Abstract
DNA synthesis is considered a defining feature in the movement of transposable elements. In determining the mechanism of piggyBac transposition, an insect transposon that is being increasingly used for genome manipulation in a variety of systems including mammalian cells, we have found that DNA synthesis can be avoided during piggyBac transposition, both at the donor site following transposon excision and at the insertion site following transposon integration. We demonstrate that piggyBac transposon excision occurs through the formation of transient hairpins on the transposon ends and that piggyBac target joining occurs by the direct attack of the 3′OH transposon ends on to the target DNA. This is the same strategy for target joining used by the members of DDE superfamily of transposases and retroviral integrases. Analysis of mutant piggyBac transposases in vitro and in vivo using a piggyBac transposition system we have established in Saccharomyces cerevisiae suggests that piggyBac transposase is a member of the DDE superfamily of recombinases, an unanticipated result because of the lack of sequence similarity between piggyBac and DDE family of recombinases.
Keywords: piggyBac, precise excision, transposase, transposition
Introduction
Transposable elements (transposons) are mobile DNA segments present in the genomes of all organisms that can move between many different positions in the genome. They have considerable influence on genome structure and function and thus are natural agents of genome evolution (Kazazian, 2004). Transposable elements are also extensively used as laboratory tools for genome manipulation by insertional mutagenesis and transgenesis (Boeke, 2002).
A problem common to the movement of all transposable element that undergo cut and paste transposition is the regeneration of intact duplex DNA both at the donor site and at the target site. Compared to transposon excision, only rarely is the donor site restored to its pre-transposon state, that is, only rarely does ‘precise excision' occur. One strategy for donor site repair is by end-joining, a pathway that leaves ‘footprints' in the donor site reflecting joining of target sequences that are duplicated upon transposon insertion and remain attached to the donor site after element excision (Coen et al, 1986; Weil and Kunze, 2000). The other pathway for donor site repair is homology-dependent gene conversion using a sister chromatid or homologue as a template (Engels et al, 1990; Plasterk, 1991). Although homology-dependent repair using a homologous site that lacks a copy of the element can result in precise excision, this is apparently an infrequently used mechanism (Perkins-Balding et al, 1999). Imprecise excision may actually be a useful strategy as it can introduce variation into the host genome (Kidwell and Lisch, 2000).
With DNA cut and paste elements, the single-strand gaps that flank the transposon in the new insertion site resulting from the joining of transposon ends to staggered positions on the target DNA must be repaired. It is this strategy of joining to staggered positions on the target DNA followed by DNA synthesis mediated by host repair proteins that results in the target site duplications, the hallmark of transposon insertion (Mizuuchi, 1983). This same strategy for target joining and gap repair also accounts for the target site duplications that flank integrated retroviral-like elements (Brown et al, 1987). DNA synthesis is also involved in the synthesis of the integrated DNA copies of non-LTR elements (Luan et al, 1993), rolling-circle transposons such as IS91 (del Pilar Garcillan-Barcia et al, 2001) and helitrons (Kapitonov and Jurka, 2001).
Thus, DNA synthesis has been considered a defining feature of transposable elements in contrast to the breakage and joining of element phage lambda, which is mediated by sequence-specific topoisomerases that use covalent protein–DNA intermediates and do not involve DNA synthesis (Grindley et al, 2006).
Intriguingly, the DNA cut and paste transposon piggyBac from cabbage looper moth Trichoplusia ni (T.ni) consistently shows precise excision upon element transposition (Cary et al, 1989; Fraser et al, 1995). Also unique for piggyBac transposition is the exclusive use of TTAA target sites (Fraser et al, 1995). Moreover, there is little obvious sequence similarity between transposases of the piggyBac and other transposon superfamilies (Sarkar et al, 2003). The mobility of piggyBac in various insects, mammalian cells including human cell lines (Wilson et al, 2007), mice (Ding et al, 2005) and a number of heterologous systems including planarian Girardia tigrina (Gonzalez-Estevez et al, 2003), the human pathogens Plasmodium falciparum (Balu et al, 2005) and Schistosoma mansoni (Morales et al, 2007) has made piggyBac an attractive genetic tool.
Here, we describe an in vitro system using piggyBac transposase purified from Escherichia coli and establish the mechanism of the DNA breakage and joining reactions that underline piggyBac transposition. We found that the pattern of cleavage during piggyBac excision results in complementary TTAA overhangs on the ends of the donor DNA, allowing the simple ligation of these ends to restore the donor site to its pre-transposon sequence, accounting for the precise excision of piggyBac. This transposon excision occurs through a hairpin intermediate on the transposon ends and leaves TTAA overhangs on the 5′ ends of the excised linear transposon.
The central step in piggyBac transposition, that is, the joining of the excised transposon to the target DNA, occurs by the direct attack of the 3′OH ends of the transposon to staggered positions at the 5′ ends of a TTAA target sequence. This is the same strategy used by the widespread DDE family of bacterial and eukaryotic transposases and retroviral integrases (Rice and Baker, 2001). This strategy of target joining means that the TTAA overhangs on the 5′ ends of the transposon can base-pair with the 5′ TTAA single-strand gaps on the target DNA that flank the positions of transposon joining. These target gaps can then be sealed simply by ligation rather than by DNA synthesis.
Thus, the target site exclusivity for TTAA sites and the strategy for excision from the donor site with 5′ TTAA overhangs means that piggyBac need not involve DNA synthesis, a defining feature of all other characterized transposition reactions.
In addition to a common mechanism, the DDE recombinases share a common catalytic core of particular protein folds that juxtapose acidic residues that provide binding sites for catalytically essential Mg2+ ions (Rice and Baker, 2001; Zhou et al, 2004; Richardson et al, 2006). We show, using secondary structure prediction and mutational analysis in vitro and in vivo in genetically tractable Saccharomyces cerevisiae, that piggyBac elements have catalytically essential DDD acidic residues that apparently lie on a partial RNaseH fold and thus are likely members of the DDE superfamily. Our studies have thus revealed that the piggyBac system is not only related to but also has unique aspects distinct from other studied transposon systems.
Results
piggyBac transposase promotes double-strand breaks to excise piggyBac from the flanking donor DNA
The ends of piggyBac are closely related; both contain a 13 bp terminal inverted repeat and a 19 bp internal inverted repeat (Cary et al, 1989) (Figure 1A). These repeat segments are, however, separated by a 3 bp spacer on the left end and a 31 bp spacer on the right end. In all the experiments reported below, we have used DNA segments containing 70 and 72 bp of the piggyBac left and right ends, respectively. These segments contain the 35 bp L-TIR13-3-19 (L-TIR) and 63 bp R-TIR13-31-19 (R-TIR) sequences that are efficient substrates for excision and interplasmid transposition in vivo (Elick et al, 1997). These end segments, however, lack several other internal repeats necessary for in vivo integration into the host genome (Li et al, 2005).
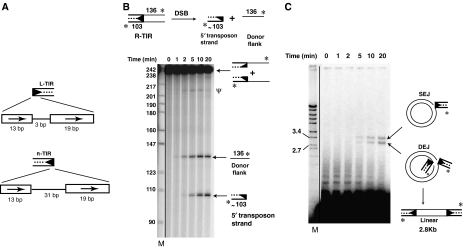
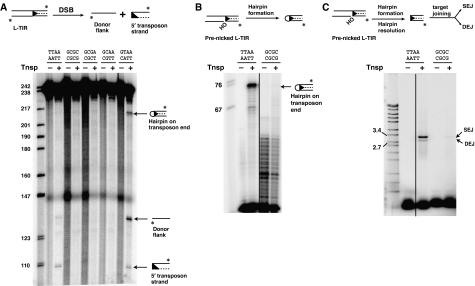
Figure 1.
piggyBac transposase catalyses DSBs and target joining. (A) Schematic representation of the piggyBac ends. The piggyBac left end (L-TIR) consists of a 13 bp terminal inverted repeat and a 19 bp internal inverted repeat separated by a 3 bp spacer; the right end (R-TIR) has a 31 bp spacer. The arrows indicate repeat sequences. (B) piggyBac transposase promotes DSBs. piggyBac transposase releases the transposon end from the flanking donor DNA by DSB, generating new products on a denaturing acrylamide gel. Ψ indicates hairpin intermediate. Here and in all other figures, * indicates the position of radiolabel. M indicates marker (radiolabelled MspI-digested pBR322 DNA in all denaturing acrylamide gels). Here and in all other figures, the solid line indicates the different portions of the same scan that were combined to form the relevant panel. (C) piggyBac transposase promotes target joining. piggyBac transposase joins the excised transposon to the target DNA generating products SEJ (nicked circular plasmid formed by joining of one transposon end to one plasmid strand) and DEJ (linearized plasmid formed by concerted joining of two transposon ends to two plasmid strands), which are displayed on a native agarose gel. Slower migrating species reflect joining to oligomeric plasmids. Here and in all other figures, M indicates marker (BglII+EcoRI-digested λ DNA in the agarose gels).
We cloned, expressed and purified T. ni piggyBac transposase as a His-tagged derivative, from E. coli (see Materials and methods). Using band shift assays, we found that purified transposase binds specifically to piggyBac L-TIR and R-TIR end fragments that are flanked by donor DNA (Supplementary Figure 1) and also to the end fragments lacking flanking donor DNA (data not shown).
piggyBac excises precisely from a donor DNA and inserts into a TTAA target site in vivo (Cary et al, 1989). To probe the mechanism of these reactions, we incubated piggyBac transposase with DNA fragments containing the piggyBac R-TIR (Figure 1) or L-TIR (Supplementary Figure 2) flanked by donor DNA, a plasmid target DNA and Mg2+, which is an essential cofactor (data not shown). Mn2+ was a much less effective cofactor in all the assays performed here (data not shown).
The 3′-end-labelled piggyBac R-TIR substrate was a 103 bp segment containing the 72 bp R-TIR sequence flanked by 136 bp of donor DNA. When the R-TIR reaction products were displayed on a denaturing gel (Figure 1A), we observed two prominent new species reflecting a double-strand break (DSB) separating the transposon end from the flanking donor DNA. The size of the slower migrating species of the two most prominent new species is consistent with a nick at the 3′ end of the transposon that would liberate the end-labelled 136 nt flanking donor top strand. The size of the faster migrating species of the two new species is consistent with a break between the 5′ transposon end and the donor DNA that releases the end-labelled 103 nt bottom strand of the piggyBac R-TIR from the flanking donor DNA. Determination of the exact positions of these cleavages is described below.
It is notable that cleavage at the 3′ and 5′ transposon ends did not occur simultaneously. The cleavage at the 3′ transposon end that generated the slower migrating species occurred before cleavage at the 5′ end that generated the faster-migrating species. Similar DSB reaction products were obtained using piggyBac L-TIR DNA (Supplementary Figure 2A), although at lower efficiency compared to piggyBac R-TIR. Thus, nicking at the 3′ transposon end appears to initiate transposition.
Also faintly visible is a very slow migrating species marked ‘ψ'. As discussed in detail below, this species is a hairpin that includes both the 5′ and 3′ strands of the piggyBac R-TIR end and is a transposition intermediate.
piggyBac transposase joins the ends of the transposon to the target DNA
When the products of the above reactions were displayed on a native agarose gel, we observed joining of the cleaved R-TIR (Figure 1C) and L-TIR (Supplementary Figure 2B) fragments to a circular plasmid target DNA to form two different products. The joining of one transposon end to one strand of the target DNA forms a single-end join (SEJ) in which one target strand is broken by the covalent joining of one transposon end and the other plasmid strand is intact, resulting in a nicked circular plasmid. Concerted joining of two transposon ends to separate strands of the same target DNA forms a double-end join (DEJ) in which each strand of the target DNA is covalently linked to one transposon end, forming a linear, double-stranded DNA molecule. Formation of such coupled DEJ products is consistent with formation of a transpososome complex in which the transposon ends pair and interact with the target DNA.
The majority of the SEJ and DEJ products appear at late times (10–20 min) when the transposon ends have already undergone DSBs separating them from the flanking donor DNA. Little target joining is observed at early times (2 min) when only cleavage at the 3′ end of the transposon has occurred, suggesting that target joining can occur only after the complete excision of the transposon end from the flanking donor DNA. We show below that DSBs are rapidly formed once nicking at the 3′ transposon ends occurs, suggesting that the nicking step can be a rate-limiting step in piggyBac transposition.
With the R-TIR substrate, we observed both SEJ and DEJ products (Figure 1C); with the L-TIR substrate, we observed only the SEJ product (Supplementary Figure 2B). The greater amount of target joined products with the R-TIR substrate is consistent with the greater amount of R-TIR end cleavage.
piggyBac DSBs proceed through a hairpin intermediate on the transposon end
The above experiments suggested that nicking at the 3′ transposon end initiates piggyBac transposition. Thus, we analysed the transposition reaction using a 35 bp piggyBac L-TIR13-3-19 substrate flanked by donor DNA where the 3′OH end of the transposon is already exposed, that is, a ‘pre-nicked' substrate. Analysis of the reaction products on a denaturing gel revealed the rapid accumulation of a new species about 74 nt in length (Figure 2A). The size of this product is consistent with the formation of a hairpin on the transposon end that includes several nucleotides of the flanking donor DNA; its exact sequence is considered below.
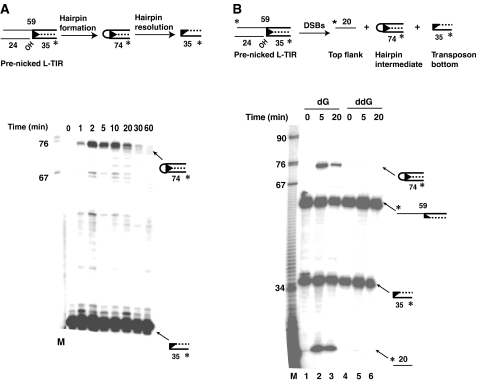
Figure 2.
piggyBac DSBs occur by means of a hairpin intermediate on the transposon end. (A) DNA hairpin formation is visualized using a pre-nicked end. A pre-nicked piggyBac L-TIR was incubated with piggyBac transposase in the presence of a target DNA for various times and then displayed on a denaturing acrylamide gel. (B) Hairpin formation requires a 3′OH transposon end. A pre-nicked piggyBac L-TIR with 3′G-OH (deoxy) or 3′G-H (dideoxy) was incubated with piggyBac transposase in the presence of a target DNA for 5 or 20 min and then displayed on a denaturing acrylamide gel. Lanes 1–3, piggyBac L-TIR with 3′G-OH (deoxy); lanes 3–5, piggyBac L-TIR with 3′G-H (dideoxy); lanes 1 and 3, ‘no protein' controls.
Hairpin formation reflects cleavage at the 5′ end of the transposon and concomitant liberation of the transposon from the flanking donor DNA. The 5′ transposon end cleavage is much faster with the pre-nicked substrate than with intact ends; some hairpin is evident at 1 min with the pre-nicked substrate (Figure 2A), whereas only 3′ end nicking is evident by 5 min with the intact end with the flanking donor substrate (Figure 1B). With the pre-nicked substrate, a high level of hairpin product is present until 10–20 min and the amount decreases thereafter, suggesting that the hairpin species can be resolved by the transposase (Figure 2A). DSBs resulting from hairpin formation on the transposon end are already known to occur with the prokaryotic elements Tn10 (Kennedy et al, 1998) and Tn5 (Bhasin et al, 1999); piggyBac is the first example of a eukaryotic element that uses this mechanism.
To test whether piggyBac hairpin formation, leading to transposon excision, requires an exposed 3′OH transposon end, we compared hairpin formation of pre-nicked piggyBac L-TIR substrates that had either a 3′ deoxyguanosine or a 3′ dideoxyguanosine at their 3′ ends (Figure 2B). If hairpin formation requires a 3′OH transposon end, the dideoxy-containing substrate should be unable to support hairpin formation and hence a DSB. In contrast to the pre-nicked 3′ deoxyguanosine substrate, which efficiently formed the 74 nt hairpin species with the concomitant release of the top 20 nt flanking donor DNA (Figure 2B, lanes 2 and 3), the 3′ dideoxyguanosine substrate failed to form the hairpin and undergo DSB (Figure 2B, lanes 4 and 5). Thus, hairpin formation is an essential step in piggyBac excision.
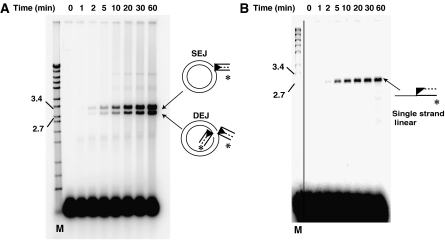
When the products of transposition reactions (Figure 2A) using the pre-nicked piggyBac L-TIR substrate are displayed on a native agarose gel (Figure 3A), joining of the excised transposon ends to the target plasmid forming the SEJ and DEJ products is observed. Significant levels of target joining species are seen late (5–10 min) in the time course of the reactions, although hairpin formation on the transposon end is evident early (1 min). Such timing suggests that the transposition reaction proceeds through end nicking and hairpin formation, leading to DSBs, that is, transposon excision, which is followed by hairpin resolution to expose the transposon ends, after which target joining occurs. The much higher levels of end cleavage and target joining observed with the pre-nicked substrate (Figure 3A) as compared to the intact substrate (Figure 1C and Supplementary Figure 2B) suggests that nicking at the 3′ transposon end is a key determinant of the rate of transposition.
Figure 3.
piggyBac joins the 3′OH of a pre-nicked transposon end substrate to the target DNA. (A) A pre-nicked end can join to target DNA. The products of the reactions described in Figure 2A are displayed on a native agarose gel. (B) The 3′OH transposon end joins to the target DNA. The products of the reactions described in Figure 2A are displayed on a denaturing agarose gel.
The 3′OH end of piggyBac joins to the target DNA
To determine directly which strand of a piggyBac end joins to the target DNA, we examined the products formed between a pre-nicked piggyBac L-TIR fragment labelled at its internal 5′ end, that is, at the end of the transposon strand containing a 3′OH end, and a plasmid target DNA, on a denaturing agarose gel (Figure 3B). Whereas target joining leads to the formation of two products, that is, SEJs and DEJs, on a native gel (Figure 3A), only a single product was evident on a denaturing agarose gel, consistent with the joining of one piggyBac L-TIR segment to a single target strand (Figure 3B). Detection of these single-stranded products in reactions using a piggyBac L-TIR in which the strand containing the 3′ terminal transposon end is labelled at its 5′ internal end reveals the chemistry of target joining: the 3′OH terminal end of the piggyBac L-TIR joins covalently to the target DNA. In a separate experiment, we have shown directly that the 5′ ends of piggyBac do not join to target DNA (Supplementary Figure 3).
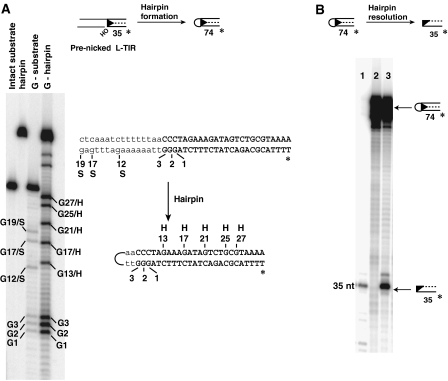
A 4-nt TTAA hairpin is formed on the piggyBac end
To define the sequence identity of the transposon end hairpin at the nucleotide level, we isolated the hairpin species generated from a transposition reaction using the pre-nicked piggyBac L-TIR substrate and then determined its sequence using the Maxim–Gilbert G-reaction (Figure 4A). The G sequence of the hairpin was identical to that of the bottom strand of the transposon DNA flanked by the donor DNA with respect to 3′ terminal end of piggyBac at positions G1, G2 and G3. However, the positions of the following G's in the hairpin and an intact transposon end differed. Whereas the G's in the intact transposon end when the flanking donor DNA is attached to the transposon were at positions G12/S, G17/S and G19/S, the next G in the hairpin was at position G13/H, followed by G17/H, G21/H, G25/H and G27/H, reflecting the sequence of the top strand of the transposon DNA (Figure 4A).
Figure 4.
Sequence of the transposon end hairpin and resolution by the transposase. (A) Sequence of the transposon end hairpin. Maxim–Gilbert G-reaction of the hairpin intermediate formed from a pre-nicked piggyBac L-TIR substrate displayed on a denaturing acrylamide gel. G-substrate: G-reaction of the intact piggyBac L-TIR fragment with flanking DNA; G-hairpin: G-reaction of the piggyBac L-TIR hairpin; G/S: G-reaction of substrate; G/H: G-reaction of hairpin. The L-TIR is shown in uppercase and the flanking donor DNA in lowercase; the top and bottom strands of piggyBac L-TIR are joined by means of a 4 nt hairpin derived from the donor strand flanking the 5′ end of the transposon. Only part of L-TIR substrate and corresponding hairpin are shown. (B) piggyBac transposase can resolve a pre-formed transposon end hairpin. Transposase was incubated with L-TIR13-3-19 oligonucleotide containing a TTAA hairpin for 20 min. Lane 1, the 3′ strand of L-TIR13-3-19 as a marker; lane 2, hairpin DNA without piggyBac transposase incubation; lane 3, hairpin DNA with piggyBac transposase incubation.
This pattern is consistent with a hairpin that includes the 3′ end of the piggyBac L-TIR DNA including the G at the 3′ terminus of the transposon, 4 nt from the flanking donor DNA—T, T, A, A—joined to the C at the 5′ end of the transposon and the rest of the 5′ strand of the piggyBac L-TIR. Thus, four nucleotides of flanking donor DNA are included in the hairpin. As piggyBac always inserts into TTAA target sites, the element will always be flanked by TTAA in the donor site and the hairpin will always include the TTAA sequence. We explore the effects of changing this sequence below.
piggyBac transposase can resolve a hairpin transposon end
We also directly analysed the ability of piggyBac transposase to resolve a pre-formed hairpin. Using a self-complementary oligonucleotide, we generated a piggyBac hairpin L-TIR species containing a 4 nt TTAA loop. In the presence of the transposase, the hairpin DNA is resolved to generate a 35 nt DNA fragment corresponding to cleavage at the 3′ end of piggyBac on the bottom strand of the piggyBac L-TIR (Figure 4B, lane 3). Thus, the lack of hairpin accumulation in Figure 1B likely results from rapid resolution of the hairpin intermediate.
piggyBac correctly inserts into TTAA target sites in vitro
piggyBac inserts exclusively into TTAA target sites in vivo and thus TTAA target sequence duplications flank a newly inserted transposon (Cary et al, 1989). To determine the fidelity of piggyBac target joining in vitro, we used PCR to generate a mini-piggyBac element in which a gene for kanamycin resistance is flanked on both ends by piggyBac L-TIR13-3-19 sequences with their 3′OH already exposed; this element lacked the TTAA extensions present on the 5′ ends of an authentic excised piggyBac element. This mini-piggyBac element was used as a substrate in an in vitro transposition reaction containing pUC19 plasmid DNA as a target. Transposition products were recovered by selection for kanamycin-resistant E. coli after transformation and the transposon–plasmid junctions were sequenced. Four independent transposition reactions were performed and five products from each reaction were recovered and analysed. In all cases, insertion occurred at a TTAA site and the element was always flanked by a 4 bp TTAA target sequence duplication; insertions into 8 of the 13 different TTAA sites on the pUC19 plasmid were recovered (Supplementary Figure 4). Thus, insertion into TTAA target sites is an intrinsic property of the piggyBac transposase and the TTAA extensions present on the 5′ ends of an excised piggyBac transposon are not required for TTAA target site selection.
Thus, our in vitro transposition system directs insertion of piggyBac into its preferred TTAA target sequence, which is a true representation of piggyBac transposition in vivo.
There are four steps in piggyBac transposition
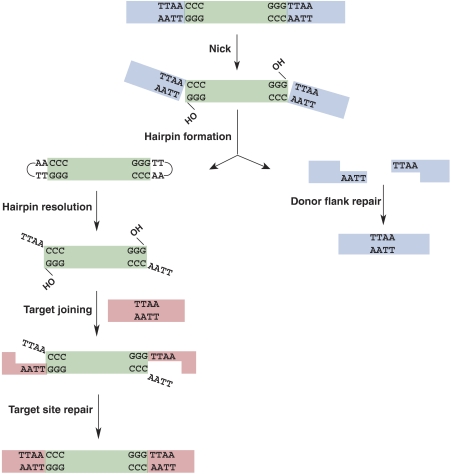
We conclude that piggyBac transposition involves four distinct chemical steps (Figure 5). Step 1: a DSB initiates with a nick at the junction of the 3′ end of the transposon and the flanking donor DNA, exposing a reactive 3′OH on the transposon end. Step 2: the 3′OH then acts as a nucleophile and attacks 4 nt into the flanking donor DNA on the complementary strand, generating a hairpin on the transposon end and simultaneously releasing the transposon DNA from the donor backbone. The flanking donor DNA contains complementary 5′ TTAA extensions that can rejoin to repair the donor gap to give a precise excision. Step 3: The hairpin on the transposon end is resolved by the transposase, leaving a 4 nt overhang at the 5′ end of the excised linear transposon and re-exposing the 3′OH at the transposon end. Step 4: the 3′OH at the transposon end then covalently joins to the target DNA.
Figure 5.
Schematic representation of the piggyBac cut and paste transposition. piggyBac transposition initiates with nicks at the 3′ ends of the transposon, exposing 3′OHs. These 3′OHs then attack the complementary strand 4 nt into the flanking donor DNA, thereby forming hairpins on the transposon ends with the concomitant release of the transposon ends. Donor site repair can occur by ligation of the complementary 5′ TTAA overhangs on the flanking donor DNA ends, precisely reforming the TTAA target sequence. Transposon end hairpins are resolved by transposase, re-exposing the 3′OH transposon ends and generating 4 nt TTAA overhangs on the 5′ ends of the excised transposon. The 3′OH transposon ends join to the staggered positions at the 5′ T's of the TTAA/AATT target sequence. Repair of the single-strand gaps flanking the newly inserted transposon gives rise to the 4 bp TTAA target sequence duplication.
piggyBac excision is influenced by the flanking donor DNA sequence
A newly inserted piggyBac element is flanked by the TTAA target sequence duplications resulting from the staggered attack on the TTAA target sequence. After excision, the TTAA nucleotides from the flanking donor DNA remain attached to the 5′ ends of piggyBac. Does the identity of the flanking donor site sequences influence piggyBac excision and target joining?
We examined the ability of piggyBac transposase to promote DSBs at piggyBac L-TIRs flanked by GCGC, a sequence different at all positions from the standard TTAA sequence. Display of the reaction products on a denaturing gel reveals that no nicks or DSBs occur with the GCGC flank (Figure 6A). Furthermore, no hairpinning is observed with a pre-nicked GCGC flank L-TIR (Figure 6B) nor does resolution of a GCGC-containing hairpin L-TIR occur (Supplementary Figure 5A). Thus, with a flanking GCGC, there is a defect in end processing at every step, that is nicking, hairpin formation and hairpin resolution, revealing that the TTAA flanking sequence has a key role in transposon excision.
Figure 6.
The flanking donor sequence influences transposon end processing. (A) Flanking sequence and 3′OH end nicking. piggyBac transposase was incubated with L-TIR fragments flanked by donor DNA of different sequences, as indicated, for 20 min and reactions displayed on a denaturing acrylamide gel. (B) Flanking donor DNA influences hairpin formation. Pre-nicked piggyBac L-TIRs with either a TTAA or GCGC flank were incubated with piggyBac transposase in the presence of a target plasmid and displayed on a denaturing gel. (C) Influence of flanking sequence on target joining. The reactions using the pre-nicked substrates were displayed on a native gel.
It is also notable that little (<10% of wild type) target joining is observed with a pre-nicked GCGC L-TIR substrate (Figure 6C), indicating that although the 3′OH at this transposon end is already exposed, it is a poor substrate for target joining.
We also analysed donor cleavage with piggyBac L-TIR transposon ends in which the standard TTAA flanking sequence was changed at three positions (GCGA), two positions (GCAA) and one position (GTAA) (Figure 6A). Nicking and DSBs were detected only with the GTAA substrate that differed at only one position from the 5′ end from the standard TTAA flank (Figure 6A). Accumulation of the hairpin intermediate was also observed with this GTAA flanking sequence substrate, suggesting that hairpin resolution is also defective when the 3′ end of the transposon is linked to a G rather than the natural T (Supplementary Figure 5B). These experiments demonstrate that the efficiency of end cleavage is highly influenced by the flanking donor DNA.
We have also analysed the influence on target joining of the sequence of the 4 nt extension on the 5′ ends of excised piggyBac (Supplementary Figure 6). An L-TIR end lacking any extension on the 5′ transposon end was a better substrate than an end having a 5′ TTAA extension. Target joining by an end with the usual 5′ TTAA extension was much better than that with a 5′ GCGC extension. Thus, although piggyBac transposase can join transposons with flush ends to target DNA, the usual TTAA overhangs on the 5′ transposon ends efficiently couple excision and integration.
Identification of the catalytic core of piggyBac transposase
The above experiments revealed that the key chemical steps in piggyBac transposition are the Mg2+-dependent excision of the transposon to yield a transposon with 3′OH ends, followed by the direct nucleophilic attack of these 3′OH ends on the target DNA. The chemistry of these steps is identical to that of the DDE recombinase family, which contains many transposases and retroviral integrases (Rice and Baker, 2001; Zhou et al, 2004; Richardson et al, 2006). In these recombinases, highly conserved acidic amino acids (DDE or DDD) are closely juxtaposed on an RNaseH-like fold and coordinate essential metal ions. There is, however, little primary sequence homology between piggyBac and the DDE/DDD recombinases (Sarkar et al, 2003).
Secondary structure prediction of T. ni piggyBac by PSIPRED (Jones, 1999b) suggests that a portion of piggyBac likely has part of the RNaseH-like fold that is conserved in DDE recombinases such as HIV1 integrase (Dyda et al, 1994) (Supplementary Figure 7). Included within this region of piggyBac are D268 and D346, which are invariant among a number of piggyBac-like ORFs that are associated with TIRs from a variety of insects and animals (Supplementary Figure 8) (Sarkar et al, 2003; Arkhipova and Meselson, 2005). The positions of these invariant D's on the putative piggyBac RNaseH-like fold are equivalent to those of the two D's on the RNaseH-like DDE transposases and retroviral integrases (Supplementary Figure 7).
To probe the involvement of these conserved acidic amino acids in piggyBac transposition, we purified piggyBac transposases with alanine substitutions at D268 and D346 and evaluated their ability to promote specific binding to the TIRs and the catalytic steps of transposition.
Although still capable of binding specifically to piggyBac L-TIR and R-TIR (Supplementary Figure 9), the D268A and D346A mutants were defective in generating the DSBs that separate the transposon ends from the flanking donor DNA (Figure 7A), resolution of a pre-formed hairpin to expose the 3′OH transposon (Figure 7B) and joining of a substrate TIR with an exposed 3′OH to a target DNA (Figure 7C). These findings suggest that these conserved acidic amino acids have critical roles in all the catalytic steps of transposition, likely through their interactions with Mg2+. Moreover, the fact that a single mutation can block these multiple catalytic steps suggests that a single active site can mediate all the chemical steps of recombination.
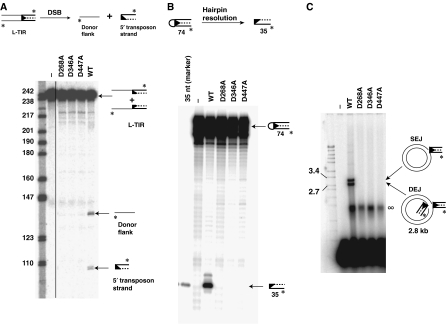
Figure 7.
Mutation of conserved DDD amino acids blocks the catalytic activity of piggyBac transposase in vitro. (A) Mutation of the conserved D's blocks 3′OH nicking. Wild-type and mutant piggyBac transposases were incubated with a piggyBac L-TIR and a target plasmid for 20 min and then displayed on a denaturing acrylamide gel. (B) Mutation of the conserved D's blocks hairpin formation. Wild-type and mutant piggyBac transposases were incubated with a pre-formed piggyBac L-TIR hairpin oligonucleotide and then displayed on a denaturing gel. (C) Mutation of the conserved D's blocks target joining. Wild-type and mutant transposases were incubated with a pre-cleaved piggyBac L-TIR lacking the usual 4 nt overhangs at 5′ transposon end and a target plasmid and then displayed on a native agarose gel. ∞ indicates nucleoprotein complexes formed by transposase binding to the labelled piggyBac L-TIRs.
There are several conserved D's outside of the RNaseH-like domain that are conserved among the piggyBac transposases: D227, D228, D239, D447 and D450 (Supplementary Figure 8). We also made alanine substitutions at these positions and tested the activities of the mutant proteins. Notably, D447A was still capable of binding to the TIRs (Supplementary Figure 9) but was defective in all the catalytic steps of transposition, suggesting that it also lies within the catalytic core (Figure 7A–C and Supplementary Figure 10).
D227A, D228A, D239A and D450A were all catalytically active in vitro; thus, these positions do not appear to be part of the catalytic core of the transposase (data not shown).
Other functional determinants of piggyBac transposase
Another highly conserved residue among piggyBac transposases is W465 (Supplementary Figure 8). Planar interactions between such an aromatic amino acid and DNA bases can have an important role in base-flipping steps, which can be integral to DNA distortions necessary for the formation and resolution of the altered DNA structures such as hairpins, as demonstrated with Tn5 and Tn10 (Davies et al, 2000; Bischerour and Chalmers, 2007), Hermes (Zhou et al, 2004) and RAG recombinase (Lu et al, 2006). It has been suggested (Arkhipova and Meselson, 2005) that piggyBac W465 is involved in DNA hairpinning. W465A, however, has much reduced nicking activity and is compromised in all subsequent catalytic steps of transposition although it still binds specifically to the piggyBac TIRs (Supplementary Figures 9 and 11). Thus, W465 cannot have a role only in DNA hairpin formation and resolution.
Another conserved feature of piggyBac transposases is the C-terminal C2C2CHC2 motif. Analysis of this region by GenTHREADER (Jones, 1999a) suggests that this region forms a Zn2+-binding PHD domain (Supplementary Figure 8); PHD domains bind to chromatin (Bienz, 2006). The C-terminus is, however, dispensable for piggyBac recombination in vitro: piggyBac1–558, which lacks the C2C2CHC2 motif, is as active in vitro as the wild-type piggyBac1–594 (data not shown), perhaps because we have used naked DNA rather than chromatin as a target substrate.
piggyBac can transpose in S. cerevisiae
To establish a simple genetic assay for the excision of piggyBac, we adapted a modified version of the yeast URA3 gene as a transposon donor (Yu and Gabriel, 1999). In this modified URA3 gene, the yeast actin intron has been introduced into the URA3 gene to form a URA3∷actin intron gene. The actin intron can be efficiently spliced from mRNA of this gene, so that a strain carrying the URA3∷actin intron is a uracil prototroph. However, if a large DNA segment such as a several kilobase transposon is introduced into the actin intron, the resulting intron is too large to be spliced from mRNA, making the strain a uracil auxotroph. Thus, excision of the transposon and restoration of the donor site to the parental URA3∷actin intron configuration can be followed by assaying for reversion of uracil auxotrophy to uracil prototrophy (Figure 8A). The restoration of the gapped donor site in yeast could occur by end-joining or gene conversion using the chromosomal actin gene intron as a template (Paques and Haber, 1999).
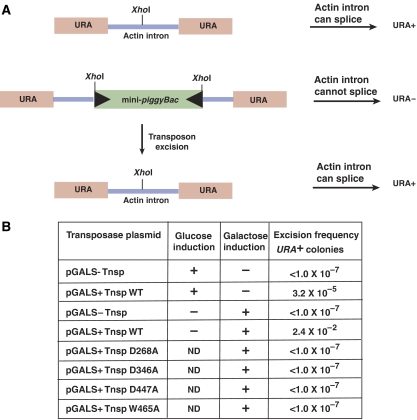
Figure 8.
piggyBac can transpose efficiently in S. cerevisiae. (A) Schematic representation of the piggyBac transposition assay. piggyBac excision from a URA3 gene containing the actin intron in S. cerevisiae was evaluated by analysing the reversion of the donor site from uracil auxotrophy to uracil prototrophy after piggyBac excision. (B) Frequency of piggyBac excision.
In our yeast two-plasmid piggyBac system, a transposon donor plasmid contains a mini-piggyBac transposon composed of 328 bp of the piggyBac left end and 361 bp of the piggyBac right end flanking a kanamycin resistance gene in the URA3∷actin intron cassette. The transposase is supplied by a second plasmid containing the piggyBac transposase gene under the galactose-inducible control of the GALS promoter (Mumberg et al, 1994).
In the absence of transposase, the frequency of URA3 reversion was very low, about 10−7 (Figure 8B). Upon galactose induction of the piggyBac transposase gene, however, the frequency of URA3 reversion was very much higher, about 10−2, indicating a high level of transposon excision (Figure 8B). Considerable excision, that is, uracil prototrophy, was observed even without galactose induction, indicating that the low level of transposase present because of leakiness of the GALS promoter can promote transposition.
We demonstrated above that certain highly conserved acidic amino acids are necessary for piggyBac activity in vitro. Are they also essential for piggyBac activity in vivo? We evaluated transposition in yeast promoted by mutant transposases substituted with alanines at D268, D346, D447 and W465. In all cases, the frequency of uracil prototrophy promoted by these mutant transposases was more than five orders of magnitude less than that observed with wild type and not significantly different from that observed in the absence of transposase, that is, about 10−7 (Figure 8B).
To demonstrate that the mutant proteins are stably produced, we have shown in a yeast transposon integration assay (Supplementary data) that the presence of the mutant transposases inhibits transposition by wild-type transposase, that is, the mutants are dominant negatives and are thus stably produced (Supplementary Figure 12). The yeast system thus supports the hypothesis based on in vitro data that these amino acids are part of the piggyBac active site.
Discussion
piggyBac: a widespread eukaryotic DNA transposon
Cabbage looper moth piggyBac is a DNA transposable element and a member of the widespread piggyBac family of transposons (Sarkar et al, 2003) with recently active elements have been identified in Xenopus (Hikosaka et al, 2007). We have developed a faithful in vitro transposition system that defines the chemical steps by which piggyBac undergoes cut and paste transposition. Our findings account for several distinctive features of piggyBac transposition and particularly notable is that piggyBac can transpose without DNA synthesis, a process involved in all other characterized transposition reactions. The systems we have described here provide valuable tools for further development of piggyBac-based systems for insertional mutagenesis and transgenesis as well as for dissection of piggyBac transposition at the molecular level.
piggyBac excision involves the formation and resolution of hairpins on the transposon ends
To undergo cut and paste transposition, piggyBac transposase induces DSBs that separate the transposon ends from the flanking donor DNA. piggyBac DSB initiates with a nick at the 3′ end of the transposon. The free 3′OH then attacks the complementary strand to form a hairpin on the transposon end, concomitantly releasing the transposon from the flanking donor DNA. As we find that more hairpin intermediate is formed at earlier times with a pre-nicked substrate compared to an intact one in which nicking must first occur, introduction of the nick may be a rate-limiting step in transposition. The slow nicking step could, however, reflect the slow assembly of an active transpososome on an intact substrate compared to rapid assembly of an active transpososome on a pre-nicked substrate rather than a limitation in the chemical steps. It is also notable that little difference in hairpin formation is observed between pre-nicked left and right ends or between pre-cleaved left and right ends whereas an intact left end is a much less efficient substrate than an intact right end. These observations support the view that the formation of an active transpososome is more stringently regulated on a DNA with a transposon end still flanked than with cleaved DNAs that are recombination intermediates. These results also suggest that transpososome assembly occurs differently on each end.
The transposase then opens the hairpin to expose the 3′OH transposon end that can then attack the target DNA. A similar ‘hairpin on the transposon ends' mechanism has been observed with the prokaryotic elements Tn5 and Tn10 (Kennedy et al, 1998; Bhasin et al, 1999) and contrasts with the ‘hairpin on the donor DNA' observed with the hAT transposon Hermes (Zhou et al, 2004) and RAG recombinase (van Gent et al, 1996). DSBs through hairpins on the transposon ends in piggyBac transposition are the first to be reported in a eukaryotic transposon system.
The intramolecular hairpin reaction occurs by the attack of the transposon end 3′OH on the donor phosphodiester bond that is 5′ to the TTAA sequences that always flank the 5′ ends of the transposon. These flanking TTAAs are present in all piggyBac donor sites because piggyBac inserts specifically at TTAA target sites. Indeed we have shown that the presence of non-TTAA flanking base pairs can greatly inhibit piggyBac excision. Despite being 4 bp away, the phosphodiester bond on the complementary strand that is attacked by the 3′OH on the transposon end is actually brought relatively near the 3′ end of the transposon by the twist of the helix. The requirement for the TTAA flank may reflect a requirement for DNA distortability to juxtapose the 3′OH transposon end and its position of attack on the complementary strand. Similar distortions have been shown by phasing analysis of the end sequences upon binding of Tn5 transposase (York and Reznikoff, 1997; Ason and Reznikoff, 2004). The hairpin species is then resolved by cleavage at the 3′ end of the transposon.
Thus, the 5′ transposon ends of the excised transposon are flanked by 4 nt, TTAA overhang, that derive from the flanking donor site DNA, unlike the hairpin formed flush at the 5′ transposon end with no flanking DNA in the Tn5 and Tn10 transposons (Kennedy et al, 1998; Bhasin et al, 1999).
piggyBac transposition is accompanied by precise excision at the donor site
After transposon excision, repair of the broken donor chromosome that contains a gap at the position from which the element was excised must occur. With most eukaryotic transposons, the original target sequence is not reformed during donor site rejoining but rather a ‘footprint' remains that reflects the joining of the flanking donor ends that contain the duplicated target sequence (Coen et al, 1986; Weil and Kunze, 2000). A distinctive feature of piggyBac transposition is that transposition is always followed by reconstitution of a single TTAA sequence at the donor site, that is, precise excision occurs (Cary et al, 1989). We have shown that piggyBac excises by a mechanism that cleanly frees the 3′ ends of the transposon and results in 4 nt TTAA overhangs on the 5′ ends of the transposon. Consequently, there are also TTAA overhangs on both 5′ ends of the broken donor DNA. As these overhangs are present on complementary strands, they can readily pair and be ligated by the host repair machinery, yielding a TTAA at the donor site equivalent to the original target site (Figure 5). Thus, gene conversion using a homologue as a template need not be invoked to account for precise excision in the case of piggyBac transposition (Paques and Haber, 1999).
It should also be noted that target site-specific insertion into a palindromic sequence is not sufficient to ensure precise excision. Tc1 mariner elements always insert at TA sites. However, cleavage of the 5′ transposon ends actually occurs inside the element, leaving non-complementary overhangs that cannot be simply ligated together such that footprints are seen most frequently upon element excision (Plasterk, 1991).
The sequence TTAA coordinates piggyBac excision and target site insertion
Previous in vivo experiments (Fraser et al, 1995) and our in vitro experiments have shown that piggyBac inserts preferentially into a TTAA target site such that the newly inserted transposon is flanked by TTAAs. Although excised piggyBac is flanked by TTAA on the 5′ transposon ends, these flanking nucleotides are not essential for correct target joining: a piggyBac element with flush 5′ ends still chooses TTAA as a target. We have found that the flanking TTAA sequence is also critical to transposon excision; changing the flanking TTAA can block nicking at the 3′ transposon end, hairpin formation and hairpin resolution. Flanking donor sequence also influences the excision of other elements (Wu and Chaconas, 1992; Williams et al, 1999).
This requirement for the same sequence for target integration and DSB formation to promote excision suggests that this TTAA sequence is recognized by the transposase during both excision and integration.
Target joining and the generation of intact DNA at the target site
We have found that piggyBac joins to the target DNA by the direct attack of the 3′OH ends of the transposon to staggered positions on to the TTAA target DNA sequence. This direct nucleophilic attack is the hallmark of target joining by the DDE superfamily that contains many prokaryotic and eukaryotic transposases and retroviral integrase (Rice and Baker, 2001; Hickman et al, 2005; Richardson et al, 2006). As the 5′ ends of the newly integrated transposon are flanked by 5′ TTAA, the generation of intact duplex DNA at the site of insertion can be accomplished by pairing and ligation of the complementary TTAA sequences on the 5′ transposon ends and at the gaps that flank the newly inserted transposon. This pairing and ligation strategy will result in TTAA base pairs flanking the newly inserted transposon. This strategy is distinct from the repair of target gaps by host polymerase and ligase as occurs in other transposon systems (Mizuuchi, 1984).
The ability of piggyBac to repair both the broken donor backbone and the target site by ligation of TTAA sequences flanking the gapped donor sites and the transposon ends rather than DNA synthesis could help make piggyBac independent of the host repair machinery and contribute to its functionality in a wide variety of organisms.
piggyBac transposase is likely a DDE recombinase
piggyBac transposase binds specifically to both the left and right ends of piggyBac transposon and promotes the Mg2+-dependent joining of a 3′OH transposon end to a target DNA. Thus, piggyBac uses the same target joining mechanism, that is, direct nucleophilic attack, as do the DDE recombinases (Rice and Baker, 2001). The catalytic cores of DDE recombinases share a common structure: an RNaseH-like fold on which the conserved acidic DDE/DDD amino acids are juxtaposed to coordinate Mg2+ ions that are essential cofactors for the chemical steps of transposition (Rice and Baker, 2001; Hickman et al, 2005; Richardson et al, 2006). However, there is no obvious sequence similarity between piggyBac and DDE recombinases (Sarkar et al, 2003).
From the results of secondary structure prediction and mutational analysis, we suggest that part of the piggyBac active site has a section of an RNaseH-like fold on which the D residues D268 and D346 that are essential for DNA breakage and joining are located in positions comparable to those of the N-terminal D residues of the DDE recombinases. We have also identified another essential C-terminal D, D447, that we suggest performs the same function as the C-terminal E in the DDE recombinases. Further analysis of these mutants will be required to establish if these amino acids have a role in transpososome formation as has been reported in the case of Mu transposase (Kim et al, 1995).
These results provide evidence that piggyBac is a member of the DDE recombinase superfamily, extending the range of protein sequences that can form this essential catalytic core.
Tn5 and Tn10 transposase carries out four distinct chemical reactions in Tn5 and Tn10 transposition: nicking at the 3′OH transposon end, hairpin formation, hairpin resolution and target joining. The available evidence argues that a single active site contributed from a single transposase monomer at each transposon end is used repeatedly to catalyse all four steps (Bolland and Kleckner, 1996; Reznikoff, 2003). The use of a single active site to carry out alternating hydrolysis and transesterification has been supported by studies on other proteins that use two metal ions in their active site that are held in place by acidic amino acids (Nowotny et al, 2005). As piggyBac transposase performs similar chemical reactions and mutations in the catalytic DDD residues block all of these steps, we are attracted to the view that piggyBac transposition similarly involves the activity of a single active site at each transposon end.
Genetic analysis of piggyBac transposition in S. cerevisiae
We have developed a piggyBac transposition system in the highly genetically tractable S. cerevisiae. In this system, we can directly follow the excision of a mini-piggyBac element from a Ura− derivative of the URA3 gene containing the transposon by measuring the frequency of uracil prototrophy in the presence of piggyBac transposase. piggyBac excision can occur at high frequency, that is, about 1/100 cells are uracil prototrophs in a single colony grown on media inducing the expression of the transposase. We have used this system to show that amino acids that we identified as being part of the catalytic site in vitro are also necessary for activity in vivo.
In addition to facilitating the analysis of piggyBac transposition in vivo, including the isolation of hyperactive mutants that will aid insertional mutagensis in mammalian cells, this piggyBac system is also well suited for the in vivo manipulation of the yeast genome.
Domesticated piggyBac transposases
Proteins related to piggyBac transposases are present in many eukaryotic genomes, including insects, fish and mammals. In the human genome, there are five piggyBac-derived genes, PGBD1–5 (Sarkar et al, 2003). Our work shows that PGBD1–3 and 5 are very unlikely to have transposase activity as at least one of the active site amino acids we have identified in T. ni piggyBac is mutated in each of these proteins. In PGBD4, however, the catalytically essential amino acids are intact, although this provides no proof that PGB4 is actually a transposase. Expressed cellular genes that are significantly related to transposases but are not flanked by terminal inverted repeats or target site duplications have been observed for many DNA transposons, some of which are ‘domesticated' transposases co-opted for by the cell for processes that do not involve DNA breakage and joining such as transcriptional regulation (Volff, 2006).
Materials and methods
piggyBac transposase expression and purification
The piggyBac transposase ORF (594 amino acids) was PCR amplified from the plasmid pBhelper (Lobo et al, 1999) and cloned between the NcoI and KpnI sites of plasmid pBAD Myc-HisC (Invitrogen) to generate a pBAD piggyBac–Myc–His6 fusion construct (p-transposase). E. coli Top10 (Invitrogen) cells containing p-transposase were grown with shaking at 30°C in LB medium containing 100 μg/ml ampicillin till an OD600 of 0.6. The culture was then induced with 0.1% L-arabinose for 18 h at 16°C. Following induction, cells were lysed with a French Pressure cell (Fisher Scientific) in TSG buffer (20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 10% v/v glycerol). The lysate was then loaded onto a pre-equilibrated Ni2+ Sepharose column (GE Healthcare). The column was washed with 10 column volumes of TSG buffer followed by 6 column volumes of TSG+50 mM imidazole buffer. piggyBac–Myc–His fusion protein was eluted with TSG+200 mM imidazole buffer, dialysed against storage buffer (20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 25% v/v glycerol) and stored at −80°C.
piggyBac DSB and target joining reactions
150 nM of piggyBac transposase was incubated with 1.5 nM of radiolabelled piggyBac L-TIR or R-TIR DNA (see Supplementary data) in 25 mM HEPES (pH 8.0), 3 mM Tris (pH 8.0), 75 mM NaCl, 2 mM DTT, 10 mM MgCl2, 0.01% BSA, 3.75% glycerol and 10 nM pUC19 in a final volume of 20 μl at 30°C for different time intervals. Reactions were stopped by incubation in 1% SDS and 20 mM EDTA for 30 min at 65°C and displayed on a 1% native agarose–1 × TAE gel. For analysis of DNA nicking and hairpin formation, the reaction products were phenol/chloroform extracted, ethanol precipitated and displayed on a 5% acrylamide–7 M urea–1 × TBE denaturing acrylamide gel. All gels described here were dried and exposed to phosphoimager plates and analysed by Imagequant software (GE Healthcare).
Analysis of hairpin formation using a pre-nicked piggyBac left end
A 35 bp oligonucleotide corresponding to the 3′ end of piggyBac L-TIR13-3-19 was radiolabelled at its 5′ end with [γ-32P]ATP (GE Healthcare) and T4 polynucleotide kinase. After purification on a G25 column, the end-labelled oligonucleotide was annealed with equimolar amounts of a 59 bp oligonucleotide containing the 5′ end of piggyBac L-TIR13-3-19 flanked by 24 nt flanking donor DNA and a 24 nt oligonucleotide corresponding to the bottom strand of flanking donor DNA. The annealed oligonucleotide mixture was then directly used as the substrate in DSB and target joining reactions as described above. The reaction products were displayed on native agarose, denaturing acrylamide and 1% agarose–50 mM NaOH gels.
Supplementary Material
Supplementary Figures 1–12
Supplementary data
Acknowledgments
We thank David O'Brochta (University of Maryland, College Park) for providing the piggyBac transposon plasmids, and Abram Gabriel (Rutgers University) and Jef Boeke (JHU-SOM) for providing yeast plasmids and strains. We also thank the Craig lab members for their valuable insights into this project. This work was partly supported by National Institute of Health (NIH) grant NC90020064 to NLC. NLC is an Investigator of the Howard Hughes Medical Institute.
References
- Arkhipova IR, Meselson M (2005) Diverse DNA transposons in rotifers of the class Bdelloidea. Proc Natl Acad Sci USA 102: 11781–11786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ason B, Reznikoff WS (2004) A high-throughput assay for Tn5 Tnp-induced DNA cleavage. Nucleic Acids Res 32: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu B, Shoue DA, Fraser MJ Jr, Adams JH (2005) High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proc Natl Acad Sci USA 102: 16391–16396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin A, Goryshin IY, Reznikoff WS (1999) Hairpin formation in Tn5 transposition. J Biol Chem 274: 37021–37029 [DOI] [PubMed] [Google Scholar]
- Bienz M (2006) The PHD finger, a nuclear protein-interaction domain. Trends Biol Sci 31: 35–40 [DOI] [PubMed] [Google Scholar]
- Bischerour J, Chalmers R (2007) Base-flipping dynamics in a DNA hairpin processing reaction. Nucleic Acids Res 35: 2584–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD (2002) Putting mobile DNA to work: the toolbox. In Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds), pp 24–37. Washington, DC: American Society for Microbiology Press [Google Scholar]
- Bolland S, Kleckner N (1996) The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell 84: 223–233 [DOI] [PubMed] [Google Scholar]
- Brown PO, Bowerman B, Varmus HE, Bishop JM (1987) Correct integration of retroviral DNA in vitro. Cell 49: 347–356 [DOI] [PubMed] [Google Scholar]
- Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ (1989) Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172: 156–169 [DOI] [PubMed] [Google Scholar]
- Coen ES, Carpenter R, Martin C (1986) Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell 47: 285–296 [DOI] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289: 77–85 [DOI] [PubMed] [Google Scholar]
- del Pilar Garcillan-Barcia M, Bernales I, Mendiola MV, de la Cruz F (2001) Single-stranded DNA intermediates in IS91 rolling-circle transposition. Mol Microbiol 39: 494–501 [DOI] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T (2005) Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122: 473–483 [DOI] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Elick TA, Lobo N, Fraser MJ Jr (1997) Analysis of the cis-acting DNA elements required for piggyBac transposable element excision. Mol Gen Genet 255: 605–610 [DOI] [PubMed] [Google Scholar]
- Engels WR, Johnson-Schlitz DM, Eggleston WB, Sved J (1990) High-frequency P element loss in Drosophila is homolog dependent. Cell 62: 515–525 [DOI] [PubMed] [Google Scholar]
- Fraser MJ, Cary L, Boonvisudhi K, Wang HG (1995) Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology 211: 397–407 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Estevez C, Momose T, Gehring WJ, Salo E (2003) Transgenic planarian lines obtained by electroporation using transposon-derived vectors and an eye-specific GFP marker. Proc Natl Acad Sci USA 100: 14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley ND, Whiteson KL, Rice PA (2006) Mechanisms of site-specific recombination. Annu Rev Biochem 75: 567–605 [DOI] [PubMed] [Google Scholar]
- Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F (2005) Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol 12: 715–721 [DOI] [PubMed] [Google Scholar]
- Hikosaka A, Kobayashi T, Saito Y, Kawahara A (2007) Evolution of the Xenopus piggyBac transposon family TxpB: domesticated and untamed strategies of transposon subfamilies. Mol Biol Evol 12: 2648–2656 [DOI] [PubMed] [Google Scholar]
- Jones DT (1999a) GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J Mol Biol 287: 797–815 [DOI] [PubMed] [Google Scholar]
- Jones DT (1999b) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J (2001) Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA 98: 8714–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr (2004) Mobile elements: drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Kennedy A, Guhathakurta A, Kleckner N, Haniford DB (1998) Tn10 transposition via a DNA hairpin intermediate. Cell 95: 125–134 [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR (2000) Transposable elements and host genome evolution. Trends Ecol Evol 15: 95–99 [DOI] [PubMed] [Google Scholar]
- Kim K, Namgoong S-Y, Jayaram M, Harshey RM (1995) Step-arrest mutants of phage Mu transposase. J Biol Chem 270: 1472–1479 [DOI] [PubMed] [Google Scholar]
- Li X, Harrell RA, Handler AM, Beam T, Hennessy K, Fraser MJ Jr (2005) piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol Biol 14: 17–30 [DOI] [PubMed] [Google Scholar]
- Lobo N, Li X, Fraser MJ Jr (1999) Transposition of the piggyBac element in embryos of Drosophila melanogaster, Aedes aegypti and Trichoplusia ni. Mol Gen Genet 261: 803–810 [DOI] [PubMed] [Google Scholar]
- Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB (2006) Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat Struct Mol Biol 13: 1010–1015 [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605 [DOI] [PubMed] [Google Scholar]
- Mizuuchi K (1983) In vitro transposition of bacteriophage Mu: a biochemical approach to a novel replication reaction. Cell 35: 785–794 [DOI] [PubMed] [Google Scholar]
- Mizuuchi K (1984) Mechanism of transposition of bacteriophage Mu: polarity of the strand transfer reaction at the initiation of transposition. Cell 39: 395–404 [DOI] [PubMed] [Google Scholar]
- Morales ME, Mann VH, Kines KJ, Gobert GN, Fraser MJ Jr, Kalinna BH, Correnti JM, Pearce EJ, Brindley PJ (2007) piggyBac transposon mediated transgenesis of the human blood fluke, Schistosoma mansoni. FASEB J 21: 3479–3489 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins-Balding D, Duval-Valentin G, Glasgow AC (1999) Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J Bacteriol 181: 4937–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RH (1991) The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J 10: 1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff WS (2003) Tn5 as a model for understanding DNA transposition. Mol Microbiol 47: 1199–1206 [DOI] [PubMed] [Google Scholar]
- Rice PA, Baker TA (2001) Comparative architecture of transposase and integrase complexes. Nat Struct Biol 8: 302–307 [DOI] [PubMed] [Google Scholar]
- Richardson JM, Dawson A, O'Hagan N, Taylor P, Finnegan DJ, Walkinshaw MD (2006) Mechanism of Mos1 transposition: insights from structural analysis. EMBO J 25: 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Sim C, Hong YS, Hogan JR, Fraser MJ, Robertson HM, Collins FH (2003) Molecular evolutionary analysis of the widespread piggyBac transposon family and related ‘domesticated' sequences. Mol Genet Genomics 270: 173–180 [DOI] [PubMed] [Google Scholar]
- van Gent DC, Mizuuchi K, Gellert M (1996) Similarities between initiation of V(D)J recombination and retroviral integration. Science 271: 1592–1594 [DOI] [PubMed] [Google Scholar]
- Volff JN (2006) Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays 28: 913–922 [DOI] [PubMed] [Google Scholar]
- Weil CF, Kunze R (2000) Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nature 26: 187–190 [DOI] [PubMed] [Google Scholar]
- Williams TL, Jackson EL, Carritte A, Baker TA (1999) Organization and dynamics of the Mu transpososome: recombination by communication between two active site. Genes Dev 13: 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Coates CJ, George AL Jr (2007) piggyBac transposon-mediated gene transfer in human cells. Mol Ther 15: 139–145 [DOI] [PubMed] [Google Scholar]
- Wu Z, Chaconas G (1992) Flanking host sequences can exert an inhibitory effect on the cleavage step of the in vitro mu DNA strand transfer reaction. J Biol Chem 267: 9552–9558 [PubMed] [Google Scholar]
- York D, Reznikoff WS (1997) DNA binding and phasing analysis of Tn5 transposase and monomeric variant. Nucleic Acids Res 25: 2153–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Gabriel A (1999) Patching broken chromosomes with extranuclear cellular DNA. Mol Cell 4: 873–881 [DOI] [PubMed] [Google Scholar]
- Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL (2004) Transposition of hAT elements links transposable elements and V(D)J recombination. Nature 432: 995–1001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–12
Supplementary data