Abstract
ATP synthase converts the electrochemical potential at the inner mitochondrial membrane into chemical energy, producing the ATP that powers the cell. Using electron cryo-tomography we show that the ATP synthase of mammalian mitochondria is arranged in long ∼1-μm rows of dimeric supercomplexes, located at the apex of cristae membranes. The dimer ribbons enforce a strong local curvature on the membrane with a 17-nm outer radius. Calculations of the electrostatic field strength indicate a significant increase in charge density, and thus in the local pH gradient of ∼0.5 units in regions of high membrane curvature. We conclude that the mitochondrial cristae act as proton traps, and that the proton sink of the ATP synthase at the apex of the compartment favours effective ATP synthesis under proton-limited conditions. We propose that the mitochondrial ATP synthase organises itself into dimer ribbons to optimise its own performance.
Keywords: ATP synthase, ATP synthesis, electron cryo-tomography, mitochondrial membranes, structure
Introduction
The mitochondrial respiratory chain converts metabolic energy into an electrochemical proton gradient across the inner mitochondrial membrane. This potential gradient is then utilised by the F1F0 ATP synthase, the molecular machine that generates ATP from ADP and phosphate by rotary catalysis. Mitochondrial ATP synthase consist of up to 18 different subunits (Meyer et al, 2007) with a total molecular mass of ∼600 kDa (Rubinstein et al, 2003), and overall dimensions of 19.1 nm by 13.7 nm by 11.0 nm (Abrahams et al, 1994; Stock et al, 1999). The globular F1 head protrudes into the mitochondrial matrix, whereas the hydrophobic F0 part is embedded in the inner mitochondrial membrane. The two parts are connected by a central and a peripheral stalk (Walker and Dickson, 2006). The F1 heads were discovered as abundant ∼10-nm particles on mitochondrial vesicles by negative stain electron microscopy (Fernandez-Moran, 1962; Kagawa and Racker, 1966a), and are shown to be the sites of ATP synthesis (Kagawa and Racker, 1966b). Atomic structures of the F1 head of ATP synthase from bovine heart mitochondria with parts of the central stalk (Abrahams et al, 1994) and of the c-ring rotor from the F0 part of a prokaryotic F-type ATP synthase (Meier et al, 2005) have been determined.
While prokaryotic ATP synthase is thought to occur exclusively as a monomer, dimers and, occasionally, higher oligomers of ATP synthase have been found in mitochondria of Chlamydomonas (van Lis et al, 2003), Polytomella (Dudkina et al, 2005), yeast (Arnold et al, 1998), various plant species (Eubel et al, 2003), and bovine (Schägger and Pfeiffer, 2000) or rat heart (Wittig et al, 2006) by mild detergent treatment and native gel electrophoresis, and shown to be catalytically active (Wittig et al, 2006). In yeast, subunits b, h, and i (Fronzes et al, 2006), as well as the dimer-specific subunits e and g (Arnold et al, 1998) appear to be involved in dimer formation. Deletion of subunits e or g resulted in abnormal mitochondria with layered onion-like inner membranes instead of cristae (Paumard et al, 2002). With the exception of h, all these subunits contain one or two predicted transmembrane helices, suggesting that the monomers are connected by hydrophobic contacts in the membrane. Although isolated mitochondrial ATP synthase is fully active as a monomer, the ubiquitous occurrence of ATP synthase dimers suggests that they may have a critical role in eukaryotic oxidative phosphorylation.
Supramolecular assemblies of other mitochondrial respiratory chain complexes, notably complex I, III, and IV, have been described (Schägger, 2002) and their structures studied by electron microscopy (Schäfer et al, 2006, 2007). These supercomplexes have been implicated in the compartmentalisation of the respiratory chain for efficient electron transport (Schäfer et al, 2006, 2007; Boekema and Braun, 2007). The macromolecular complexes that work together in oxidative phosphorylation may thus be organised at a higher level in the membrane to optimise this fundamental process. If this is indeed the case, it should be possible to visualise their spatial arrangement in situ by electron microscopy. The ATP synthase should be particularly suitable for such studies, as its large F1 part acts as a conspicuous, built-in marker. We therefore investigated the organisation of the ATP synthase in the inner mitochondrial membrane by electron cryo-tomography, the method of choice for three-dimensional visualisation of such large macromolecular complexes. We found that the ATP synthase in mitochondrial membranes is organised in long ribbons of dimers, and conclude that this arrangement is at least partly responsible for shaping the cristae.
Results and discussion
Mitochondria contain active oligomers of ATP synthase
Mitochondria isolated from beef heart and rat liver are highly active in ATP hydrolysis (Glaser et al, 1980). We first investigated the oligomeric state of ATP synthase biochemically by extracting whole mitochondria with low amounts of digitonin. Native gels of the crude extracts stained for ATPase activity indicated a characteristic ladder of bands (Figure 1), shown to represent monomers, dimers, and higher oligomers of the ATP synthase (Wittig and Schägger, 2005). Mild sonication produces large fragments and vesicles that sediment at a g force as used for whole mitochondria. Mitochondrial membranes from rat liver and bovine heart both show high levels of dicyclohexylcarbodiimide (DCCD)-sensitive ATPase activity, with rates of 0.47 and 1.95 μmol phosphate produced per milligram protein per minute (Glaser et al, 1980), respectively. The sedimented membrane fractions contained 60–70% of the ATPase activity.
Figure 1.
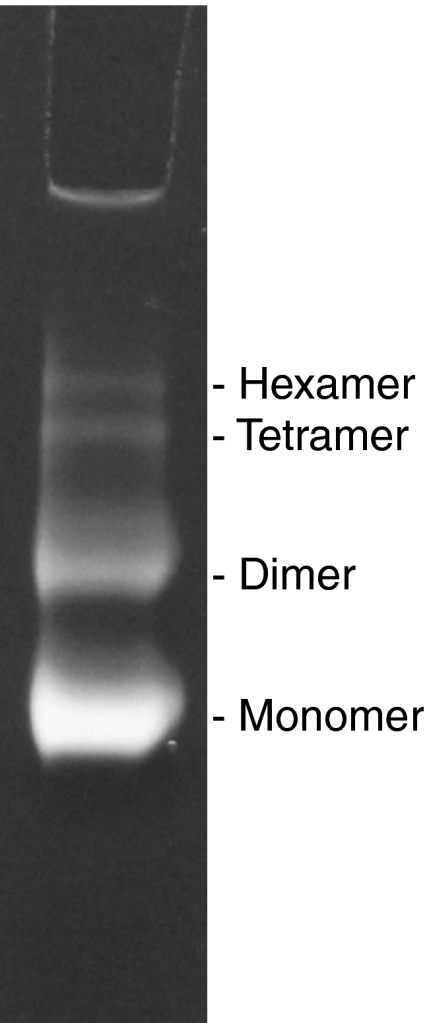
Activity of ATP synthase oligomers. Native gel of whole, digitonin-solubilised beef heart mitochondria stained for ATPase activity. The lowest band corresponds to the ATPase monomer, with higher bands indicating active oligomers up to the hexamer (Wittig and Schägger, 2005).
Mitochondrial ATP synthase forms dimer ribbons in the membrane
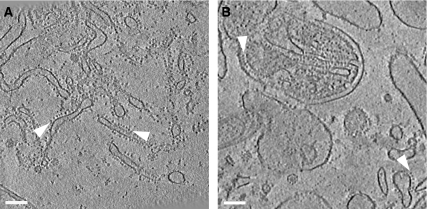
Electron tomograms of such minimally disrupted preparations from rat liver (Figure 2A) and bovine heart (Figure 2B) show mitochondrial fragments and membrane vesicles. Many of the vesicles are studded with ∼10-nm particles typical of the F1 heads of ATP synthase. All F1 heads have the same orientation with respect to the vesicle, indicating that the naturally asymmetrical protein organisation in the membrane is preserved. The orientation of the F1 heads towards the outside, as well as the tubular or lamellar shape of the vesicles indicates that they are detached inner membrane cristae. Most of the F1 heads are found in rows measuring up to 1 μm in the direction of the longest dimensions of elongated vesicles, or around the perimeter of round, disk-shaped vesicles (Figures 2 and 3).
Figure 2.
ATP synthase is arranged in rows. Slices of typical tomograms of membrane vesicles from rat liver (A) and bovine heart (B) mitochondria. Near the centre in panel B, a fragment of a mitochondrion with folded cristae and outer membrane is visible. Arrowheads indicate rows of ∼10-nm F1 ATP synthase heads. Slice thickness is 2.8 nm in panel A and 4.5 nm in panel B. Scale bars, 100 nm.
Figure 3.
Tomographic series of ATP synthase dimer rows. Slices in x–y show a disk-shaped crista vesicle from bovine heart mitochondria. Each slice has a thickness of 3 nm, and the spacing between slices is 7 nm. At given x–y positions on consecutive sections from upper left to lower right, a circular row of F1 heads appears and then disappears, and then a second, parallel row becomes visible. Scale bar, 50 nm.
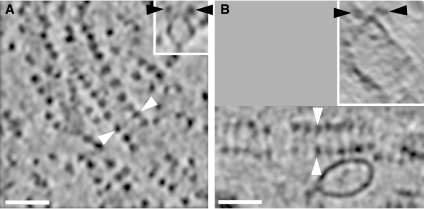
Closer examination of consecutive sections through tomograms revealed that there seem to be always two rows of F1 heads running in parallel, separated by a small, defined distance. The ATP synthase complexes in the cristae vesicles thus form long ribbons of up to 80 dimers. This striking arrangement is evident when the dimer ribbons happen to run parallel to the x–y plane of the tomogram. More often, they are obvious only when the tomograms are viewed in an oblique direction, in slices tangential or perpendicular to a dimer row (Figure 4). No rows of monomers were identified in any of the tomograms. Three typical examples of dimer ribbons are shown in Figure 5. Sub-volumes of tomograms containing either long tubular vesicles (Figure 5A) or round, disk-shaped ones (Figure 5B and C) were rendered manually with standard segmentation software. The dimer ribbons are always found in the most highly curved membrane regions. The same dimer rows are also visible in the inner membranes of small, undisrupted yeast mitochondria (unpublished results). We can therefore exclude the unlikely possibility that the dimer ribbons are an artefact of the membrane preparation.
Figure 4.
Dimer rows in bovine and rat mitochondria. Ribbons of ATP synthase dimers in tomograms of rat liver (A) and bovine heart (B) mitochondrial membranes. Black or white arrowheads indicate the same pairs of ATP synthase heads in (A) or (B), respectively. White arrowheads indicate the direction of perpendicular sections in the inserts. Slab thickness is 2.8 nm in panel A and 4.5 nm in panel B. Scale bars, 50 nm.
Figure 5.
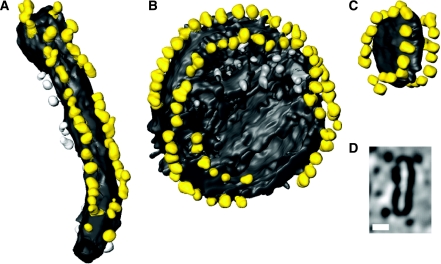
Tomographic volumes of dimer ribbons. Surface views of dimer ribbons in a tubular crista vesicle from rat liver mitochondria (A) and in round vesicles from bovine heart mitochondria (B, C). (D) A cross section through the tomographic volume of the small vesicle in panel C. F1 heads are yellow, while the membrane is grey. Particles not assigned to dimer ribbons are shown in lighter grey. The length of the tube in panel A is 280 nm, and the diameter of the vesicles in panels B and C is 180 and 51 nm, respectively. Scale bar in panel D, 20 nm.
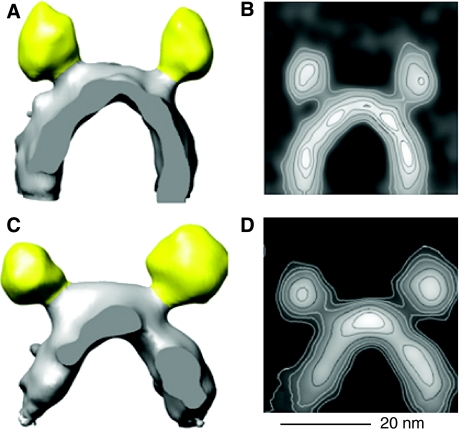
Previous electron microscopy studies of ATP synthase dimers produced projection maps of isolated, detergent-solubilised complexes (Dudkina et al, 2005, 2006). By contrast, our study shows the three-dimensional (3D) volume of dimers in situ in the inner mitochondrial membrane, and reveals that they form extensive linear supramolecular arrays. We calculated 3D volumes of the mitochondrial ATP synthase dimers by selecting 235 (rat liver) and 90 pairs (bovine heart) in the tomographic volumes and averaging them (Figure 6). An estimate obtained by Fourier shell correlation of two equal subgroups of the rat liver dimers indicated a resolution of 5 nm. The F1 head is connected by a narrow stalk to the F0 part in the membrane, which is resolved in the contour plot of Figure 6B. The distance between F1 heads is ∼28 nm, and the centres of the F0 assemblies in the membrane are ∼13 nm apart. The distance between dimers along the ribbon is ∼12 nm. The angle between the central stalks is roughly 70° in rat liver dimers, which were averaged over long, approximately straight rows. In bovine heart dimers this angle depends on the curvature of the rows and ranges from 55° to 95°, indicating that the dimer link is flexible.
Figure 6.
Averaged dimer volumes. Averaged 3D volumes of ATP synthase dimers from rat liver (A) and bovine heart membranes (C) in situ. F1 heads are coloured as in Figure 5. In panel A, 235 dimer volumes were averaged. In panel C, 90 volumes of the largest of several classes with different ribbon curvature and, hence, dimer angles were averaged. Contour plots (B, D) of volumes in panels A, C projected in the viewing direction. Scale bar, 20 nm.
Dimer ribbons are a ubiquitous feature of mitochondrial membranes
A comparison of our averaged 3D volumes of the ATP synthase dimers to the published map of the detergent-solubilised, ATP synthase monomer (Rubinstein et al, 2003) from bovine heart mitochondria indicated a good match both in dimension and shape. The peripheral stalk is not visible in our tomographic volumes due to limited resolution. The overall arrangement and dimensions of detergent-solubilised ATP synthase dimers from mitochondria of yeast (Dudkina et al, 2006), and the unicellular flagellate Polytomella (Dudkina et al, 2005) agree with our findings, with angles of ∼90° and 70° between monomers, respectively. Ribbons of ATP synthase have previously been observed by electron microscopy of deep-etched Paramecium mitochondria (Allen et al, 1989), but the same approach failed to reveal a similar arrangement in mammalian specimens (Allen et al, 1989). It was therefore unclear whether or not the dimer ribbons were a special feature of ciliate mitochondria. Our results show ATP synthase dimer ribbons in two different mammalian species. Moreover, a recent atomic force microscopy (AFM) study has reported similar but much shorter arrays in yeast (Buzhynskyy et al, 2007). We conclude that the ribbons of ATP synthase dimers are common to all eukaryotes.
The dimer rows in Paramecium (Allen et al, 1989) and yeast (Buzhynskyy et al, 2007) suggest a interdigitated, zipper-like arrangement of the ATP synthase monomers. This is not evident in the long dimer rows we observe in rat liver (Figures 4A, 5A, 6A) and beef heart (Figure 4B, 5B–D, 6C). Further studies will show if the interdigitated rows are perhaps a characteristic of lower eukaryotes. Buzhynskyy et al (2007) hypothesise that this arrangement may reflect the active rather than an inhibited state of the dimer. We have demonstrated that our preparations are highly active, even though the F1 heads do not appear to interdigitate, but we cannot exclude that the F0 parts in the membrane may be staggered, as seen by AFM of the small clusters in yeast.
Our 3D volumes indicate that the rows of dimers are found consistently in the most tightly curved membrane regions, with an outer radius of ∼17 nm. In the average volume of Figure 6C, the dimer ribbons are associated with a sharp ∼60° bend in the lipid bilayer. Considering the small vesicle shown in Figure 5C, where the dimers form a circular ribbon around the perimeter of a ∼20 nm wide disk, there is little room for complexes other than the ATP synthase to bend the membrane. Vesicles reconstituted with monomeric ATP synthase appear perfectly spherical in our tomograms (results not shown). We conclude that the dimer ribbons are responsible for imposing a high local curvature on the inner mitochondrial membrane.
Although a local bending of the membrane by ATP synthase dimers has been postulated (Dudkina et al, 2005), this has not been demonstrated before. The AFM study (Buzhynskyy et al, 2007) did not reveal membrane curvature, most likely because the yeast membranes were flattened against the substrate. The small radius of curvature at the dimer ribbons is the same as that found in tubular cristae and at the apex of lamelliform cristae (Nicastro et al, 2000), implying that the ATP synthase arrays are a key factor in shaping cristae morphology. Presumably the other complexes of the respiratory chain, which act as proton pumps, as well as other essential membrane proteins, such as the abundant nucleotide and phosphate carriers (Palmieri et al, 2006), populate the less highly curved regions.
Membrane curvature affects charge density and local pH
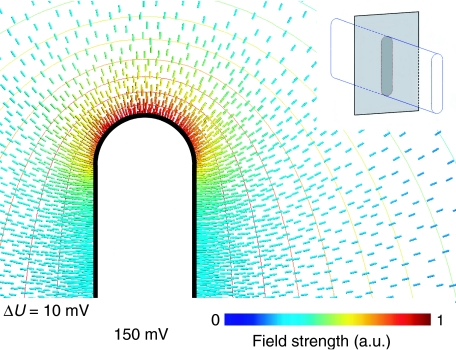
What is the effect of membrane curvature on the ionic conditions in the microenvironment within mitochondrial cristae? To address this question, we considered the number of protons that can be accommodated per unit area on a flat membrane surface, compared to one with a 17-nm outer radius of curvature. We used Finite Element Modelling to simulate the electrical field surrounding an idealised lamelliform vesicle with parallel planar membranes on the sides and half cylinders at either end, at a typical membrane potential of 150 mV (Brand, 1995). A plot calculated for an axial ratio of 10:1 (Figure 7) indicates that the electrostatic field is strongest in the most highly curved regions, as expected. The divergence of the electric field is proportional to the electric charge, so that the field strength at the membrane surface is a direct measure of the surface charge density. Our numerical simulations enable us to calculate the difference in charge density or local pH. For the assumed geometry, the charge density on the curved membrane surface is up to 3.5 times higher than on planar membranes at the same constant membrane potential. This translates into an increase in the local pH difference along the membrane by ∼0.5 units.
Figure 7.
Simulation of electric field on membrane surface. Electric field strength around a cristae-shaped membrane surface calculated for a constant potential of 150 mV. The isopotential surfaces (ΔU=10 mV) are more closely spaced around the curved edges than along the planar sides, indicating that the potential gradient is strongest in the curved regions. The corresponding charge density on the inner membrane surface (solid black line) is directly proportional to the field strength. The numerical values indicate a 3.5-fold increase in surface charge density at the apex (red field lines), compared with the flat sides (blue field lines). The insert illustrates the geometry used to solve the electrostatic Poisson equation. The apical field strength is higher for larger axial ratios, but does not vary strongly with this parameter.
Functional significance
Cristae are perhaps the most notable ultrastructural feature of active mitochondria. Whether simply to increase the surface area of the inner membrane, or to serve another purpose, such as optimising ATP synthesis, the function of these invaginations has not been definitively explained. Our results suggest that the ATP synthase dimers impose a high local curvature on the inner mitochondrial membrane, as the ribbons are found either on the crest of lamellae, or along the length of tubular cristae. Considering that the enzyme is fully functional in the monomeric form, does this arrangement confer any advantage for ATP production?
ATP synthesis is driven by the proton-motive force (PMF), which consists of a ΔpH and a ΔΨ term, accounting to the driving force due to the pH gradient and the membrane potential, respectively. The numerical solution to the Poisson equation (Figure 7) indicates a higher charge density, and hence a higher proton concentration in areas of sharp membrane curvature (Figure 8). Consequently, the ΔpH contribution to the PMF increases for highly curved regions. Assuming that the PMF along the inner membrane is constant at thermodynamic equilibrium, all protons in the mitochondrial cristae would be at the same chemical potential, but the pH contribution to the PMF will be larger for protons at the apex.
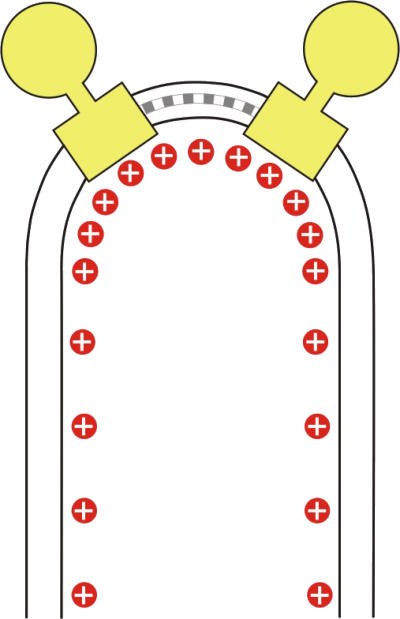
Figure 8.
Proton density at position of dimers in the membrane. Schematic drawing of an F1–F0 ATP synthase dimer (yellow) in the inner mitochondrial membrane. Monomers are connected by dimer-specific subunits (grey). The dimers associate into ribbons, which induce a tight bend in the membrane, with an ∼17-nm outer radius of curvature. The higher surface density of protons (red) in the curved membrane regions would result in a local pH difference of ∼0.5 units.
This raises the question of whether ATP synthesis benefits from an increased pH gradient. This would be the case if ΔpH is more effective in driving the enzyme than ΔΨ when the two terms are not equivalent (Junesch and Gräber, 1991; Kaim and Dimroth, 1999). Indeed, it has been shown that an increase in ΔpH increases the rate of ATP synthesis, even when a further increase of ΔΨ does not (Kaim and Dimroth, 1999). Similarly, the non-equivalence of ΔΨ and ΔpH has recently been demonstrated for the bacterial flagellar motor (Lo et al, 2007), the only other rotary machine in the membrane known to be driven by ion motive force.
Under ATP synthesis conditions, far removed from thermodynamic equilibrium, replenishing protons may become rate limiting, especially for species with the positive (p) side of the membrane at basic pH, as in alkaliphilic bacteria (Cherepanov et al, 2003) or at neutral pH, as in mitochondria. In fact, cristae might be seen as a further development of bacterial proton traps, which are invaginations in the bacterial inner membrane (Mulkidjanian et al, 2005). Placing a proton sink at the end of the invagination will enhance the efficiency of the trap. The gradient along the curved membrane would drive charges to the apex where we show the ATP synthase dimers are located. As protons move faster than other hydrated ions, particularly along the membrane surface (Cherepanov et al, 2004), they will reach the apex more quickly, and this may be an important advantage for ATP synthesis under limiting conditions.
Our conclusions are in full agreement with a recent study (Bornhövd et al, 2006), which shows that yeast mutants lacking the dimer-specific subunits e and g have a severely reduced growth rate, with a generation time up to ∼1.6 times longer than wild type, and a membrane potential that is reduced by ∼50%. As shown by Paumard et al (2002), the mitochondria of these mutants do not have cristae or tightly curved inner membrane regions. Together with these findings, our results suggest strongly that the ribbons of ATP synthase dimers are in fact responsible for cristae formation, and essential for optimal performance of the mitochondrial enzyme.
Conclusion
Using electron cryo-tomography, we have shown that ATP synthase forms ribbons of up to 80 dimers in the inner membrane of rat and bovine mitochondria. The supramolecular assemblies are very similar in both species, even though the overall membrane morphology of beef heart and rat liver mitochondria differs. Rows of ATP synthase dimers have also been observed in ciliates (Allen et al, 1989) and yeast (Buzhynskyy et al, 2007), and we conclude that they are a ubiquitous feature of mitochondrial membrane organisation. Biochemical analysis shows that the supramolecular assemblies are active. The dimer ribbons of ATP synthase seem to induce a conspicuous bend in the membrane, with a constant 17-nm radius of curvature. The shape of small inner membrane vesicles indicates that the ATP synthase dimers themselves enforce the local curvature of the membrane.
Numerical simulation indicates that, although the electrochemical potential must be the same along the entire inner membrane surface under equilibrium conditions, the local pH gradient, and hence the ΔpH contribution to the proton motive force is significantly higher in regions of high membrane curvature. As the ΔpH contribution seems to be more effective than ΔΨ in driving ATP synthesis, this would suggest that a position of the ATP synthase at the apex of mitochondrial cristae is optimal for ATP production. We propose that the ribbons of ATP dimers have evolved to bend the membrane in an optimal way, thus taking advantage of the locally increased pH at the apex of mitochondrial cristae, which act as proton traps. This arrangement would out-perform prokaryotes which do not have ATP synthase dimers, or inner membrane cristae. We propose that the mitochondrial ATP synthase provides an example of spatial self-organisation in the cell to optimise its own function, the fundamental process of ATP production.
Materials and methods
Preparation of mitochondria and native membranes biochemical characterisation
Energised mitochondria were purified from beef heart or rat liver by differential centrifugation using standard procedures (Hackenbrock, 1972), and sonicated on ice for 3–5 s to produce large submitochondrial vesicles, which sedimented at 8000–10 000 g for 10 min at 4°C.
Native gel electrophoresis and in-gel staining for ATPase activity of oligomers was performed according to Wittig and Schägger (2005). Briefly, beef heart mitochondria were extracted with 8 mg digitonin/mg protein and insoluble material was pelleted at 13 000 g for 10 min. The supernatant was separated on a native 3–10% acrylamid gel at 150 V for 6 h. The gel was incubated in assay buffer (270 mM glycine, 35 mM Tris pH 8.4) for 3 h, and ATPase activity was detected upon adding 16 mM MgCl2, 8 mM ATP and 0.2% lead nitrate as a white precipitate of lead phosphate.
Cryo-electron tomography
The sedimented fraction of mitochondrial fragments was resuspended in isolation buffer (Hackenbrock, 1972) and 20 μl of the suspension was mixed with 20 μl of 6-nm gold particles used as fiducial markers and applied to a glow discharged Quantifoil specimen support grid, blotted from one side in a humidified atmosphere for 5 s, and plunged into liquid ethane. Grids were mounted under liquid nitrogen and transferred at a temperature of 100 K into an FEI Polara electron microscope equipped with a field emission gun operated at 300 kV. Images were recorded with a 2k × 2k 863 GIF Tridiem energy filter (Gatan, Pleasanton, CA, USA) at 10–15 μm underfocus at a specimen temperature of 82 K, using a slit width of 20 eV. Uniaxial tilt series were recorded at intervals of 1°–1.5°, with a total dose of 1–2 × 104 e/nm2. Tomograms were generated using the IMOD tomography software (Kremer et al, 1996) and subjected to 20 cycles of denoising by non-linear anisotropic diffusion (Frangakis and Hegerl, 2001). The thickness of tomographic volumes ranged from 80 to 160 nm.
Volume rendering and dimer averaging
Segmentation of tomographic volumes was carried out manually with AMIRA (Mercury Systems, Düsseldorf, Germany). For averaging dimer volumes, individual ATP synthase dimers were marked in the tomogram using IMOD (Kremer et al, 1996). Dimer sub-volumes were then cut out, aligned, averaged, and refined in SPIDER (Frank et al, 1996). Surface views were produced with the UCSF Chimera (Pettersen et al, 2004) package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). Contour plots of projections were calculated with IMAGEJ (Abramoff et al, 2004).
Finite-element modelling of surface charge density
The proton distribution along curved membranes was modelled according to the physical properties of electric charges. Protons move rapidly along the membrane surface (Cherepanov et al, 2004), forming a thin charged layer. The charge distribution within this layer can be calculated from the Poisson equation for a fixed electrostatic potential by solving the differential equations for the potential U and electric field vector E: 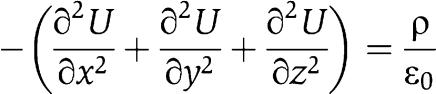 and
and 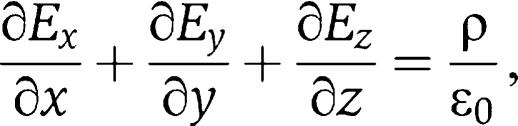 where ρ denotes the charge density and 1/ɛ0 the field strength of the unit charge. These equations were solved numerically for various model geometries by Finite Element Modelling, using the Gmsh/GetDP software package (http://www.geuz.org/gmsh/; http://www.geuz.org/getdp/) for geometric modelling, calculation of field strength and charge distribution, and for the visualisation of results.
where ρ denotes the charge density and 1/ɛ0 the field strength of the unit charge. These equations were solved numerically for various model geometries by Finite Element Modelling, using the Gmsh/GetDP software package (http://www.geuz.org/gmsh/; http://www.geuz.org/getdp/) for geometric modelling, calculation of field strength and charge distribution, and for the visualisation of results.
Acknowledgments
We thank Deryck Mills for expert assistance with operating the Polara electron microscope.
References
- Abrahams JP, Leslie AG, Lutter R, Walker JE (1994) Structure at 2.8 Å resolution of F1–ATPase from bovine heart mitochondria. Nature 370: 621–628 [DOI] [PubMed] [Google Scholar]
- Abramoff M, Magelhaes P, Ram S (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Allen RD, Schroeder CC, Fok AK (1989) An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J Cell Biol 108: 2233–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H (1998) Yeast mitochondrial F1F0–ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J 17: 7170–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema EJ, Braun HP (2007) Supramolecular structure of the mitochondrial oxidative phosphorylation system. J Biol Chem 282: 1–4 [DOI] [PubMed] [Google Scholar]
- Bornhövd C, Vogel F, Neupert W, Reichert AS (2006) Mitochondrial membrane potential is dependent on the oligomeric state of F1F0–ATP synthase supracomplexes. J Biol Chem 281: 13990–13998 [DOI] [PubMed] [Google Scholar]
- Brand MD (1995) Measurement of Mitochondrial Protonmotive Force. Oxford, England: IRL Press [Google Scholar]
- Buzhynskyy N, Sens P, Prima V, Sturgis JN, Scheuring S (2007) Rows of ATP synthase dimers in native mitochondrial inner membranes. Biophys J 93: 2870–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov DA, Feniouk BA, Junge W, Mulkidjanian AY (2003) Low dielectric permittivity of water at the membrane interface: effect on the energy coupling mechanism in biological membranes. Biophys J 85: 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov DA, Junge W, Mulkidjanian AY (2004) Proton transfer dynamics at the membrane/water interface: dependence on the fixed and mobile pH buffers, on the size and form of membrane particles, and on the interfacial potential barrier. Biophys J 86: 665–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun HP (2005) Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett 579: 5769–5772 [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Sunderhaus S, Braun HP, Boekema EJ (2006) Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett 580: 3427–3432 [DOI] [PubMed] [Google Scholar]
- Eubel H, Jansch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Moran H (1962) Cell-membrane ultrastructure. Low-temperature electron microsopy and X-ray diffraction studies of lipoprotein components in lamellar systems. Circulation 26: 1039–1065 [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R (2001) Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135: 239–250 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Fronzes R, Weimann T, Vaillier J, Velours J, Brethes D (2006) The peripheral stalk participates in the yeast ATP synthase dimerization independently of e and g subunits. Biochemistry 45: 6715–6723 [DOI] [PubMed] [Google Scholar]
- Glaser E, Norling B, Ernster L (1980) Reconstitution of mitochondrial oligomycin and dicyclohexylcarbodiimide-sensitive ATPase. Eur J Biochem 110: 225–235 [DOI] [PubMed] [Google Scholar]
- Hackenbrock CR (1972) Energy-linked ultrastructural transformations in isolated liver mitochondria and mitoplasts. Preservation of configurations by freeze-cleaving compared to chemical fixation. J Cell Biol 53: 450–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junesch U, Gräber P (1991) The rate of ATP-synthesis as a function of delta pH and delta psi catalyzed by the active, reduced H(+)-ATPase from chloroplasts. FEBS Lett 294: 275–278 [DOI] [PubMed] [Google Scholar]
- Kagawa Y, Racker E (1966a) Partial resolution of the enzymes catalyzing oxidative phosphorylation. IX. Reconstruction of oligomycin-sensitive adenosine triphosphatase. J Biol Chem 241: 2467–2474 [PubMed] [Google Scholar]
- Kagawa Y, Racker E (1966b) Partial resolution of the enzymes catalyzing oxidative phosphorylation. X. Correlation of morphology and function in submitochondrial particles. J Biol Chem 241: 2475–2482 [PubMed] [Google Scholar]
- Kaim G, Dimroth P (1999) ATP synthesis by F-type ATP synthase is obligatorily dependent on the transmembrane voltage. EMBO J 18: 4118–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Lo CJ, Leake MC, Pilizota T, Berry RM (2007) Nonequivalence of membrane voltage and ion-gradient as driving forces for the bacterial flagellar motor at low load. Biophys J 93: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier T, Polzer P, Diederichs K, Welte W, Dimroth P (2005) Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308: 659–662 [DOI] [PubMed] [Google Scholar]
- Meyer B, Wittig I, Trifilieff E, Karas M, Schägger H (2007) Identification of two proteins associated with mammalian ATP synthase. Mol Cell Proteomics 6: 1690–1699 [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, Cherepanov DA, Heberle J, Junge W (2005) Proton transfer dynamics at membrane/water interface and mechanism of biological energy conversion. Biochemistry (Mosc) 70: 251–256 [DOI] [PubMed] [Google Scholar]
- Nicastro D, Frangakis AS, Typke D, Baumeister W (2000) Cryo-electron tomography of neurospora mitochondria. J Struct Biol 129: 48–56 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Agrimi G, Blanco E, Castegna A, Di Noia MA, Iacobazzi V, Lasorsa FM, Marobbio CM, Palmieri L, Scarcia P, Todisco S, Vozza A, Walker J (2006) Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta 1757: 1249–1262 [DOI] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J (2002) The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 21: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Walker JE, Henderson R (2003) Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J 22: 6182–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer E, Dencher NA, Vonck J, Parcej DN (2007) Three-dimensional structure of the respiratory chain supercomplex I1III2IV1from bovine heart mitochondria. Biochemistry 46: 12579–12585 [DOI] [PubMed] [Google Scholar]
- Schäfer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J (2006) Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem 281: 15370–15375 [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H (2002) Respiratory chain supercomplexes of mitochondria and bacteria. Biochim Biophys Acta 1555: 154–159 [DOI] [PubMed] [Google Scholar]
- Stock D, Leslie AG, Walker JE (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286: 1700–1705 [DOI] [PubMed] [Google Scholar]
- van Lis R, Atteia A, Mendoza-Hernandez G, Gonzalez-Halphen D (2003) Identification of novel mitochondrial protein components of Chlamydomonas reinhardtii. A proteomic approach. Plant Physiol 132: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Dickson VK (2006) The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta 1757: 286–296 [DOI] [PubMed] [Google Scholar]
- Wittig I, Carrozzo R, Santorelli FM, Schägger H (2006) Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta 1757: 1066–1072 [DOI] [PubMed] [Google Scholar]
- Wittig I, Schägger H (2005) Advantages and limitations of clear-native PAGE. Proteomics 5: 4338–4346 [DOI] [PubMed] [Google Scholar]