IMS, intermembrane space; MX, matrix. (B) Growth on glucose (YPD plates) or glycerol (YPGly plates), after 3 days at 30°C. (C) Sensitivity to t-BOOH or diamide of the strains after 3 days at 30°C on YPD plates. (D) Ratio between aconitase and malate dehydrogenase activities in exponential cultures at 30°C in YPGalactose medium.
Purification and spectroscopic characterization of Fe–S cluster-containing poplar GrxS14 and AtGrxS16
The mature form of the three organellar poplar CGFS Grxs was expressed in E. coli to check their ability to incorporate Fe–S clusters. On the basis of our previous experience with GrxC1, GSH, which stabilize and ligate the [2Fe–2S] cluster, was added during the first steps of the purification (Rouhier et al, 2007). Although the presence of a brownish colouration typical of a Fe–S cluster was clearly evident in cells overexpressing poplar and Arabidopsis thaliana (At) GrxS14 and S16, almost no holoprotein was obtained at the end of an aerobic purification, even in the presence of GSH, suggesting that the cluster degrades quickly in air. In contrast, there was no indication of a Fe–S cluster prosthetic group in recombinant poplar or A. thaliana GrxS15.
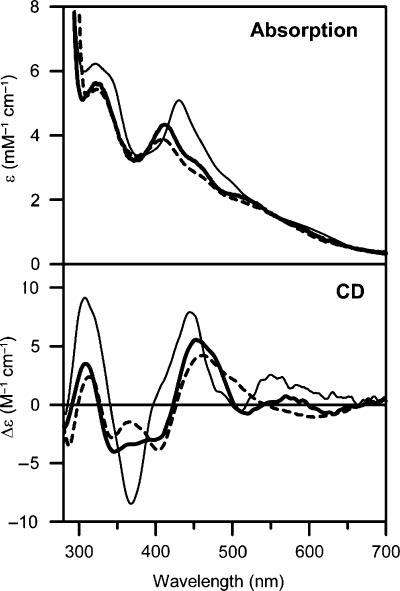
Purification of poplar GrxS14 under strictly anaerobic conditions was undertaken to address the type, stoichiometry and properties of the putative Fe–S centre. The reddish-brown purified samples contained 0.80±0.10 Fe per monomer. The UV–visible absorption and CD spectra of anaerobically purified poplar GrxS14 are shown in Figure 4 and both are characteristic of a [2Fe–2S]2+ centre (Stephens et al, 1978; Dailey et al, 1994). On the basis of the theoretical and experimental ɛ280 values for the apo protein (9.9 mM−1 cm−1), the ɛ280 and ɛ411 values for the [2Fe–2S]2+ centre are estimated to be 3.9 and 4.4 mM−1 cm−1, respectively, and the A411/A280 was found to be 0.31±0.04. In accord with the analytical data, these extinction coefficients are indicative of 0.4–0.5 [2Fe–2S]2+ clusters per monomer. Hence the analytical, absorption and CD data are consistent with approximately one [2Fe–2S]2+ per dimeric GrxS14. Anaerobically purified At GrxS16 contained an analogous [2Fe–2S]2+ centre as judged by very similar UV–visible absorption and CD spectra (Figure 4).
Figure 4.
Comparison of the UV–visible absorption and CD spectra of [2Fe–2S] cluster-bound forms of poplar GrxS14 (thick line), At GrxS16 (broken line) and poplar GrxC1 (thin line).
In vitro reconstitution of aerobically purified apo GrxS14 was attempted under strictly anaerobic conditions in the presence of 5 mM GSH and 2 mM DTT, using Fe(II), L-cysteine and catalytic amounts of E. coli IscS. After chromatographic removal of excess reagents, the resulting cluster-loaded form of GrxS14 was essentially identical to anaerobically purified [2Fe–2S] GrxS14, as judged by Fe analyses and UV–visible absorption and CD spectra (data not shown). GSH was required for successful reconstitution of a [2Fe–2S] cluster on GrxS14. Samples of apo GrxS14 reconstituted using the same procedure in a reaction mixture containing 2 mM DTT, but no GSH, showed no evidence of the presence on a bound Fe–S cluster following repurification. Hence, in vitro Fe–S cluster reconstitution studies confirm the potential of poplar GrxS14 to act as a scaffold for the assembly of [2Fe–2S] clusters in a cysteine desulphurase-mediated reaction and indicate that GSH is required for cluster assembly.
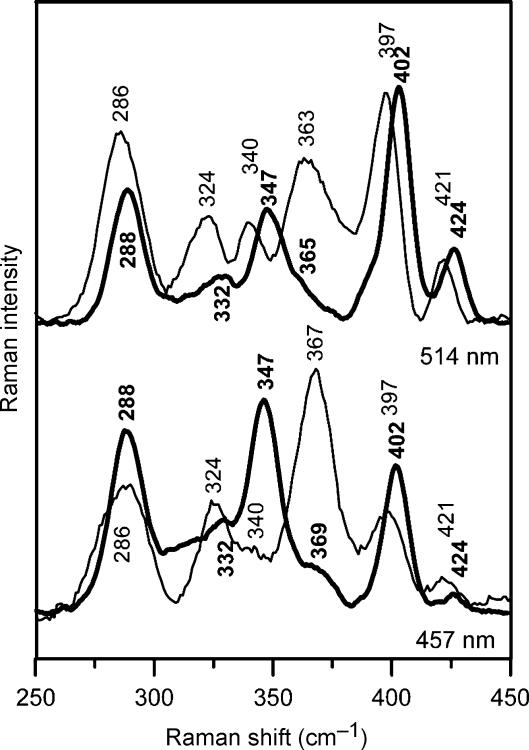
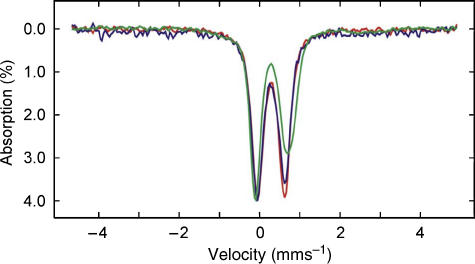
Resonance Raman and Mössbauer studies of anaerobically purified poplar GrxS14 confirm the presence of a [2Fe–2S]2+ centre and provide insight into the cluster ligation. Resonance Raman spectra obtained using 457- and 514-nm excitation reveal Fe–S stretching modes at 288, 332, 347, 365, 402 and 424 cm−1 (Figure 5). The vibrational frequencies are generally similar to those of structurally characterized [2Fe–2S] ferredoxins with complete cysteinyl cluster ligation and are readily assigned to vibrational modes of the Fe2Sb2St4 unit (Sb=bridging S and St=terminal or cysteinyl) by direct analogy with published data (Han et al, 1989; Fu et al, 1992). Figure 6 compares the Mössbauer spectra of poplar GrxS14 with those of the all cysteinyl-ligated [2Fe–2S]2+ cluster in poplar GrxC1 and the IscU [2Fe–2S]2+ cluster which has one non-cysteinyl ligand (Agar et al, 2000). Each spectrum is indicative of a S=0 [2Fe–2S]2+ centre that results from antiferromagnetic coupling of two high-spin Fe(III) ions and is simulated as the sum of quadrupole doublets from each Fe site using the parameters listed in the figure legend. The similarity and values of the isomer shift (δ) and quadrupole splitting (ΔEQ) parameters for each Fe site of the [2Fe–2S]2+ clusters in GrxC1 and GrxS14 are consistent with approximately tetrahedral S ligation at each Fe site. Non-cysteinyl ligation is generally manifested by anomalous isomer shifts and quadrupole splittings for the unique Fe site, which results in marked asymmetry in the observed spectrum, as is apparent in the spectrum of [2Fe–2S]2+ centre in IscU (Agar et al, 2000). Hence, the Mössbauer data indicate a [2Fe–2S]2+ cluster as the sole Fe-containing prosthetic group in anaerobically purified poplar GrxS14, and the Mössbauer and resonance Raman data taken together provide support for complete cysteinyl ligation.
Figure 5.
Comparison of the resonance Raman spectra of [2Fe–2S] cluster-bound forms of poplar GrxS14 (thick line) and GrxC1 (thin line) with 514- and 457-nm laser excitation. Samples were ∼4 mM in Grx and were in the form of a frozen droplet at 17 K. Each spectrum is the sum of 100 scans, with each scan involving counting photons for 1 s each 0.5 cm−1 with 6 cm−1 spectral resolution. Lattice modes of ice have been subtracted.
Figure 6.
Comparison of the Mössbauer spectra of [2Fe–2S] cluster-bound forms of poplar GrxS14 (blue), poplar GrxC1 (red) and A. vinelandii IscU (green). The GrxS14 and C1 Mössbauer samples were prepared by growing cells on 57Fe-enriched media and the IscU sample was prepared by IscS-mediated reconstitution using 57Fe(II) (Agar et al, 2000). The Mössbauer spectra were recorded at 4.2 K with a magnetic field of 50 mT applied parallel to the γ-beam. Each spectrum is best simulated as the sum of two overlapping quadrupole doublets with the following parameters: ΔEQ=0.56 and δ=0.26 mm s−1 for doublet 1, and ΔEQ=0.76 and δ=0.28 mm s−1 for doublet 2 of GrxS14; ΔEQ=0.54 and δ=0.27 mm s−1 for doublet 1, and ΔEQ=0.76 and δ=0.28 mm s−1 for doublet 2 of GrxC1; ΔEQ=0.66 and δ=0.27 mm s−1 for doublet 1, and ΔEQ=0.94 and δ=0.32 mm s−1 for doublet 2 of IscU.
Cluster ligation in GrxS14 and S16
The two structurally characterized [2Fe–2S]2+ centres in dithiol Grxs, human Grx2 (CSYC active site) and poplar GrxC1 (CGYC active site) have very similar absorption and CD spectra and have analogous coordination environments, involving the catalytic cysteine of two Grxs and the cysteines of two GSH (Johansson et al, 2007; Rouhier et al, 2007). On the basis of UV–visible absorption and CD spectra shown in Figure 4, a distinct type of [2Fe–2S]2+ centre may be present in GrxS14 and S16. Marked differences in the excited-state electronic properties and ground-state vibrational properties of the [2Fe–2S]2+ centres in poplar GrxC1 and GrxS14 are evident in comparing the UV–visible absorption/CD and resonance Raman spectra shown in Figures 4 and 5, respectively. Differences in the relative intensities of corresponding Raman bands reflect changes in excitation profiles resulting from perturbation of the excited-state electronic structure. Nevertheless, it is clear that corresponding Fe–S stretching frequencies are upshifted by 2–8 cm−1 in GrxS14 compared with GrxC1, suggesting stronger Fe–S bonds for both terminal and bridging S.
As the spectroscopic properties of the [2Fe–2S]2+ centres in monothiol Grxs indicate complete cysteinyl ligation, mutagenesis studies were undertaken to address the possibility that GSH is replaced as a ligand by an intrinsic cysteine residue in GrxS14 and S16. Three cysteine residues are present in GrxS14 at positions 33, 87 and 108 (recombinant poplar GrxS14 numbering). The active site cysteine (Cys33) is conserved in all CGFS Grxs. Cys87 is present in all S14- and S16-type plant Grxs, but not in S15-type plant Grxs, whereas Cys108 is even not conserved in all S14-type plant Grxs (Supplementary Figure 1). Hence, to address the cluster ligation in GrxS14 and check whether the inability for GrxS15 to incorporate a Fe–S cluster is a consequence of the absence of the second cysteine residue, cysteine mutants both on poplar and AtGrxS14 and on poplar GrxS15 have been generated. AtGrxS14 was used for these mutation studies because introducing these mutations in poplar GrxS14 led mostly to insoluble proteins. On the basis of the colour of the cells and supernatant following sonication and centrifugation, it was clear that AtGrxS14 C33S was no longer able to incorporate the cluster, whereas AtGrxS14 C87S, AtGrxS14 C108S and AtGrxS14 C87/108S were still able to incorporate it. Further confirmation that the second cysteine residue is not a ligand came from the observation that poplar GrxS15 S87C was still unable to accommodate a Fe–S cluster. Taken together, the mutagenesis and spectroscopic data, coupled with the requirement for GSH in cluster reconstitution experiments, strongly support similar cluster ligation in monothiol and dithiol Grxs (as typified by GrxS14 and GrxC1, respectively), involving the catalytic cysteine of two monomers and two external GSH molecules. The structural origin of the observed differences in spectroscopic properties of the [2Fe–2S]2+ clusters in monothiol and dithiol Grxs is therefore likely to result from differences in ligand arrangement or cluster environment.
In vitro cluster transfer from [2Fe–2S] GrxS14 to apo chloroplast ferredoxin
The results presented above provide in vitro evidence in support of a role for monothiol Grxs as scaffolds for the assembly of [2Fe–2S] clusters. However, functional Fe–S cluster scaffold proteins need to be capable both of assembling clusters and transferring them to apo forms of physiologically relevant acceptor proteins. We have therefore investigated the ability of [2Fe–2S] GrxS14, which is localized in chloroplasts, to transfer its cluster to an apo form of Synechocystis [2Fe–2S] ferredoxin, one of the most abundant and highly conserved of all chloroplastic Fe–S proteins.
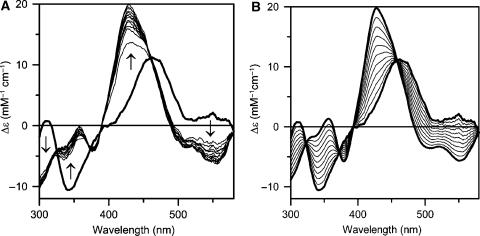
Direct evidence for rapid and quantitative cluster transfer from [2Fe–2S] GrxS14 to apo Fd was provided by anaerobic CD studies as a function of time using a reaction mixture involving stoichiometric [2Fe–2S] GrxS14 and apo Fd (Figure 7). The marked difference in the CD spectra of [2Fe–2S]2+ centres in GrxS14 and holo Fd in the reaction mixture facilitates direct monitoring of cluster transfer and assessment of the extent of intact cluster transfer via concomitant decrease and increase of the CD spectra of the cluster donor and acceptor, respectively. Comparison of the time course of CD changes in the reaction mixture (Figure 7A) with simulated data for 0–100% intact cluster transfer (Figure 7B) indicates quantitative cluster transfer from [2Fe–2S] GrxS14 to apo Fd that is 60% complete after 5 min and 100% complete after approximately 60 min. The agreement between the observed and simulated data and the rapid rate of Fd reconstitution suggests quantitative and intact cluster transfer from [2Fe–2S] GrxS14 to apo Fd and argues strongly against cluster degradation and reassembly of apo Fd. This conclusion is further supported by three additional pieces of evidence. First, absorption and CD studies of the reaction mixture in the absence of the apo Fd showed <10% degradation of the [2Fe–2S] cluster on GrxS14 after 60 min. Second, parallel CD studies of apo Synechocystis Fd reconstitution with equivalent amounts of S2− and Fe3+ or Fe2+ under identical conditions resulted in ∼5% cluster assembly over a 60-min period. Third, the addition of 1 mM EDTA to the reaction mixture completely inhibited cluster reconstitution using S2− and Fe3+ or Fe2+, but had no significant effect on the time course of [2Fe–2S] GrxS14-mediated cluster assembly on apo Fd. Taken together, these observations indicate rapid, quantitative and intact cluster transfer from [2Fe–2S] GrxS14 to apo Fd. In contrast, parallel cluster transfer studies using the poplar dithiol [2Fe–2S] GrxC1 showed no indication of cluster transfer to apo Synechocystis Fd after 120 min (data not shown). Hence, the ability to transfer [2Fe–2S] clusters to acceptor proteins appears to be limited to monothiol Grxs. Comparative stability studies of GrxS14 and GrxC1 indicate that this is likely to be a consequence of increased accessibility and lability for the [2Fe–2S] clusters in monothiol Grxs compared with dithiol Grxs, see Supplementary Figure 2.
Figure 7.
Time course of cluster transfer from poplar GrxS14 to apo Synechocystis Fd monitored by UV–visible CD spectroscopy at 23°C in 1 cm cuvettes. (A) CD spectra were recorded at 5-min intervals for a period of 60 min for a reaction mixture that was initially 15 μM in GrxS14 [2Fe–2S] clusters and 15 μM apo Fd. The spectrum at zero time (thick line) corresponds to [2Fe–2S] GrxS14 in the same reaction mixture in the absence of apo Fd. The arrows indicate the direction of intensity change with time at selected wavelengths. (B) Predicted changes in the CD spectra for quantitative cluster transfer. Thick lines correspond to holo forms of Synechocystis [2Fe–2S] Fd and [2Fe–2S] GrxS14 and thin lines correspond to simulated CD spectra corresponding to 10–90% [2Fe–2S] cluster transfer from GrxS14 to Fd in 10% increments. In both panels, Δɛ values are expressed per [2Fe–2S]2+ cluster.
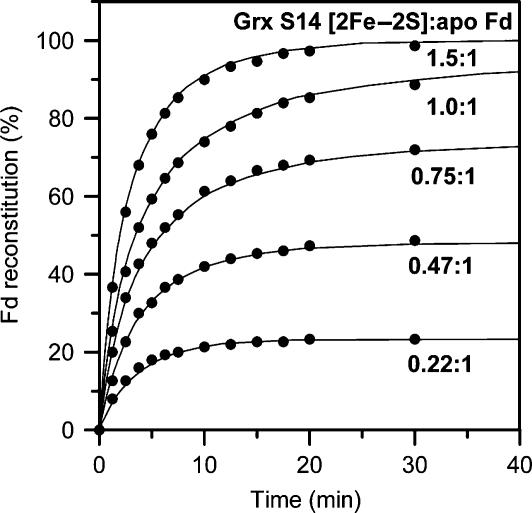
Quantitative assessment of the rate of cluster transfer as a function of [2Fe–2S] GrxS14 to apo Fd stoichiometry (0.22:1–1.5:1 based on cluster content of GrxS14) was obtained by continuous monitoring of the CD changes at 423 nm (Figure 8). On the basis of the initial concentrations of GrxS14 [2Fe–2S] clusters and apo Fd, the data in each case are well fit by second-order kinetics corresponding to stoichiometric cluster transfer with a rate constant of 20 000 M−1 min−1. To put this into context, the fastest intact [2Fe–2S] cluster transfer reported so far is for HscA/HscB/ATP-mediated [2Fe–2S] cluster transfer from [2Fe–2S] IscU to apo-IscFdx, which had a second-order rate constant of 800 M−1 min−1 (Chandramouli and Johnson, 2006). Hence, the results demonstrate that [2Fe–2S] cluster transfer from [2Fe–2S] GrxS14 to apo Fd is stoichiometric, quantitative and occurs at a rate that is 25 × faster than in vitro cluster transfer studies using the IscU scaffold protein. Clearly these in vitro studies demonstrate that CGFS Grxs have the potential to function as scaffold proteins for the assembly and efficient delivery of [2Fe–2S] clusters.
Figure 8.
Kinetics of cluster transfer from poplar GrxS14 to apo Synechocystis Fd at 23°C as a function of the stoichiometry of GrxS14 [2Fe–2S] clusters to apo Fd. The experimental conditions are as described in Figure 7, except that the concentration of GrxS14 [2Fe–2S] clusters was varied to give the indicated GrxS14 [2Fe–2S] to apo Fd ratios. Reactions were continuously monitored using the CD intensity at 423 nm and converted to percent Fd reconstitution based on simulated data (as illustrated in Figure 7 for a 1:1 stoichiometry). Solid lines correspond to second-order kinetics with k=20 000 M−1 min−1 based on the initial concentrations of GrxS14 [2Fe–2S] clusters and apo Fd.
Discussion
Involvement of chloroplastic CGFS Grxs in iron–sulphur cluster assembly
Grxs with a CGFS active site constitutes a recently described subgroup of the Grx family, whose functions are largely unknown at the present time (Herrero and de la Torre-Ruiz, 2007). In yeast, Grx5 was originally identified as playing a central role in protecting against oxidative damage and subsequent investigations refined a role in mitochondrial Fe–S cluster biogenesis (Rodríguez-Manzaneque et al, 1999, 2002). Immunoprecipitation studies suggested that Grx5 facilitates transfer of clusters assembled on the Isu scaffold protein to acceptor proteins (Mühlenhoff et al, 2003). In contrast, the other two CGFS Grxs in yeast, Grx3 and Grx4, are nuclear proteins involved in the nuclear localization regulation of the transcriptional iron regulators Aft1 and Aft2 (Ojeda et al, 2006). In plants, the CGFS Grx subgroup is expanded to four members, but the only functional information stems from in vivo analysis of A. thaliana GrxS14 (aka AtGRXcp), which was shown to be a chloroplastic protein probably involved in the oxidative stress response (Cheng et al, 2006).
In this study, the subcellular locations of each of the poplar CGFS Grxs were determined and yeast Δgrx5 complementation studies of mitochondrial-targeted forms were used to assess the possibility that plant CGFS Grxs are also involved in Fe–S cluster assembly. All the CGFS Grxs, except GrxS15, but not dithiol Grxs (even GrxC1, which contains a [2Fe–2S] cluster), were able to complement the defects of the yeast Δgrx5 mutant, although not always to a similar extent. These results were somewhat surprising because the two chloroplastic Grxs (GrxS14 and S16) are the most efficient proteins, whereas the mitochondrial GrxS15 is essentially not effective. In addition, the proteins involved in plant and yeast mitochondrial Fe–S cluster assembly belong to the ISC type of machinery, whereas plant chloroplasts contain essentially the SUF system. Nevertheless, it is evident that both GrxS14 and S16 have the ability to assume a role analogous to that of Grx5 in mitochondrial Fe–S cluster biosynthesis. Whether GrxS15 is involved in plant mitochondria Fe–S cluster assembly is still uncertain, but only one or two other Grxs, displaying a CCMS active site, are predicted to be present in mitochondria and could fulfil an analogous role. As discussed below, a major difference between GrxS14 or S16 and S15, that could explain their different behaviour, is the capacity of the two chloroplastic Grxs to incorporate a Fe–S cluster when expressed in E. coli.
GrxS14 and S16 as Fe–S cluster scaffold proteins
The observation that both plant GrxS14 and S16 contain analogous [2Fe–2S]2+ centres when heterologously expressed in E. coli raises the possibility of a role for CGFS Grxs as scaffolds for the assembly of chloroplastic Fe–S clusters. Additional support for a scaffolding role comes from the ability to assemble spectroscopically identical clusters on apo GrxS14 in a cysteine desulphurase-mediated reaction in the presence of L-cysteine, Fe2+ ion and GSH. Cysteine mutagenesis studies, analytical and spectroscopic data, and the requirement of GSH to effect cluster assembly on apo GrxS14, indicate the presence of a subunit-bridging [2Fe–2S]2+ cluster ligated by the first active site cysteine of two Grxs and the cysteines of two GSH molecules. A similar ligation has also been structurally established in the dithiol poplar GrxC1 (Rouhier et al, 2007). However, pronounced differences in the lability or accessibility and the ground-state vibrational and excited-state electronic properties of the [2Fe–2S] centres in monothiol and dithiol plant Grxs indicate differences in the cluster environment or the arrangement of coordinating cysteine residues. Possible differences could involve the extent and arrangement of aromatic residues in the vicinity of the cluster or cis rather than trans GSH cluster ligation (Rouhier et al, 2007). Crystallographic studies of cluster-bound forms of monothiol Grxs will be required to address these differences and to assess the cluster release mechanism in monothiol Grxs.
A functional Fe–S cluster scaffold protein must also be effective in transferring clusters to physiologically relevant acceptor proteins. Hence, the observation of rapid, stoichiometric and intact cluster transfer from [2Fe–2S] GrxS14 to apo Fd provides in vitro evidence for a role of CGFS Grxs as [2Fe–2S] cluster donors for maturation of chloroplast Fe–S proteins. In contrast, analogous cluster transfer experiments using the structurally characterized [2Fe–2S] cluster-bound dithiol GrxC1 failed to show any evidence of cluster transfer to apo Fd. Clearly, there is a pressing need to investigate the ability of [2Fe–2S] GrxS14 and [2Fe–2S] GrxS16 to effect maturation of the apo forms of a variety of [2Fe–2S] cluster-containing chloroplastic proteins, for example, sirohydrochlorin ferrochelatase, dihydroxyacid dehydratase and Rieske-type centres in oxygenases and the cytochrome b6f, complex, and to assess the possibility that these proteins also participate in the maturation of [3Fe–4S] and [4Fe–4S] clusters in chloroplastic proteins. Such studies will address the specificity of these Grxs in chloroplastic Fe–S cluster biogenesis and are currently in progress in our laboratories. The proposed role for CGFS Grxs as Fe–S cluster scaffold proteins also provides a rationalization for the apparent disparity concerning the role of the second partly conserved cysteine residue in yeast Grx5 in in vivo and in vitro activity data. Mutagenesis results indicated that the second conserved cysteine is required for in vitro deglutathionylation activity, but is not required for in vivo Fe–S cluster biogenesis activity (Belli et al, 2002) or the assembly of a [2Fe–2S] cluster on GrxS14. Nevertheless, among all natural or mutated plant CGFS Grxs tested, those which do not possess this additional cysteine (GrxS15 and GrxC4 CGFS) were not able to complement the S. cerevisiae Δgrx5 mutant. This indicates that, although not essential for cluster incorporation, this additional cysteine may be required for an efficient complementation with plant Grxs.
Two pieces of evidence argue against a scaffolding role for monothiol CGFS Grxs in de novo Fe–S cluster assembly. First, gene disruptions of known chloroplastic scaffold proteins such as the Nfu proteins and other intrinsic components of the chloroplastic Fe–S assembly machinery are generally associated with a dwarf phenotype or abnormal development (Touraine et al, 2004; Yabe et al, 2004; Balasubramanian et al, 2006; Xu and Moller, 2006; Van Hoewyk et al, 2007). In the case of GrxS14, the phenotype consists of a defect in early seedling growth under oxidative stress (Cheng et al, 2006) and is not as strong as that associated with nfu gene disruptions. Taking into account the large number of Fe–S proteins in the chloroplast, one possibility is that GrxS14 has specific target proteins whose functions are not essential for plant development. Alternatively, the absence of GrxS14 may be compensated by other potential scaffold proteins such as GrxS16 or SufA in the loss-of-function mutant. To address this issue, it will be necessary to investigate the phenotypes of gene disruptions resulting in depletion of other chloroplast proteins, both individually and together, for example, GrxS16, GrxS14/GrxS16, SufA/GrxS14 and SufA/GrxS16. Second, the role of Grx5 in yeast mitochondrial Fe–S cluster biogenesis does not appear to be dependent on GSH, which is required for the assembly of a [2Fe–2S] cluster on GrxS14. Depletion of GSH was found to affect the maturation of cytosolic Fe–S proteins, but had no significant effect on mitochondrial Fe–S cluster biogenesis (Sipos et al, 2002). However, the significance of this observation remains to be established as there is currently no reliable information on the type of cluster or the requirements for cluster assembly on Grx5, or the level of GSH that is required to support mitochondrial Fe–S cluster assembly. A recent paper by Picciocchi et al (2007) reported in vitro assembly of a Fe–S cluster in yeast Grx5 and other CGFS Grxs in the presence of GSH. However, variability in the reported UV–visible absorption spectra, coupled with the absence of Mössbauer, CD, resonance Raman or quantitative EPR data, leaves the cluster content of these samples unresolved.
Alternative functions for monothiol CGFS Grxs
The ability of CGFS Grxs to accommodate [2Fe–2S] clusters and transfer them to acceptor proteins is also consistent with a role in storage of preformed Fe–S clusters or as Fe–S cluster delivery systems that mediate cluster transfer from other potential scaffold proteins (i.e. SufA, SufB and Nfu in chloroplasts and Isa/IscA, Nfu and Isu in mitochondria) to specific acceptor proteins. Such a role is consistent with yeast Δgrx5 immunoprecipitation studies, which suggested that Grx5 facilitates transfer of clusters assembled on the Isu scaffold protein to acceptor proteins (Mühlenhoff et al, 2003). This would also explain why a strong phenotype may not be associated with gene disruption, because the transfer would still occur although at a lower rate. In addition, yeast two-hybrid studies indicate an in vivo interaction between Grx5 and Isa1 (Vilella et al, 2004) and the crystal structure of an IscA protein with an asymmetrical subunit-bridging [2Fe–2S] cluster has recently been published (Morimoto et al, 2006). Hence, it is possible that IscA/SufA-type proteins hand off clusters to monothiol Grxs for delivery to acceptor proteins and that this cluster assembly pathway would only be completely shut down by deletion of both genes. In vitro cluster transfer experiments involving chloroplast [2Fe–2S] SufA and [2Fe–2S] Nfu proteins and apo GrxS14 and kinetic studies of the effect on apo GrxS14 on the rates of cluster transfer from chloroplast [2Fe–2S] SufA and [2Fe–2S] Nfu to apo Fd are in progress to test this hypothesis.
In light of the proposed role for Grx3 and Grx4 in the regulation of Fe homeostasis in yeast (Ojeda et al, 2006), an alternative rationalization for the cluster-binding ability of chloroplast Grxs is that they play a role in the regulation of the SUF Fe–S cluster biogenesis machinery by facilitating assessment of the chloroplast Fe–S cluster status. Using bioinformatics, Huynen et al (2005) proposed that BolA proteins act as reductases that interact with monothiol Grxs in the oxidative stress response. Nevertheless, such a function seems unlikely in plants as BolA proteins do not possess conserved cysteines. Nevertheless, among the three genes encoding SufE proteins in A. thaliana, SufE1 (At4g26500) contains a C-terminal BolA domain (Xu and Moller, 2006; Ye et al, 2006a). SufE proteins are sulphur transferases that serve as activators for the NifS and SufS cysteine desulphurases in the initial steps of chloroplast Fe–S cluster biosynthesis (Xu and Moller, 2006). Hence, it is possible that GrxS14 acts as a Fe–S cluster-dependent regulator of the SUF machinery by interacting with SufE1 via the BolA domain. Under this scenario, Fe–S cluster incorporation into GrxS14 would occur under Fe–S cluster-replete conditions, resulting in enhanced interaction with SufE1 to limit the activity of the SUF Fe–S cluster biogenesis machinery.
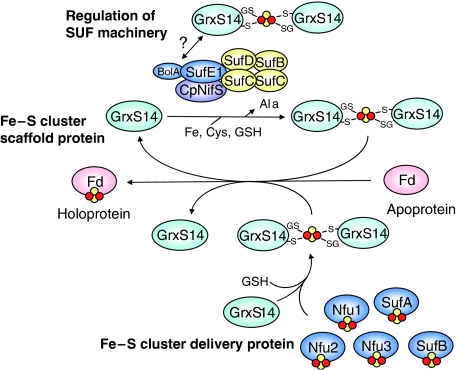
The three possible roles for CGFS Grxs in chloroplast Fe–S cluster biogenesis are summarized in Figure 9. It is important to emphasize that these three roles are not mutually exclusive and that all three could be operative in chloroplasts with GrxS14 and S16 having different specificity for acceptor proteins, cluster-donor proteins or cysteine desulfurases. Such functional variability and specificity differences may also be responsible for anomalous behaviour of GrxS15 and the truncated form of GrxS17 in yeast Grx5 complementation studies.
Figure 9.
Working model for the potential roles of GrxS14 and S16 in chloroplastic Fe–S cluster assembly. GrxS14 and S16 could function as scaffold proteins for de novo synthesis and transfer of Fe–S clusters, as Fe–S cluster delivery proteins for mediating the transfer of Fe–S clusters from other potential scaffold proteins (Nfu1, 2 and 3, SufA and SufB) to acceptor proteins, or as regulators of the SUF machinery by interacting with the BolA domain of SufE1 in the cluster-bound form.
Materials and methods
Heterologous expression and purification of CGFS Grxs in E. coli
The cloning and aerobic purification procedures of CGFS Grxs are described as Supplementary data. Anaerobic purification of poplar GrxS14 and AtGrxS16 was carried out under Ar in a Vacuum Atmospheres glove box at oxygen levels <2 p.p.m. The cell pellet was resuspended in 100 mM Tris–HCl pH 8 with 1 mM GSH (buffer B), sonicated and centrifuged at 39 700 g for 1 h at 4°C to remove the cell debris. The reddish-brown cell-free extract containing holo GrxS14 was subjected to 40% ammonium sulphate cut followed by centrifugation. The brown pellet was resolubilized in buffer B and loaded onto a 10 ml Phenyl Sepharose column (GE Healthcare). The protein was eluted with a 1–0 M (NH4)2SO4 gradient using buffer B. The purest fractions, as judged by SDS–PAGE analysis, were pooled and (NH4)2SO4 was removed by ultrafiltration dialysis using a YM10 membrane and buffer B.
Reconstitution of a Fe–S cluster in apo poplar GrxS14
Reconstitution of poplar apo GrxS14, 0.4 mM in buffer B, was accomplished in a glove box under anaerobic conditions by incubating at room temperature for 150 min with 5 mM GSH, 2 mM DTT, 12-fold excess of Fe(II) (ferrous ammonium sulphate), L-cysteine and catalytic amounts of E. coli IscS (20 μM). Reagents in excess were removed by loading onto a High-Trap Q-Sepharose column (GE Healthcare) and eluting with a 0–1 M NaCl gradient in buffer B. The holo protein was eluted between 0.45 and 0.55 M NaCl and was desalted using ultrafiltration dialysis.
Yeast plasmids and strains
Grx sequences were cloned in-frame in the yeast plasmid pMM221, which contains the S. cerevisiae mitochondrial targeting sequence of Grx5 plus a C-terminal 3HA/His6 tag, under the control of the doxycycline-regulatory tetO2 promoter (Molina et al, 2004) (Table I). pMM54 (Rodríguez-Manzaneque et al, 2002) contains a yeast GRX5-3HA construction under its own promoter. S. cerevisiae strains are described in Table II. Plasmids were linearized by ClaI digestion previous to chromosomal integration.
Table 1.
New plasmids employed in this study
| Plasmids | Characteristics |
|---|---|
| pMM628 | Sequence coding from amino acid 2–117 of GrxC1 cloned between NotI–PstI sites of pMM221 |
| pMM630 | Sequence coding from amino acid 2–117 of GrxC1(G32P) cloned between NotI–PstI sites of pMM221 |
| pMM632 | Sequence coding from amino acid 2–113 of GrxC4 cloned between NotI–PstI sites of pMM221 |
| pMM634 | Sequence coding from amino acid 37–174 of GrxS15 cloned between NotI–PstI sites of pMM221 |
| pMM657 | Sequence coding from amino acid 66–173 of GrxS14 cloned between NotI–PstI sites of pMM221 |
| pMM676 | Derivative of pMM628 with the sequence coding for CGFS instead of CGYC in GrxC1 |
| pMM712 | Sequence coding from amino acid 85–296 of GrxS16 cloned between NotI–PstI sites of pMM221 |
| pMM713 | Sequence coding from amino acid 1–492 of GrxS17 cloned between NotI–PmeI sites of pMM221 |
| pMM714 | Sequence coding from amino acid 398–492 of GrxS17 cloned between NotI–PmeI sites of pMM221 |
Table 2.
Yeast strains employed in this study
| Strains | Relevant phenotype | Comments |
|---|---|---|
| W303-1A | MATa ura3-1 ade2-1 leu2-3,112 trp1-1 his3-11,15 | Wild type |
| W303-1B | As W303-1A but MATα | Wild type |
| MML100 | MATa grx5∷kanMX4 | Rodríguez-Manzaneque et al (2002) |
| MML240 | MATa grx5∷kanMX4 [pMM54(GRX5-3HA)]∷LEU2 | Rodríguez-Manzaneque et al (2002) |
| MML755 | MATα [pMM628(GrxC1-3HA)]∷LEU2 | Integration of linear pMM628 in W303-1B |
| MML756 | MATα [pMM630(GrxC1G32P-3HA)]∷LEU2 | Integration of linear pMM630 in W303-1B |
| MML757 | MATα [pMM632(GrxC4-3HA)]∷LEU2 | Integration of linear pMM632 in W303-1B |
| MML758 | MATα [pMM634(GrxS15-3HA)]∷LEU2 | Integration of linear pMM634 in W303-1B |
| MML759 | MATα [pMM657(GrxS14-3HA)]∷LEU2 | Integration of linear pMM657 in W303-1B |
| MML761 | MATa grx5∷kanMX4 [pMM628(GrxC1-3HA)]∷LEU2 | Spore from a cross MML100 × MML755 |
| MML763 | MATa grx5∷kanMX4 [pMM630(GrxC1G32P-3HA)]∷LEU2 | Spore from a cross MML100 × MML756 |
| MML765 | MATa grx5∷kanMX4 [pMM632(GrxC4-3HA)]∷LEU2 | Spore from a cross MML100 × MML757 |
| MML767 | MATa grx5∷kanMX4 [pMM634(GrxS15-3HA)]∷LEU2 | Spore from a cross MML100 × MML758 |
| MML769 | MATa grx5∷kanMX4 [pMM657(GrxS14-3HA)]∷LEU2 | Spore from a cross MML100 × MML759 |
| MML779 | MATα [pMM676(GrxC1CGFS-3HA)]∷LEU2 | Integration of linear pMM676 in W303-1B |
| MML780 | MATa grx5∷kanMX4 [pMM676(GrxC1CGFS-3HA)]∷LEU2 | Spore from a cross MML100 × MML779 |
| MML786 | MATα [pMM712(GrxS16-3HA)]∷LEU2 | Integration of linear pMM712 in W303-1B |
| MML787 | MATα [pMM713(GrxS17-3HA)]∷LEU2 | Integration of linear pMM713 in W303-1B |
| MML788 | MATα [pMM714(GrxS17398–492-3HA)]∷LEU2 | Integration of linear pMM714 in W303-1B |
| MML806 | MATa grx5∷kanMX4 [pMM712(GrxS16-3HA)]∷LEU2 | Spore from a cross MML100 × MML786 |
| MML808 | MATa grx5∷kanMX4 [pMM713(GrxS17-3HA)]∷LEU2 | Spore from a cross MML100 × MML787 |
| MML810 | MATa grx5∷kanMX4 [pMM714(GrxS17398–492-3HA)]∷LEU2 | Spore from a cross MML100 × MML788 |
Growth conditions for S. cerevisiae cells
S. cerevisiae cultures were grown as described in Molina et al (2004). Samples were taken from cultures grown exponentially for at least 10 generations at 30°C. Sensitivity to oxidants was determined onto YPD plates containing the indicated concentration of the agent, by spotting 1:5 serial dilutions of exponential cultures and recording growth after 2 days of incubation at 30°C.
Other methods
Mitochondria were purified and subfractionated (Diekert et al, 2001) from exponential yeast cultures in YPLactate medium at 30°C. Aconitase and malate dehydrogenase were assayed as described in Robinson et al (1987), in extracts prepared (Molina-Navarro et al, 2006) from cells growing exponentially in YPGalactose medium.
In vivo subcellular localization by GFP fusions
Full-length open reading frames were cloned in 5′ of the GFP sequence under the control of a double 35S promoter into the plasmid pCK S65C between NcoI and BamHI sites (underlined) using primers described in Supplementary Table I. Nicotiana benthamiana cells were transfected by bombardment of leaves with tungsten particles coated with plasmid DNA and images were obtained with a Zeiss LSM510 confocal microscope.
Analytical and spectroscopic methods
Protein concentrations were determined by the DC protein assay (Bio-Rad), using BSA as a standard. Iron concentrations were determined using bathophenanthroline under reducing conditions, after digestion of the protein in 0.8% KMnO4/0.2 M HCl (Fish, 1988). Sample concentrations and extinction coefficients are based on protein monomer and samples were in 100 mM Tris–HCl pH 8 with 1 mM GSH, unless indicated otherwise. Samples for spectroscopic studies were prepared under Ar in a Vacuum Atmospheres glove box (O2<2 p.p.m.). UV–visible absorption and CD spectra were recorded at room temperature using a Shimadzu UV-3101PC spectrophotometer and Jasco J-715 spectropolarimeter, respectively. Resonance Raman spectra were recorded as previously described (Cosper et al, 2004), using an Instruments SA Ramanor U1000 spectrometer coupled with a Coherent Sabre argon ion laser, with 20 μl frozen droplets of 2–3 mM sample mounted on the cold finger of an Air Products Displex Model CSA-202E closed cycle refrigerator. Mössbauer spectra were recorded by using the previously described instrumentations (Ravi et al, 1994). The zero velocity of the spectra refers to the centroid of a room temperature spectrum of a metallic Fe foil. Analysis of the Mössbauer data was performed with the WMOSS program (Web Research).
Fe–S cluster transfer experiments
Synechocystis Fd for cluster transfer experiments was heterologously expressed in E. coli and purified according to published procedures (Glauser et al, 2004). Apo Fd was prepared by treating the holo protein with EDTA and potassium ferricyanide at room temperature under anaerobic conditions and removing excess reagents by ultrafiltration dialysis using a YM10 membrane and buffer B. The time course of cluster transfer from [2Fe–2S] GrxS14 to apo Fd was monitored under anaerobic conditions in 1 cm cuvettes at 23°C using UV–visible CD spectroscopy. The reactions were carried out in buffer B with 5 mM DTT and apo Fd was added 60 min prior to initiation of the cluster transfer reactions by addition of [2Fe–2S] GrxS14. Changes in the CD spectra at 423 nm were used to assess the concentration of holo Fd formed as a function of time. The time course of holo Fd formation was analysed by fitting to second-order kinetics, based on the initial concentrations of GrxS14 [2Fe−2S] clusters and apo-Fd in the reaction mixture, using the Chemical Kinetics Simulator software package (IBM).
Supplementary Material
Supplementary data
Acknowledgments
This research was supported by grants from the National Institutes of Health (GM62524 to MKJ and GM47295 to BHH), from BFU2004-03167 (Ministerio de Educación y Ciencia) and 2005SGR-00677 (Generalitat de Catalunya) to EH and from the ANR programs (GNP05010G and JC07_204825) to NR, FG and JPJ.
References
- Achebach S, Tran QH, Vlamis-Gardikas A, Mullner M, Holmgren A, Unden G (2004) Stimulation of Fe–S cluster insertion into apoFNR by E. coli glutaredoxins 1, 2 and 3 in vitro. FEBS Lett 565: 203–206 [DOI] [PubMed] [Google Scholar]
- Agar JN, Krebs B, Frazzon J, Huynh BH, Dean DR, Johnson MK (2000) IscU as a scaffold for iron–sulfur cluster biosynthesis: sequential assembly of [2Fe–2S] and [4Fe–4S] clusters in IscU. Biochemistry 39: 7856–7862 [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Shen G, Bryant DA, Golbeck JH (2006) Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 88: 3182–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Lobreaux S (2005) Biogenesis of iron–sulfur proteins in plants. Trends Plant Sci 10: 324–331 [DOI] [PubMed] [Google Scholar]
- Belli G, Polaina J, Tamarit J, De La Torre MA, Rodriguez-Manzaneque MT, Ros J, Herrero E (2002) Structure–function analysis of yeast Grx5 monothiol glutaredoxin defines essential amino acids for the function of the protein. J Biol Chem 277: 37590–37596 [DOI] [PubMed] [Google Scholar]
- Chandramouli K, Johnson MK (2006) HscA and HscB stimulate [2Fe–2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 45: 11087–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli K, Unciuleac M-C, Naik S, Dean DR, Huynh BH, Johnson MK (2007) Formation and properties of [4Fe–4S] clusters on the IscU scaffold protein. Biochemistry 46: 6804–6811 [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Brock A, Nelson RS, Hirschi KD (2006) AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem 281: 26280–26288 [DOI] [PubMed] [Google Scholar]
- Cosper MM, Krebs C, Hernandez H, Jameson GNL, Eidsness MK, Huynh BH, Johnson MK (2004) Characterization of the cofactor content of Escherichia coli biotin synthase. Biochemistry 43: 2007–2021 [DOI] [PubMed] [Google Scholar]
- Dailey HA, Finnegan MG, Johnson MK (1994) Human ferrochelatase is an iron–sulfur protein. Biochemistry 33: 403–407 [DOI] [PubMed] [Google Scholar]
- Diekert K, de Kroon AIPM, Kispal G, Lill R (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol 65: 37–51 [DOI] [PubMed] [Google Scholar]
- Feng Y, Zhong N, Rouhier N, Hase T, Kusunoki M, Jacquot JP, Jin C, Xia B (2006) Structural insight into poplar glutaredoxin C1 with a bridging iron–sulfur cluster at the active site. Biochemistry 45: 7998–8008 [DOI] [PubMed] [Google Scholar]
- Fish WW (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol 158: 357–364 [DOI] [PubMed] [Google Scholar]
- Fu W, Drozdzewski PM, Davies MD, Sligar SG, Johnson MK (1992) Resonance Raman and magnetic circular dichroism studies of reduced [2Fe–2S] proteins. J Biol Chem 267: 15502–15510 [PubMed] [Google Scholar]
- Gelling C, Dawes IA, Richhardt N, Lill R, Mühlenhoff U (2008) Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol Cell Biol 28: 1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser DA, Bourquin F, Manieri W, Schürmann P (2004) Characterization of ferredoxin:thioredoxin reductase modified by site-directed mutagenesis. J Biol Chem 279: 16662–16669 [DOI] [PubMed] [Google Scholar]
- Han S, Czernuszewicz RS, Kimura T, Adams MWW, Spiro TG (1989) Fe2S2 protein resonance Raman revisited: structural variations among adrenodoxin, ferredoxin, and red paramagnetic protein. J Am Chem Soc 111: 3505–3511 [Google Scholar]
- Herrero E, de la Torre-Ruiz MA (2007) Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci 64: 1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MA, Spronk CA, Gabaldon T, Snel B (2005) Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett 579: 591–596 [DOI] [PubMed] [Google Scholar]
- Johansson C, Kavanagh KL, Gileadi O, Oppermann U (2007) Reversible sequestration of active site cysteines in a 2Fe–2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem 282: 3077–3082 [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK (2005) Structure, function, and formation of biological iron–sulfur clusters. Annu Rev Biochem 74: 247–281 [DOI] [PubMed] [Google Scholar]
- Johnson MK, Smith AD (2005) Iron–sulfur proteins. In Encyclopedia of Inorganic Chemistry, King RB (ed), 2nd edn, pp 2589–2619. Chichester, UK: John Wiley & Sons [Google Scholar]
- Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW (2007) SufE transfers sulfur from SufS to SufB for iron–sulfur cluster assembly. J Biol Chem 282: 13342–13350 [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U (2005) Iron–sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci 30: 133–141 [DOI] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Vergnolle O, Lonn ME, Hudemann C, Bill E, Holmgren A (2005) Characterization of human glutaredoxin 2 as iron–sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci USA 102: 8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina MM, Bellí G, de la Torre MA, Rodríguez-Manzaneque MT, Herrero E (2004) Nuclear monothiol glutaredoxins of S. cerevisiae can function as mitochondrial glutaredoxins. J Biol Chem 279: 51923–51930 [DOI] [PubMed] [Google Scholar]
- Molina-Navarro MM, Casas C, Piedrafita L, Bellí G, Herrero E (2006) Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett 580: 2273–2280 [DOI] [PubMed] [Google Scholar]
- Morimoto K, Yamashita E, Kondou Y, Lee SJ, Arisaka F, Tsukihara T, Nakai M (2006) The asymmetric IscA homodimer with an exposed [2Fe–2S] cluster suggests the structural basis of the Fe–S cluster biosynthetic scaffold. J Mol Biol 360: 117–132 [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron–sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda L, Keller G, Mühlenhoff U, Rutherford JC, Lill R, Winge DR (2006) Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in S. cerevisiae. J Biol Chem 281: 17661–17669 [DOI] [PubMed] [Google Scholar]
- Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F (2007) CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe–2S] cluster. Biochemistry 46: 15018–15026 [DOI] [PubMed] [Google Scholar]
- Ravi N, Bollinger JM, Huynh BH, Edmondson DE, Stubbe J (1994) Mechanism of assembly of the tyrosyl radical-diiron(III) cofactor of E. coli ribonucleotide reductase. 1. Mössbauer characterization of the diferric radical precursor. J Am Chem Soc 116: 8007–8014 [Google Scholar]
- Robinson JB Jr, Brent LG, Sumegi B, Srere PA (1987) An enzymatic approach to the study of the Krebs tricarboxylic acid cycle. In Mitochondria: a Practical Approach, Darley-Usmar WM, Rickwood D, Wilson MT (eds), pp 153–170. Oxford, UK: IRL Press [Google Scholar]
- Rodríguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E (1999) Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in S. cerevisiae. Mol Cell Biol 19: 8180–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Manzaneque MT, Tamarit J, Bellí G, Ros J, Herrero E (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur clusters. Mol Biol Cell 13: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP (2004) Plant glutaredoxins: still mysterious reducing systems. Cell Mol Life Sci 61: 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Unno H, Bandyopadhyay S, Masip L, Kim SK, Hirasawa M, Gualberto JM, Lattard V, Kusunoki M, Knaff DB, Georgiou G, Hase T, Johnson MK, Jacquot JP (2007) Functional, structural and spectroscopic characterization of a glutathione-ligated [2Fe−2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci USA 104: 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos K, Lange H, Fekete Z, Ullmann P, Lill R, Kispal G (2002) Maturation of cytosolic iron–sulfur proteins requires glutathione. J Biol Chem 277: 26944–26949 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Thomson AJ, Dunn JBR, Keiderling TA, Rawlings J, Rao KK, Hall DO (1978) Circular dichroism and magnetic circular dichroism of iron–sulfur proteins. Biochemistry 17: 4770–4778 [DOI] [PubMed] [Google Scholar]
- Tamarit J, Bellí G, Cabiscol E, Herrero E, Ros J (2003) Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem 278: 25745–25751 [DOI] [PubMed] [Google Scholar]
- Touraine B, Boutin JP, Marion-Poll A, Briat JF, Peltier G, Lobreaux S (2004) Nfu2: a scaffold protein required for [4Fe–4S] and ferredoxin iron–sulfur cluster assembly in Arabidopsis chloroplasts. Plant J 40: 101–111 [DOI] [PubMed] [Google Scholar]
- Van Hoewyk D, Abdel-Ghany SE, Cohu CM, Herbert SK, Kugrens P, Pilon M, Pilon-Smits EA (2007) Chloroplast iron–sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proc Natl Acad Sci USA 104: 5686–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella F, Alves R, Rodríguez-Manzaneque MT, Bellí G, Swaminathan S, Sunnerhagen P, Herrero E (2004) Evolution and cellular function of monothiol glutaredoxins: involvement in iron–sulfur cluster assembly. Comp Funct Genom 5: 328–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K (2005) Deficiency of glutaredoxin 5 reveals Fe–S clusters are required for vertebrate haem synthesis. Nature 436: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Xu XM, Moller SG (2006) AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J 25: 900–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M (2004) The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron–sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16: 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Abdel-Ghany SE, Anderson TD, Pilon-Smits EA, Pilon M (2006a) CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe–S cluster formation. J Biol Chem 281: 8958–8969 [DOI] [PubMed] [Google Scholar]
- Ye H, Pilon M, Pilon-Smits EA (2006b) CpNifS-dependent iron–sulfur cluster biogenesis in chloroplasts. New Phytol 171: 285–292 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data