Abstract
Autophagy is a newly recognized innate defense mechanism, acting as a cell-autonomous system for elimination of intracellular pathogens. The signals and signalling pathways inducing autophagy in response to pathogen invasion are presently not known. Here we show that autophagy is controlled by recognizing conserved pathogen-associated molecular patterns (PAMPs). We screened a PAMP library for effects on autophagy in RAW 264.7 macrophages and found that several prototype Toll-like receptor (TLR) ligands induced autophagy. Single-stranded RNA and TLR7 generated the most potent effects. Induction of autophagy via TLR7 depended on MyD88 expression. Stimulation of autophagy with TLR7 ligands was functional in eliminating intracellular microbes, even when the target pathogen was normally not associated with TLR7 signalling. These findings link two innate immunity defense systems, TLR signalling and autophagy, provide a potential molecular mechanism for induction of autophagy in response to pathogen invasion, and show that the newly recognized ability of TLR ligands to stimulate autophagy can be used to treat intracellular pathogens.
Keywords: autophagy, HIV, LC3, TLR, tuberculosis
Introduction
Autophagy is a fundamental cellular homeostatic process, where cells ingest and digest portions of their own cytoplasm, thus periodically cleansing their interiors (Levine and Klionsky, 2004). Autophagy is based on formation within the cytosol of double-membrane organelles termed autophagosomes to sequester portions of the cytoplasm earmarked for autophagic removal or turnover (Mizushima et al, 2002). Autophagosomes fuse with lysosomes to form autolysosomes, resulting in degradation of the captured cytosolic constituents, including (i) long-lived proteins and other stable macromolecules and (ii) membranous structures such as damaged, spent or surplus organelles (Levine and Klionsky, 2004). Autophagy endows cells with a capability to adjust down their biomass and turn over their own constituents at times of starvation (Lum et al, 2005). This provides amino acids and other components for the synthesis of essential proteins and other macromolecules allowing the cells to survive. Autophagy also removes faulty organelles such as spuriously damaged or leaky mitochondria lest cells undergo unscheduled apoptosis and die. All eukaryotic cells from yeast to man are capable of undergoing autophagy, and most cells in the human body can activate this process (Levine and Klionsky, 2004) or even undergo a considerable level of constitutive autophagy (Schmid et al, 2007).
Since autophagy affects many cell types, it has a broad effect on a wide range of normal physiological processes, including ageing and diseases such as cancer (Levine, 2007) and neurodegeneration (Alzheimer's, Huntington's and Parkinson's diseases and ataxias; Rubinsztein, 2006). It has recently been recognized that akin to the role of autophagy in eliminating toxic protein aggregates and thus protecting against neurodegeneration (Nixon, 2006), autophagy also plays a role in innate immunity against intracellular pathogens (Deretic, 2005; Levine and Deretic, 2007; Schmid and Munz, 2007), by clearing microbes directly via ingestion into autophagosomes for subsequent degradation in autolysosomes (Gutierrez et al, 2004; Nakagawa et al, 2004; Ogawa et al, 2005; Andrade et al, 2006; Orvedahl et al, 2007). Agonist-induced autophagosomes capture and destroy intracellular pathogens even when they are safely ensconced in protective vacuoles, such as the immature phagosome harbouring Mycobacterium tuberculosis (Singh et al, 2006), or parasitophorous vacuoles containing Toxoplasma gondii (Andrade et al, 2006; Ling et al, 2006). In addition to these cell-autonomous protective functions against invading pathogens, autophagy participates in other aspects of immunity (Deretic et al, 2006; Levine and Deretic, 2007; Schmid and Munz, 2007) and is not limited to a role in direct elimination of invading bacteria, protozoans and viruses. For example, autophagy supports sequestration of endogenously synthesized viral or self-antigens into autophagosomes and their delivery to major histocompatibility complex (MHC) class II loading compartments, leading to MHC class II-restricted presentation of cytoplasmic antigens (Schmid and Munz, 2007). Autophagy has a role in T-cell homeostasis (Li et al, 2006), for example by controlling T-cell lifespan once they exit the thymus (Pua et al, 2007), and is an effector of Th1–Th2 polarization in defense against intracellular pathogens (Harris et al, 2007). The role of autophagy in immunity has been further underscored by the recent recognition of a genetic association between a Chron's disease-susceptibility locus with one of the core autophagy genes, Atg16 (Hampe et al, 2007), and with the human immunity-related GTPase IRGM (Parkes et al, 2007) implicated in autophagy (Singh et al, 2006).
Two of the remaining top-tier questions in the field of autophagy are how the presence of potential autophagic targets is detected by the cell and how these targets become earmarked for autophagic degradation. These issue are complicated by the inherent diversity of autophagic targets, represented by a variety of macromolecules, protein aggregates and an assortment of organelles, including mitochondria (Lemasters, 2005), peroxisomes (Farre and Subramani, 2004) and endoplasmic reticulum (Bernales et al, 2006). In yeast, unique proteins select the targets for autophagy-related processes known as the Cvt pathway (a specialized zymogen transport and maturation pathway; Kim et al, 2001) and pexophagy (autophagy of peroxisomes; Rayapuram and Subramani, 2006). In mammalian cells, the initial information points towards ubiquitylation, suggesting that ubiquitin may be a part of the molecular recognition at least in the case of protein aggregates (Bjorkoy et al, 2005). While this issue has only begun to be addressed for endogenous cellular targets, even less is known regarding signals that lead to the recognition of intracellular pathogens. In this work, we have considered the possibility that innate immunity pattern recognition receptors (PRRs) have a role in activation of autophagy upon detection of pathogen molecules. PRRs such as Toll-like receptors (TLRs), Nods and Nod-like receptors (NLRs), RIG-I and RIG-I-like receptors (RLRs) detect the presence of microbial invaders by recognizing pathogen-associated molecular patterns (PAMPs; Medzhitov, 2007). PRRs specialize for a subset of cellular compartments that they patrol and for unique classes of microbial products that they recognize. It is generally believed that topologically external spaces, including vacuolar compartments, are covered by one or a combination of different TLRs, whereas the cytosol is under the surveillance by NLR and RLR molecular complexes. It has been shown that autophagic machinery can deliver PAMPs to endosomal TLRs (Li et al, 2006), but the question of whether the reverse is true, that is, whether PAMPs can stimulate autophagy once recognized by TLR molecules, has not been addressed. Here we report a screen with a panel of PAMPs for their capacity to induce autophagy and show that a subset of TLR ligands can activate autophagy. The TLR7 ligand single-stranded RNA (ssRNA) and other TLR7 agonists are potent inducers of autophagy. This validates the hypothesis that PRR-based recognition of PAMPs serves as a detector for microbial presence and induces autophagy as a defense mechanism against invading pathogens, thus bridging the two seemingly separate innate-immunity systems.
Results
TLR ligands induce formation of LC3 puncta in macrophages
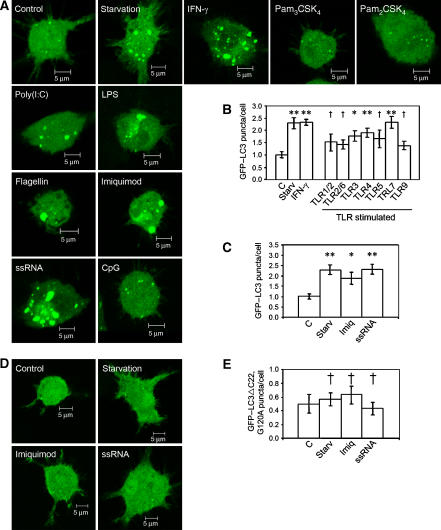
Autophagy induction in various systems is invariably monitored by an assay that depends on translocation of the autophagosome protein LC3 (Atg8) from the cytosol (diffuse cytosolic distribution) to newly formed autophagosomes, which appear as cytoplasmic puncta (Kabeya et al, 2000). To test whether recognition of microbial PAMPs via TLR molecules can induce an autophagic response, we screened a panel of standard PAMP ligands at standard concentrations (see Supplementary data) in macrophages for induction of LC3 puncta. RAW 264.7 macrophages were transfected with green fluorescent protein–LC3 (GFP–LC3) and 24 h later incubated in starvation media (positive control for autophagy induction), or in complete media with or without individual PAMPs. As a measure of autophagy, GFP–LC3 puncta (⩾1 μm) were quantified in >100 cells per sample (Figure 1; Supplementary Figure S1), in the absence of any further manipulation (Figure 1A–C) or in the presence of autolysosomal protease inhibitors, to prevent degradation of formed GFP–LC3 puncta (Supplementary Figure S1). A significant induction of GFP–LC3 puncta formation was observed with TLR3, TLR4 and TLR7 ligands. No significant increase in LC3 puncta was detected with synthetic bacterial lipopeptides, Pam3CSK4 (TLR1/TLR2 ligand) and Pam2CSK4 (TLR2/TLR6 ligand), Salmonella typhimurium flagellin (TLR5 ligand) and a CpG oligonucleotide (TLR9 ligand; Figure 1A and B; Supplementary Figure S1). Poly(I:C), a TLR3 ligand, and lipopolysaccharide (LPS), a TLR4 ligand, evoked an increase in GFP–LC3 puncta (Figure 1A and B). To ascertain TLR's presence and their responsiveness to all ligands tested, and thus validate those results that were negative, we carried out an nuclear factor-κB (NF-κB) activation assay using a NF-κB–luciferase reporter construct. All TLR ligands tested, with the exception of poly(I:C) and bacterial flagellin, induced NF-κB activation (Supplementary Figure S2A). Poly(I:C) was nevertheless able to stimulate the cells, as detected by induction of interferon-β (IFN-β) secretion (Supplementary Figure S2B), although no IκB-α degradation or Janus kinase (JNK) phosphorylation were detected (Supplementary Figure S2D and E). Whereas TLR5 (flagellin receptor) was present in RAW 264.7 macrophages, as shown by western blotting (Supplementary Figure S2C), no NF-κB activation (Supplementary Figure S2A), IκB-α degradation or JNK phosphorylation was detected with flagellin (Supplementary Figure S2D and E). Activation of NF-κB, detected by direct measurement (Supplementary Figure S2A) or in correlation with IκB-α degradation (Supplementary Figure S2D), or transient JNK phosphorylation (Supplementary Figure S2D and E), detected for all other TLR ligands, was however not sufficient to induce LC3 puncta formation (Figure 1A and B; Supplementary Figure S1). Although no induction of GFP–LC3 puncta was observed after TLR2 activation with individual lipopeptides (Figure 1B; Supplementary Figure S3A), zymosan (a more complex TLR2 agonist engaging additional receptors) was a strong inducer of GFP–LC3 puncta (Supplementary Figure S3A and B), indicating that in some instances a combination of receptors was required for induction of autophagy.
Figure 1.
TLR7 ligands are strong inducers of LC3 puncta. (A) RAW 264.7 macrophages cells were transfected with GFP–LC3 and after 24 h cells were incubated for 2 h in starvation media or for 4 h in complete media alone or in the presence of 500 U/ml IFN-γ, 1 μg/ml Pam3CSK4, 100 ng/ml Pam2CSK4, 25 μg/ml poly(I:C), 500 ng/ml LPS, 1 μg/ml S. typhimurium flagellin, 10 μg/ml Imiquimod, 10 μg/ml ssRNA or 3 μM CpG oligonucleotide 1826. Bar, 5 μm. (B, C) Quantification of GFP–LC3 puncta (⩾1 μm) in RAW 264.7 macrophages in panel A. Data are means±s.e.m. (n⩾3); **P<0.01, *P<0.05, †P⩾0.05 (analysis of variance (ANOVA)). (D) RAW 264.7 macrophages cells were transfected with GFP–LC3ΔC22,G120A and after 24 h cells were incubated for 2 h in starvation media (Starv) or 4 h in complete media alone (C) or in the presence of 10 μg/ml imiquimod (Imiq) or 10 μg/ml ssRNA. Bar, 5 μm. (E) Quantification of GFP–LC3ΔC22,G120A puncta (⩾1 μm) in RAW 264.7 macrophages in panel D. Data are means±s.e.m. (n=6); †P⩾0.05 (ANOVA).
ssRNA, a TLR7 ligand, induced the most prominent increase in LC3 puncta formation (Figure 1A and B). Stimulation with ssRNA was comparable to starvation used as a gold standard for autophagy induction (Seglen and Bohley, 1992), or to the previously reported (Gutierrez et al, 2004) induction with an immunologically relevant agonist IFN-γ (Figure 1A and B). We further examined whether stimulation of TLR7 induced autophagy, by employing a different TLR7 ligand, imiquimod (R837), an imidazoquinoline amine guanosine analogue often used as a conventional TLR7 ligand (Hemmi et al, 2002; Lee et al, 2003). Increased formation of LC3 puncta was detected upon stimulation with imiquimod (Figure 1A and C). This was additionally confirmed by four-dimensional live confocal microscopy (Supplementary Movie 1).
To confirm that GFP–LC3 puncta formation induced by ssRNA and imiquimod is a reflection of autophagy activation and not nonspecific protein aggregation (Kuma et al, 2007), we employed a control based on GFP fusion with a mutant LC3 form, LC3ΔC22,G120A (Kabeya et al, 2000). This LC3 mutant lacks the last 22 residues and a C-terminal glycine, disabling it for C-terminal lipidation with phosphatidylethanolamine (PE) by the autophagic Atg7–Atg3–PE conjugation system (Kabeya et al, 2000). The PE posttranslational modification of the C-terminal Gly residue in LC3 is required for LC3 association with autophagic membranes and autophagosome elongation (Kabeya et al, 2000). Figure 1D and E show that the number of GFP–LC3Δ22,G120A puncta per cell did not change upon stimulation with imiquimod, ssRNA or starvation, demonstrating that GFP–LC3 puncta formation following addition of TLR7 ligands in our experiments was dependent on a functional LC3 association with growing autophagosomes.
TLR7 ligands induce autophagy
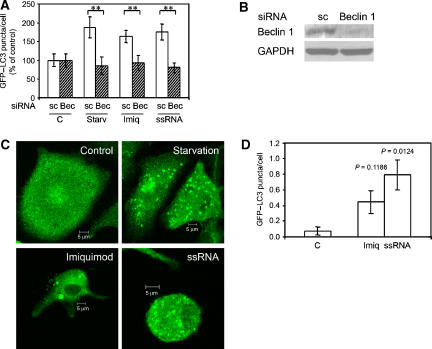
To demonstrate that the process detected as an increase in LC3 puncta was indeed a bona fide autophagy, we carried out a panel of additional experiments and assays. Beclin 1 (Liang et al, 1999) is a critical autophagic protein (Atg6) required for induction of autophagy and execution of the various stages along the autophagic pathway. The TLR7 ligands imiquimod and ssRNA, as well as the starvation control, induced fewer GFP–LC3 puncta (Figure 2A) when Beclin 1 levels were knocked down by short interfering RNA (siRNA) (Figure 2B), Thus, imiquimod and ssRNA induced autophagy in a Beclin 1-dependent manner.
Figure 2.
TLR7 ligands induce autophagy. (A) Quantification of GFP–LC3 puncta (⩾1 μm) in RAW 264.7 macrophages cells co-transfected with GFP–LC3 and either control scrambled siRNA (sc) or Beclin 1 siRNA. After 22 h cells were incubated for 4 h with complete media alone (C) or in the presence of 10 μg/ml imiquimod (Imiq) or 10 μg/ml ssRNA, or incubated for the last 2 h in starvation medium (Starv). Data are means±s.e.m. (n=6); **P<0.01 (ANOVA). (B) RAW 264.7 macrophages were transfected with control scrambled siRNA (sc) or Beclin 1 siRNA and after 24 h cells were lysed and analysed by western blotting using anti-Beclin 1 or anti-GAPDH antibodies. (C) Primary BMMs derived from GFP–LC3 transgenic mice were incubated for 2 h with starvation media or 4 h with complete media alone (control) or in the presence of 10 μg/ml imiquimod or 10 μg/ml ssRNA. Bar, 5 μm. (D) Quantification of GFP–LC3 puncta (⩾1 μm) in BMMs expressing GFP–LC3 in panel C. Cells were stimulated with complete media alone (C), imiquimod (Imiq) or ssRNA. Data are means±s.e.m. (n=3). Exact P-values (ANOVA) are shown. A full-colour version of this figure is available at The EMBO Journal Online.
We next examined the autophagic response in primary cells, using murine bone marrow macrophages (BMMs) derived from knock-in transgenic mice expressing GFP–LC3 (Mizushima et al, 2004). Unlike the transiently transfected cells, primary macrophages expressing endogenous GFP–LC3 displayed none or very few background GFP–LC3 puncta when not stimulated for autophagy (Figure 2C and D). When autophagy was stimulated by starvation, BMMs showed increase in GFP–LC3 puncta (Figure 2C). Imiquimod and ssRNA elicited mild GFP–LC3 puncta response (Figure 2C and D). These data indicate that although primary macrophages can react to TLR stimulation by increase in autophagy markers, this response may be under a tighter control in primary cells relative to RAW 264.7 cells.
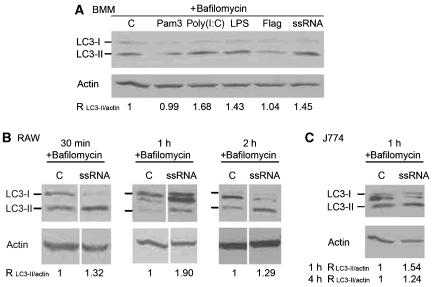
LC3-I lipidation, which is a hallmark of autophagy induction, can be monitored by electrophoretic mobility shift from the slower migrating, non-lipidated LC3-I form to the lipidated and faster moving LC3-II form (Kabeya et al, 2000). We used this autophagy assay to examine TLR ligand effects on murine primary macrophages (BMM) (Figure 3A) and murine macrophage cell lines (RAW 264.7 and J774; Figure 3B and C). Immunoblot analysis of LC3 bands was carried out in the presence of Bafilomycin A1 (Mizushima and Yoshimori, 2007), an inhibitor of autophagic degradation/flux. The TLR ligands Poly(I:C) (TLR3), LPS (TLR4) and ssRNA (TLR7) induced LC3-II conversion, whereas Pam3CSK4 (TLR1/TLR2) and flagellin (TLR5) did not (Figure 3A), in keeping with the results of the LC3 puncta assay (Figure 1). A positive response to the TLR7 ligand ssRNA was also detected in RAW 264.7 (Figure 3B) and J774 (Figure 3C) cells monitored at different times following ssRNA challenge. The response to TLR7 ligands was further confirmed in the absence of inhibitors of autophagic degradation with ssRNA (Supplementary Figure S4A) and imiquimod (Supplementary Figure S4B). As positive control for autophagy induction, rapamycin (Supplementary Figure S4A and B) and IFN-γ (Supplementary Figure S4B) were used to treat RAW 264.7 cells. The identity of LC3-I and LC3-II bands was confirmed (due to appearance of an additional band in Figure 3B and C) by siRNA knockdown of LC3B (Supplementary Figure S4A, lane 1), one of the five murine Atg8 paralogues (MAP1LC3A, MAP1LC3B, GABARAP, GABARAPL1, GABARAP); LC3B is the mammalian Atg8 invariably monitored in autophagic studies (Kabeya et al, 2004). In conclusion, the TLR7 ligands induced conversion of LC3 from LC3-I to LC3-II, consistent with induction of autophagy.
Figure 3.
Immunoblot analysis of LC3-I to LC3-II conversion in cells stimulated with TLR ligands. (A) Primary BMMs were incubated in the presence of 100 nM Bafilomycin A for 2 h in complete media alone (C) or in the presence of 1 μg/ml Pam3CSK4, 25 μg/ml poly(I:C), 500 ng/ml LPS, 1 μg/ml S. typhimurium flagellin or 10 μg/ml ssRNA. Cells were lysed and analysed by immunoblotting using anti-LC3 or anti-actin antibodies. Densitometric LC3-II/actin ratios are shown (average from three experiments). (B) RAW 264.7 macrophages were incubated in the presence of 100 nM Bafilomycin A for 30 min, 1 h or 2 h in complete media alone (C) or in the presence of 10 μg/ml ssRNA. Cells were lysed and analysed by immunoblotting using anti-LC3 or anti-actin antibodies. Densitometric LC3-II/actin ratios are shown as averages from two (30 min and 2 h) or three independent experiments (1 h). (C) J774 macrophages were incubated in the presence of 100 nM Bafilomycin A for 1 h in complete media alone (C) or in the presence of 10 μg/ml ssRNA. Cells were lysed and analysed by immunoblotting using anti-LC3 or anti-actin antibodies. Blot, 1 h sample. Densitometric LC3-II/actin ratios for 1 and 4 h incubation time points are shown underneath the blot.
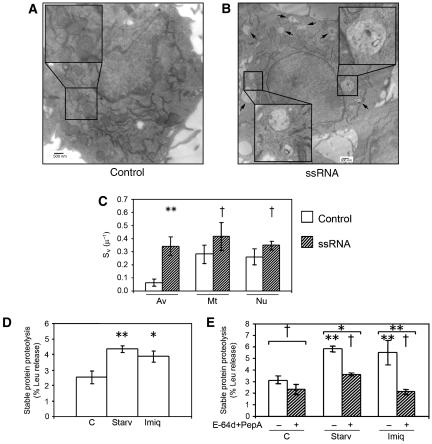
Next, we monitored PAMP-induced autophagy by two LC3-independent criteria, (i) detection of autophagic organelles by ultrastructural analysis using electron microscopy and (ii) autophagic proteolysis of stable polypeptides. An ultrastructural analysis of RAW 264.7 cells treated with ssRNA for 2 or 4 h revealed presence of typical autophagosomal profiles at later stages of maturation, as shown in Figure 4 and in Supplementary Figure S5A and B and enlarged sections, where electron-dense material (denser than the cytoplasm surrounding the vacuole) was seen, characteristically present in autolysosomes due to ribosomal degradation (Eskalinen, 2008). These organelles were absent from the control untreated cells (Figure 4A). A stereometric quantification of organellar surface density in a volume (Sv) (Weibel and Bolender, 1973) showed that, upon stimulation with ssRNA, only autophagic vacuoles increased their Sv value from 0.06±0.03 to 0.34±0.07 μ−1, whereas mitochondria or nuclei did not show a statistically significant change in their Sv values (Figure 4C).
Figure 4.
Assessment of autophagy induction with TLR7 ligands by ultrastructural analysis and by monitoring degradation of long-lived proteins. (A, B) Electron microscopy of RAW 264.7 macrophages incubated for 4 h with (A) complete media alone (control) or (B) in the presence of 10 μg/ml ssRNA. Arrows and enlarged areas indicate autophagic organelles. (C) Quantification of organellar surface in a volume (Sv) (Weibel and Bolender, 1973) for autophagic vacuoles (Eskalinen, 2008), mitochondria and nuclei. **P<0.05, †P⩾0.05 (ANOVA). (D) Proteolysis of long-lived proteins was measured in RAW 264.7 cells labelled for 24 h in media containing [3H]leucine. Cells were washed, incubated for 24 h in complete medium (containing cold leucine) and incubated in starvation media (Starv) for 4 h or in full media alone (C) or in complete media supplemented with 10 μg/ml imiquimod (Imiq) for 24 h. Leucine release was calculated from radioactivity in the tricarboxylic acid-soluble form relative to total cell radioactivity. Data are means±s.e.m. (n=9); **P<0.01, *P<0.05 (ANOVA). (E) Proteolysis of long-lived proteins was measured in RAW 264.7 cells labelled as in panel D, with 1 h of preincubation with 10 μg/ml E-64d and 10 μg/ml pepstatin A in complete media before stimulation, and stimulated as in panel D but in the presence of 10 μg/ml E-64d and 10 μg/ml pepstatin A. Data are means±s.e.m. (n=3); **P<0.01, *P<0.05, †P⩾0.05 (ANOVA) relative to the corresponding control. Symbols placed over the lines indicate significance between samples from same condition group with and without E-64d+pepstatin A; symbols under the lines indicate statistical significance relative to control (C).
Degradation of long-lived proteins represents an end-point autophagy assay, relying on macroautophagic proteolysis of stable polypeptides. It is often used to examine whether a given process represents autophagy and to test whether the autophagic pathway has been executed in full, as it depends on conversion of autophagosomes into degradative autolysosomal organelles (Meley et al, 2006). Increased proteolysis of stable polypeptides was observed when autophagy was induced by starvation (Figure 4D). Upon stimulation of cells with the TLR7 ligand imiquimod, a similar increase in degradation of stable proteins was detected (Figure 4D). In control samples, employing autophagic proteolysis inhibitors E-64d and pepstatin A, both starvation- and imiquimod-induced degradation of stable proteins were equally inhibited by this treatment (Figure 4E). A full (with the exception of LPS and zymosan) PAMP panel analysis for induction of stable protein degradation (Supplementary Figure S6) was in keeping with the results obtained with LC3 assays. The results of ultrastructural analyses, proteolysis assays, LC3 puncta and LC3 lipidation tests, and dependence on Beclin 1 demonstrate that TLR7 ligands induce autophagy.
ssRNA induces autophagy through TLR7
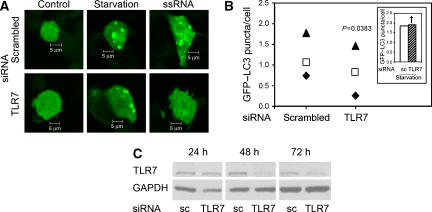
After demonstrating that imiquimod and ssRNA induce autophagy, we examined whether these compounds activated autophagy via TLR7. TLR7 was knocked down in RAW 264.7 cells with siRNA (Figure 5A–C). Macrophages were stimulated with a TLR7 ligand and autophagy induction quantified by GFP–LC3 puncta assay (Figure 5A and B). Cells were incubated in complete media in the absence or presence of ssRNA and compared with starved cells. Although necessary manipulations increased background levels of LC3 puncta in some experiments, there was significant induction of autophagy upon starvation or ssRNA stimulation in cells transfected with the control scrambled siRNA compared with cells transfected with TLR7 siRNA (Figure 5A and B). Importantly, TLR7 knockdown did not change LC3 response to starvation (Figure 5B, inset), demonstrating that TLR7 was required specifically for TLR ligand-induced autophagy, but not for starvation-induced autophagy. As a further control, cells transfected with scrambled siRNA or with TLR7 siRNA had the same level of GFP–LC3 puncta in the complete medium (not shown). TLR7 knockdown, relative to scrambled siRNA control, also diminished LC3-II levels assessed by immunoblotting upon stimulation with ssRNA (Supplementary Figure S7A) or imiquimod (Supplementary Figure S7B). Electron microscopic morphometry of autophagosomal organelles induced by ssRNA indicated that TLR7 knockdown by siRNA diminished Sv from 0.49±0.1 to 0.19±0.05 μ−1 (Supplementary Figure S5D). Thus, induction of autophagy by ssRNA and imiquimod is TLR7-dependent.
Figure 5.
TLR7 is responsible for ssRNA-induced autophagy. (A) RAW 264.7 macrophages cells were co-transfected with GFP-LC3 and control scrambled (sc) or TLR7 siRNA. After 46 h, cells were incubated for 4 h with complete media alone (control), in the presence of 10 μg/ml ssRNA (ssRNA), or incubated for the last 2 h in the starvation medium (starvation). Bars, 5 μm. (B) Quantification of GFP–LC3 puncta (⩾1 μm) in RAW 264.7 macrophages transfected as in panel A and stimulated with 10 μg/ml ssRNA. Inset, quantification of GFP–LC3 puncta in RAW 264.7 macrophages transfected in panel A and stimulated 2 h in starvation media. Symbols denote paired data from same experiments: ▴, experiment 1; □ experiment 2; ♦ experiment 3. P-value, paired t-test. (C) RAW 264.7 macrophages were transfected with control scrambled siRNA (sc) or TLR7 siRNA and after 24, 48 or 72 h cells were lysed and analysed by western blotting using anti-TLR7 or anti-GAPDH antibodies.A full-colour version of this figure is available at The EMBO Journal Online.
TLR7 stimulation activates autophagy via MyD88
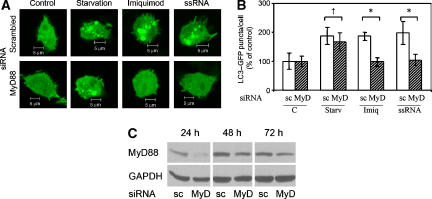
As all known effector functions of TLR7 signalling depend on the MyD88 adapter protein (Lee and Kim, 2007), we next tested whether MyD88 was required for TLR7 induction of autophagy (Figure 6A–C). Figure 6C shows a temporal analysis of MyD88 knockdown, with optimal MyD88 knockdown being achieved early, at 24 h post siRNA transfection. Macrophages were co-transfected with GFP–LC3 along with control (scrambled) siRNA or MyD88 siRNA. Following a 24-h incubation period, cells were maintained for 4 h in complete medium (control), or in complete medium supplemented with imiquimod or ssRNA. Another sample treated with siRNA was run in parallel and was subjected to starvation for the last 2 h. Treatment of cells with imiquimod, ssRNA and starvation stimulated LC3 puncta with equal potency in cells co-transfected with the scrambled siRNA (Figure 6A and B). However, in cells subjected to MyD88 knockdown, induction of autophagy was abrogated specifically in response to imiquimod or ssRNA, but not starvation (Figure 6A and B). These relationships were further confirmed by LC3 immunoblotting, using imiquimod and ssRNA in the presence of inhibitors of autolysosomal degradation (Supplementary Figure S8), with MyD88 knockdown diminishing relative amounts of LC3-II under induction conditions. These results demonstrate that induction of autophagy via TLR7 requires MyD88 as a downstream adapter, similar to other, previously known effector functions of TLR7.
Figure 6.
Autophagy induced by TLR7 ligands depends on MyD88. (A) Confocal microscopy images of RAW 264.7 macrophages co-transfected with GFP-LC3 and control scrambled siRNA (scrambled) or MyD88 siRNA (MyD88). After 22 h, cells were incubated for 4 h in complete media alone (control) or in complete media supplemented with 10 μg/ml imiquimod or 10 μg/ml ssRNA, or incubated for the last 2 h in the starvation medium. Bars, 5 μm. (B) Quantification of GFP–LC3 puncta (⩾1 μm) from experiments illustrated in panel A. Data are means±s.e.m. (n=3); *P<0.05, †P⩾0.05 (ANOVA). (C) RAW 264.7 macrophages were transfected with control scrambled siRNA (sc) or MyD88 siRNA (MyD) and after 24, 48 or 72 h cells were lysed and analysed by immunoblotting using anti-MyD88 or anti-GAPDH antibodies. A full-colour version of this figure is available at The EMBO Journal Online.
Induction of autophagy by TLR7 ligands can eliminate intracellular pathogens
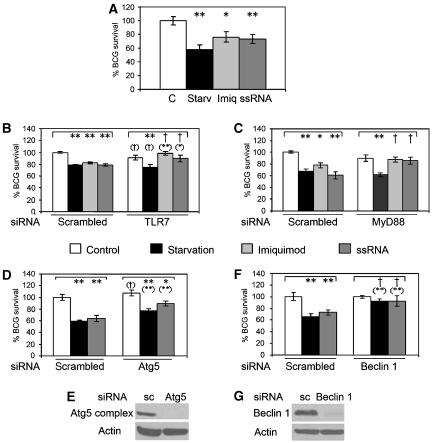
Starvation-induced autophagy can kill intracellular mycobacteria (Gutierrez et al, 2004). As our results presented here indicated that TLR7 was a potent inducer of autophagy, we wondered whether TLR7 ligands could be used as heterologous ligands to induce anti-mycobacterial effects in macrophages, with potential therapeutic implications. We tested whether stimulation of autophagy with TLR7 ligands can reduce viability of mycobacteria in infected macrophages. Following phagocytosis of M. tuberculosis var. bovis Bacille Calmette-Guérin (BCG), macrophages were stimulated for 4 h with TLR7 ligands or subjected to starvation. After incubation, cells were lysed and lysates plated for colony-forming unit counts. Imiquimod or ssRNA treatment enabled macrophages to decrease BCG survival (Figure 7A). Treatment of infected cells with Pam2CSK4 or CpG (TLR ligands not inducing autophagy in our system) did not affect BCG survival significantly (Supplementary Figure S9). We next established that stimulation with imiquimod or ssRNA caused BCG killing via TLR7, comparing cells subjected to TLR7 siRNA knockdown with those transfected with scrambled control siRNA (Figure 7B). In cells with TLR7 knockdown, differences in BCG survival following imiquimod or ssRNA stimulation were abrogated (Figure 7B). The effect of TLR7 knockdown did not affect autophagic BCG killing induced by starvation (Figure 7B). Similarly, knockdown of MyD88 also abrogated TLR7-ligand induced BCG elimination, but did not affect starvation-induced autophagic killing of BCG (Figure 7C). Finally, the effects on BCG survival in macrophages stimulated with TLR7 ligands were dependent on autophagic machinery, as Atg5 knockdown diminished BCG elimination upon ssRNA treatment, showing, as expected, effects similar to those when autophagy was induced by starvation (Figure 7D). A knockdown of another autophagy factor, Beclin 1, completely abrogated the killing effects of starvation and ssRNA treatments (Figure 7F). Thus, a TLR agonist that can induce autophagy in infected macrophages can control an infection caused by a pathogen not normally associated with stimulation of TLR7. This suggests a potential therapeutic use of TLR ligands in treatment of non-cognate infectious agents through induction of autophagy as a generic cell-autonomous mechanism for elimination of intracellular microbes.
Figure 7.
TLR-induced autophagy eliminates intracellular BCG. (A) RAW 264.7 macrophages were infected with BCG for 1 h, washed and incubated for 4 h in complete media alone (C) or in the presence of 10 μg/ml imiquimod (Imiq) or 10 μg/ml ssRNA, or incubated in starvation medium (Starv). Cells were lysed to quantify bacterial survival by counting colony-forming units. Data are means±s.e.m. (n=5); **P<0.01, *P<0.05 (ANOVA). (B) RAW 264.7 macrophages were transfected with control scrambled siRNA (sc) or TLR7 siRNA. After 46 h cells were infected, washed, incubated for 4 h and lysed as in panel A. Data are means±s.e.m. (n=3); **P<0.01, *P<0.05, †P⩾0.05 (ANOVA) relative to control. Symbols in parentheses indicate significance relative to the equally treated cells from the scrambled siRNA group. (C) RAW 264.7 macrophages were transfected with control scrambled siRNA (sc) or MyD88 siRNA. After 24 h cells were infected, washed, incubated for 4 h and lysed as in panel A. Data are means±s.e.m. (n=6); **P<0.01, *P<0.05, †P⩾0.05 (ANOVA) relative to control. (D) RAW 264.7 macrophages cells were transfected with control scrambled siRNA (sc) or Atg5 siRNA. After 24 h cells were infected, washed, incubated for 4 h and lysed as in panel A. Data are means±s.e.m. (n=6); **P<0.01, *P<0.05, †P⩾0.05 (ANOVA) relative to control. Symbols in parentheses indicate statistical significance relative to the equally treated cells from the scrambled siRNA group. (E) RAW 264.7 macrophages cells were transfected as in panel D. After 24 h cells were lysed and analysed by immunoblotting using anti-Atg5 or anti-actin antibodies. (F) RAW 264.7 macrophage cells were transfected with control scrambled siRNA (sc) or Beclin 1 siRNA. After 24 h, cells were infected, washed, incubated for 4 h and lysed as in panel A. Data are means±s.e.m. (n=6); **P<0.01, *P<0.05, †P⩾0.05 (ANOVA) relative to control. Symbols in parentheses indicate significance relative to the equally treated cells from the scrambled siRNA group. (G) RAW 264.7 macrophages cells were transfected as in panel F. After 24 h cells were lysed and analysed by immunoblotting using anti-Beclin 1 or anti-actin antibodies.
We also tested whether infection with a pathogen known to present natural ligands to TLR7 or TLR8 can induce autophagy. One such infectious agent is HIV, with HIV-derived guanosine- and uridine-rich ssRNA known to stimulate TLR7/TLR8 in human cells (Heil et al, 2004). The role for TLR7/TLR8 stimulation in HIV infection has been implicated both as an antiviral defense during acute HIV infection (Beignon et al, 2005; Schlaepfer et al, 2006) and in development of AIDS through immune activation (Meier et al, 2007) and replication of the latent virus (Schlaepfer et al, 2006). When HeLa cells were infected with vesicular stomatitis virus-G-pseudotyped HIV virus, we detected increase in relative levels of LC3-II (Supplementary Figure S10A and B). The increase in LC3-II levels, indicative of autophagy induction, was TLR8-dependent, as TLR8 knockdown with siRNA abrogated relative increase in LC3-II (Supplementary Figure S10B). Thus, autophagy induction in response to natural TLR7 or TLR8 ligands can be detected during infection with HIV.
Discussion
Autophagy has a role in innate and adaptive immunity (Levine and Deretic, 2007; Schmid and Munz, 2007) as a mechanism for elimination of intracellular bacteria, including M. tuberculosis, Listeria monocytogenes, Streptococcus pyogenes, Salmonella and Shigella (Rich et al, 2003; Gutierrez et al, 2004; Nakagawa et al, 2004; Ogawa et al, 2005; Birmingham et al, 2006); viruses such as Herpes simplex virus (Orvedahl et al, 2007) and protozoans exemplified by T. gondii (Rich et al, 2003; Andrade et al, 2006; Ling et al, 2006). This is in keeping with the cellular maintenance function of autophagy and its role in removal of harmful objects from the cytoplasm (Deretic, 2005). The signals that activate autophagy and molecular tags that guide autophagosomes to sequester the invading pathogens are a matter of current investigations. A large body of literature points to the capability of cells to recognize the presence of pathogens via PRRs such as TLRs. Here we have uncovered a connection between the two systems, TLR signalling and autophagy, thus linking pathogen recognition via PRRs and pathogen elimination through autophagy. Specifically, we found that ssRNA can activate TLR7 and induce autophagy in RAW 264.7 cells as a mechanism that can eliminate a non-cognate intracellular pathogen.
On the basis of multiple assays, a subset of TLR ligands can induce autophagy in murine macrophages, poly(I:C) (TLR3), LPS (TLR4) and ssRNA (TLR7). The strongest autophagy induction was observed with TLR7 ligands. Both ssRNA and imiquimod induced autophagy through TLR7. Showing a partially overlapping specificity pattern, the synthetic imidazoquinoline compound imiquimod activates both human TLR7 (hTLR7) and mouse TLR7 (mTLR7), but not human TLR8 (hTLR8) (Heil et al, 2003; Lee et al, 2003). In a partial contrast, GU-rich ssRNA is a ligand for mTLR7 and hTLR8, but not for hTLR7 in certain cells (Heil et al, 2004) or murine TLR8 (mTLR8), with mTLR8 responsive only to a combination of PAMPs (Gorden et al, 2006). Consistent with this reactivity pattern, we found that a marker of autophagy was induced in a TLR8-dependent manner when HeLa cells were infected with HIV, a known source of naturally occurring GU-rich ssRNA (Heil et al, 2004; Beignon et al, 2005; Schlaepfer et al, 2006; Meier et al, 2007). TLR7 is expressed in macrophages (Figure 5C), but its highest expression and its most prominent role are in plasmocytoid dendritic cells (pDCs) (Lee and Kim, 2007). Recently, it has been reported that in pDC autophagy can deliver a viral TLR7 ligand to the intracellular compartment where TLR7 localizes, thus bringing together the ligand and the receptor to initiate signalling leading to antiviral responses (Lee et al, 2007). Whereas this indicates that autophagy may enhance TLR recognition of PAMPs, the converse, that is, whether TLR induce or modulate autophagy, had not been addressed. Our present study indicates that autophagy may not be simply a peripheral pathway adding to TLR signalling, but that it could instead represent a previously unappreciated effector of TLR signalling. In support of this conclusion is a report that appeared while our study was in revision, indicating that TLR4 induces autophagy (Xu et al, 2007).
Although our screen has been focused on individual PAMPs, our own study with complex PAMP sources (e.g., zymosan) indicates that combinations of ligands may provoke autophagy in cases where individual PAMPs do not. Thus, we cannot rule out the possibility that Pam3CSK4, Pam2CSK4, bacterial flagellin or CpG oligonucleotide, the ligands that individually did not stimulate autophagy in our screen, can induce autophagy in certain cell types or in combination with additional inputs. A trivial explanation that the above ligands for TLR1/TLR2, TLR2/TLR6 and TLR9 when tested in our system simply did not activate the cells was experimentally ruled out, as these stimuli had the capacity to cause IκB-α degradation, activate NF-κB, and stimulate JNK phosphorylation. A different interpretation may apply to flagellin, a TLR5-specific ligand, as it also failed to activate NF-κB and did not induce JNK phosphorylation. It has been reported that RAW 264.7 macrophages do not express TLR5 and therefore do not respond to flagellin (Mizel et al, 2003). Although we detected TLR5 protein by western blotting (Supplementary Figure S2C), it is unlikely that TLR5 is functional in RAW 264.7 cells.
Can rules regarding which TLR is likely to induce autophagy be inferred from the downstream factors engaged by a specific TLR? TLR3 signalling is TRIF-dependent and MyD88-independent, TLR4 can be MyD88- or TRIF-dependent, whereas TLR7 is strictly MyD88-dependent (Lee and Kim, 2007). Thus, a simple correlation between the downstream signalling pathways induced by these TLRs cannot be drawn. This is further evidenced by the finding that not all MyD88-engaging TLRs (e.g., TLR9) induced autophagy in our study. A related question is how TLRs might induce autophagy. TLRs initiate common NF-κB/mitogen-activate protein kinase (MAPK) (extracellular signal-regulated kinase (ERK), p38 and JNK) and distinct IRF3/7 pathways to coordinate innate immunity and initiate adaptive immunity against diverse pathogens (Lee and Kim, 2007). TLR7 signalling can lead to MAPK activation (ERK, JNK and p38; Heil et al, 2003; Lund et al, 2004; Koziczak-Holbro et al, 2007). ERK and p38 activation has been implicated in autophagy, both as a positive and a negative factor, affecting the maturation step (Corcelle et al, 2006, 2007). A potential role for JNK in activating autophagy via modulation of Bcl-2–Beclin 1 interactions and Beclin 1 activity (Pattingre et al, 2005) cannot be excluded, despite our findings that JNK phosphorylation signal is of short duration and does not differentiate between autophagy-inducing and autophagy-non-inducing PAMPs at the time periods examined. In considering MAPK, there could be a connection between p38 activation and generation of reactive oxygen species (ROS) by NADPH oxidase activation (Laroux et al, 2005). It is known in the case of tumour necrosis factor-α that it induces autophagy via generation of ROS (Djavaheri-Mergny et al, 2006), while ROS have been implicated in induction of autophagy via Atg4 (Scherz-Shouval et al, 2007). Furthermore, interleukin-1 receptor-associated kinase-4, which is a key downstream kinase activated upon TLR7 stimulation (Koziczak-Holbro et al, 2007), phosphorylates p47phox and activates NADPH oxidase to generate ROS (Pacquelet et al, 2007). The likely relevance of ROS in autophagy induction is further underscored by our findings that zymosan (Supplementary Figure S3), in contrast to individual lipopeptides, stimulates LC3 puncta formation. Zymosan represents yeast cell wall particles that activate TLR2/TLR6, which induce inflammatory signalling, and Dectin-1, which induces phagocytosis and ROS generation (Underhill, 2003).
Among the alternative possibilities for TLR7 activation of autophagy are (a) induction of type I interferon (IFN-α and β) and (b) NF-κB activation. A pivotal role for type I interferon is unlikely as (i) RAW 264.7 macrophages stimulated with ssRNA for 4 h did not secrete IFN-β (Supplementary Figure S2B); (ii) it has been reported that IFN-α/β does not induce autophagy in RAW 264.7 macrophages (Gutierrez et al, 2004) and (iii) in experiments shown in Figure 1A and B, cells stimulated with poly(I:C) secreted 7–8 times more IFN-β than those stimulated with ssRNA (Supplementary Figure S2B), in inverse correlation relative to the more prominent autophagy induction with ssRNA than with poly(I:C). NF-κB activation downstream of TLR stimulation may more likely play an inhibitory role in autophagy regulation. A recent report has indicated that activation of NF-κB counteracts and represses autophagy (Djavaheri-Mergny et al, 2006). It is possible that robust NF-κB activation or modulation of some other factors may preclude efficient autophagy activation by TLR7 in some cells (Z Zhao and H Virgin, personal communication). Furthermore, several TLR ligands (Pam3CSK4, Pam2CSK4, CpG) caused IκB-α degradation (Supplementary Figure S2D) and activated NF-κB (Supplementary Figure S2A), but did not induce autophagy in our study.
The connections between TLR signalling and induction of autophagy shown here link two broad aspects of innate immunity, TLR signalling and autophagy. In addition, the relationships uncovered in this work open the possibility of putting them to practical use. The TLR7 ligand imiquimod is a prescription medication known under the trade name Aldara with therapeutic applications in patients with viral infections and certain cancers (Beutner et al, 1998; Edwards et al, 1998; Syed et al, 1998). Our data show that activation of macrophages with imiquimod reduces mycobacterial viability in infected macrophages. This suggests a potential application of the relationships shown in this work as a basis for treatment of early or latent M. tuberculosis infections. The link between TLR signalling and autophagy may prove to be an unanticipated but valuable application of the detailed knowledge of TLR-signalling pathways, now expanded to the elimination of pathogens through induction of autophagy.
Materials and methods
Cell and bacterial cultures
Murine RAW 264.7 and J774 macrophage cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS) and L-glutamine (complete media). HeLa cells were maintained in DMEM, 10% FBS. BMMs were derived from C57/BL6 mice or GFP–LC3 mice (Dr N Mizushima, Japan) as described previously (Via et al, 1998). M. tuberculosis var. bovis BCG was grown in Middlebrook 7H9 broth with 0.5% Tween, 0.2% glycerol and albumin–dextrose–catalase (ADC) supplement (BD Diagnostics, Franklin Lakes, NJ, USA) and homogenized to generate a single-cell suspension, or on 7H11 plates with 0.5% Tween, 0.2% glycerol and ADC.
Antibodies, TLR ligands, drugs, cytokines, siRNAs and DNA constructs
The rabbit polyclonal antibody against LC3 (T Ueno and E Kominami, Japan) was used at 1:500 dilution; mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), rabbit polyclonal anti-TLR7, rabbit polyclonal anti-MyD88, rabbit polyclonal anti-GFP, mouse monoclonal anti-TLR5 antibodies and mouse monoclonal anti-actin antibodies were from Abcam Inc. (Cambridge, MA, USA); goat polyclonal anti-Beclin 1 antibody (Santa Cruz Biotechnology Inc., CA, USA) was used at 1:200 dilution; mouse monoclonal anti-IκB-α, rabbit polyclonal anti-phospho-JNK and rabbit polyclonal anti-JNK antibodies were from Cell Signaling Technology Inc. (Danvers, MA, USA); rabbit polyclonal anti-Atg5 was from Novus Biologicals (Littleton, CO, USA); anti-rabbit Alexa-488-conjugated antibody was from Molecular Probes (Eugene, OR, USA). Lipopeptides Pam2CSK4 and Pam3CSK4, zymosan, polyinosine–polycytidylic acid poly(I:C), imiquimod (R837), ssRNA (ssRNA40/LyoVec) and the CpG oligonocleotide ODN1826 were from InvivoGen (San Diego, CA, USA). Mouse IFN-γ, LPS from Escherichia coli 026:B6, E-64d, pepstatin A, rapamycin and rabbit polyclonal anti-LC3B antibody were from Sigma-Aldrich (St Louis, MO, USA). Bafilomycin A1 was from LC Laboratories (Worburn, MA, USA). Secondary horseradish peroxidase-conjugated antibodies were from Pierce (Rockford, IL, USA). Control siRNA (siCONTROL non-targeting siRNA) and siRNAs for mouse LC3B, mouse Beclin 1, mouse Atg5, mouse TLR7, mouse MyD88 and human TLR8 (siGENOME SMARTpool siRNA) were from Dharmacon (Chicago, IL, USA). NF-κB-responsive luciferase reporter plasmid PathDetect® NF-κB cis-Reporting System was from Stratagene (La Jolla, CA, USA) and the β-galactosidase construct, pEF1-Bos, was from G Nunez (University of Michigan).
Methods
All other materials and methods are described in Supplementary data.
Supplementary Material
Supplementary Material
Supplementary movie
Acknowledgments
We thank Z Zhao and H Virgin for information on LC3-II response. This work was supported by National Institutes of Health grant AI069345 and in part by grants AI45148 and AI42999. ASD was an NIH T-32 AI07538 fellow.
References
- Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS (2006) CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest 116: 2366–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N (2005) Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor–viral RNA interactions. J Clin Invest 115: 3265–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutner KR, Spruance SL, Hougham AJ, Fox TL, Owens ML, Douglas JM Jr (1998) Treatment of genital warts with an immune-response modifier (imiquimod). J Am Acad Dermatol 38: 230–239 [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH (2006) Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 281: 11374–11383 [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcelle E, Djerbi N, Mari M, Nebout M, Fiorini C, Fenichel P, Hofman P, Poujeol P, Mograbi B (2007) Control of the autophagy maturation step by the MAPK ERK and p38: lessons from environmental carcinogens. Autophagy 3: 57–59 [DOI] [PubMed] [Google Scholar]
- Corcelle E, Nebout M, Bekri S, Gauthier N, Hofman P, Poujeol P, Fenichel P, Mograbi B (2006) Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res 66: 6861–6870 [DOI] [PubMed] [Google Scholar]
- Deretic V (2005) Autophagy in innate and adaptive immunity. Trends Immunol 26: 523–528 [DOI] [PubMed] [Google Scholar]
- Deretic V, Singh S, Master S, Harris J, Roberts E, Kyei G, Davis A, de Haro S, Naylor J, Lee HH, Vergne I (2006) Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol 8: 719–727 [DOI] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Amelotti M, Mathieu J, Besancon F, Bauvy C, Souquere S, Pierron G, Codogno P (2006) NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem 281: 30373–30382 [DOI] [PubMed] [Google Scholar]
- Edwards L, Ferenczy A, Eron L, Baker D, Owens ML, Fox TL, Hougham AJ, Schmitt KA (1998) Self-administered topical 5% imiquimod cream for external anogenital warts. HPV Study Group. Human papillomavirus. Arch Dermatol 134: 25–30 [DOI] [PubMed] [Google Scholar]
- Eskalinen E (2008) Fine structure of the autophagosome. In Autophagosome and Phagosome, Deretic V (ed), Humana Press vol. (in press) [Google Scholar]
- Farre JC, Subramani S (2004) Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol 14: 515–523 [DOI] [PubMed] [Google Scholar]
- Gorden KK, Qiu XX, Binsfeld CC, Vasilakos JP, Alkan SS (2006) Cutting edge: activation of murine TLR8 by a combination of imidazoquinoline immune response modifiers and polyT oligodeoxynucleotides. J Immunol 177: 6584–6587 [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753–766 [DOI] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M et al. (2007) A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 39: 207–211 [DOI] [PubMed] [Google Scholar]
- Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V (2007) T Helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity 27: 505–517 [DOI] [PubMed] [Google Scholar]
- Heil F, Ahmad-Nejad P, Hemmi H, Hochrein H, Ampenberger F, Gellert T, Dietrich H, Lipford G, Takeda K, Akira S, Wagner H, Bauer S (2003) The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur J Immunol 33: 2987–2997 [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S (2004) Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303: 1526–1529 [DOI] [PubMed] [Google Scholar]
- Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S (2002) Small antiviral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3: 196–200 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117: 2805–2812 [DOI] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA Jr, Klionsky DJ (2001) Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 153: 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziczak-Holbro M, Joyce C, Gluck A, Kinzel B, Muller M, Tschopp C, Mathison JC, Davis CN, Gram H (2007) IRAK-4 kinase activity is required for interleukin-1 (IL-1) receptor- and Toll-like receptor 7-mediated signaling and gene expression. J Biol Chem 282: 13552–13560 [DOI] [PubMed] [Google Scholar]
- Kuma A, Matsui M, Mizushima N (2007) LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3: 323–328 [DOI] [PubMed] [Google Scholar]
- Laroux FS, Romero X, Wetzler L, Engel P, Terhorst C (2005) Cutting edge: MyD88 controls phagocyte NADPH oxidase function and killing of Gram-negative bacteria. J Immunol 175: 5596–5600 [DOI] [PubMed] [Google Scholar]
- Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A (2007) Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315: 1398–1401 [DOI] [PubMed] [Google Scholar]
- Lee J, Chuang TH, Redecke V, She L, Pitha PM, Carson DA, Raz E, Cottam HB (2003) Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc Natl Acad Sci USA 100: 6646–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kim YJ (2007) Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem 76: 447–480 [DOI] [PubMed] [Google Scholar]
- Lemasters JJ (2005) Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 8: 3–5 [DOI] [PubMed] [Google Scholar]
- Levine B (2007) Cell biology: autophagy and cancer. Nature 446: 745–747 [DOI] [PubMed] [Google Scholar]
- Levine B, Deretic V (2007) Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 7: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477 [DOI] [PubMed] [Google Scholar]
- Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B (2006) Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol 177: 5163–5168 [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS (2006) Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med 203: 2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, DeBerardinis RJ, Thompson CB (2005) Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol 6: 439–448 [DOI] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA (2004) Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA 101: 5598–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R (2007) Recognition of microorganisms and activation of the immune response. Nature 449: 819–826 [DOI] [PubMed] [Google Scholar]
- Meier A, Alter G, Frahm N, Sidhu H, Li B, Bagchi A, Teigen N, Streeck H, Stellbrink HJ, Hellman J, van Lunzen J, Altfeld M (2007) MyD88-dependent immune activation mediated by human immunodeficiency virus type 1-encoded Toll-like receptor ligands. J Virol 81: 8180–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ (2006) AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem 281: 34870–34879 [DOI] [PubMed] [Google Scholar]
- Mizel SB, Honko AN, Moors MA, Smith PS, West AP (2003) Induction of macrophage nitric oxide production by Gram-negative flagellin involves signaling via heteromeric Toll-like receptor 5/Toll-like receptor 4 complexes. J Immunol 170: 6217–6223 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Ohsumi Y, Yoshimori T (2002) Autophagosome formation in mammalian cells. Cell Struct Funct 27: 421–429 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3: 542–545 [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T (2004) Autophagy defends cells against invading group A Streptococcus. Science 306: 1037–1040 [DOI] [PubMed] [Google Scholar]
- Nixon RA (2006) Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci 29: 528–535 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C (2005) Escape of intracellular Shigella from autophagy. Science 307: 727–731 [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib D, Levine B (2007) HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1: 23–35 [DOI] [PubMed] [Google Scholar]
- Pacquelet S, Johnson JL, Ellis BA, Brzezinska AA, Lane WS, Munafo DB, Catz SD (2007) Cross-talk between IRAK-4 and the NADPH oxidase. Biochem J 403: 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D et al. (2007) Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet 39: 830–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939 [DOI] [PubMed] [Google Scholar]
- Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW (2007) A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med 204: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram N, Subramani S (2006) The importomer—a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim Biophys Acta 1763: 1613–1619 [DOI] [PubMed] [Google Scholar]
- Rich KA, Burkett C, Webster P (2003) Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol 5: 455–468 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443: 780–786 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer E, Audige A, Joller H, Speck RF (2006) TLR7/8 triggering exerts opposing effects in acute versus latent HIV infection. J Immunol 176: 2888–2895 [DOI] [PubMed] [Google Scholar]
- Schmid D, Munz C (2007) Innate and adaptive immunity through autophagy. Immunity 27: 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Pypaert M, Munz C (2007) Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 26: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Bohley P (1992) Autophagy and other vacuolar protein degradation mechanisms. Experientia 48: 158–172 [DOI] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V (2006) Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313: 1438–1441 [DOI] [PubMed] [Google Scholar]
- Syed TA, Ahmadpour OA, Ahmad SA, Ahmad SH (1998) Management of female genital warts with an analog of imiquimod 2% in cream: a randomized, double-blind, placebo-controlled study. J Dermatol 25: 429–433 [DOI] [PubMed] [Google Scholar]
- Underhill DM (2003) Toll-like receptors: networking for success. Eur J Immunol 33: 1767–1775 [DOI] [PubMed] [Google Scholar]
- Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V (1998) Effects of cytokines on mycobacterial phagosome maturation. J Cell Sci 111: 897–905 [DOI] [PubMed] [Google Scholar]
- Weibel E, Bolender R (1973) Stereological techniques for electron microscopic morphometry. In Principles and Techniques of Electron Microscopy. Biological Applications, Hyat M (ed), Vol. 3, pp 237–296. New York: Van Nostrand Reinhold [Google Scholar]
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT (2007) Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary movie