Abstract
BTG2 is a prototype member of the BTG/Tob family of antiproliferative proteins, originally identified as a primary response gene induced by growth factors and tumour promoters. Its expression has been linked to diverse cellular processes such as cell-cycle progression, differentiation or apoptosis. BTG2 has also been shown to interact with the Pop2/Caf1 deadenylase. Here, we demonstrate that BTG2 is a general activator of mRNA decay, thereby contributing to gene expression control. Detailed characterizations of BTG2 show that it enhances deadenylation of all transcripts tested. Our results demonstrate that Caf1 nuclease activity is required for efficient deadenylation in mammalian cells and that the deadenylase activities of both Caf1 and its Ccr4 partner are required for Btg2-induced poly(A) degradation. General activation of deadenylation may represent a new mode of global regulation of gene expression, which could be important to allow rapid resetting of protein production during development or after specific stresses. This may constitute a common function for BTG/Tob family members.
Keywords: BTG, deadenylase, mRNA decay, poly(A), Tob
Introduction
Regulation of gene expression may occur at both the transcriptional and post-transcriptional level. In the recent years, it has become clear that mRNA degradation plays an important role in this process (Fan et al, 2002; Raghavan et al, 2002; Wang et al, 2002). Indeed, not only are half-lives of various mRNA very different (Herrick et al, 1990), but also the decay rates of individual mRNA can also be regulated in response to different stimuli, as is the case for transcripts encoding cytokines or proto-oncogenes bearing AU-rich elements in their 3′ untranslated region (UTR) (Chen and Shyu, 1995; Barreau et al, 2005). Accordingly, several mRNA turnover pathways with different functions have been identified including general mechanisms targeting functional mRNAs and quality control pathways affecting aberrant transcripts (Fasken and Corbett, 2005; Tang, 2005).
In eukaryotes, the cap structure and the poly(A) tail protect functional mRNA from degradation by exonucleases. In the general turnover pathways, mRNA degradation is initiated by shortening of the poly(A) tail. Most often, deadenylation induces decapping of the target mRNA through a poorly known mechanism. This triggers the rapid destruction of the mRNA body by the 5′-to-3′ exonuclease Xrn1. Alternatively, deadenylation is followed by the exosome-mediated degradation of the mRNA body in the 3′-to-5′ direction (Meyer et al, 2004). Deadenylation is a critical step for mRNA turnover, as poly(A) shortening removes the targeted mRNA from the translatable pool and initiates the mRNA degradation process. Deadenylation has also been shown to be the rate-limiting step of mRNA decay, yet little is known about how poly(A) tail shortening is initiated and regulated. Both in yeast and in mammals, the Ccr4–Pop2 complex was shown to bear the main catalytic activity responsible for cytoplasmic deadenylation (Daugeron et al, 2001; Tucker et al, 2001; Yamashita et al, 2005), whereas the second known cytoplasmic deadenylase, the Pan2–Pan3 complex, affects more particularly the initial phase of poly(A) shortening (Brown and Sachs, 1998; Yamashita et al, 2005). In mammals, two orthologues of Ccr4, Ccr4a and Ccr4b, and two orthologues of Pop2, Pop2 and Caf1, have been identified and shown to localize in the cytoplasm (Yamashita et al, 2005; Wagner et al, 2007). However, the respective contributions of these factors to deadenylation remain unclear.
Interestingly, two-hybrid screens have revealed an interaction of the Pop2/Caf1 deadenylase subunit with the mouse or human BTG2 factor (Bogdan et al, 1998; Rouault et al, 1998; Ikematsu et al, 1999; Prevot et al, 2001). The BTG2 gene is a prototype member of the BTG/Tob protein family of antiproliferative factors that are found in metazoan cells (Matsuda et al, 2001; Tirone, 2001; Duriez et al, 2004; Lim et al, 2006). The BTG/Tob family has been shown to contain six members in humans, which are divided into two distinct subfamilies, BTGs (BTG1–4) and Tob (Tob1 and 2), characterized by the presence of the conserved BTG (or APRO) domain in the N-terminal part of the proteins but differing in other specific features (Matsuda et al, 2001). The rat and mouse orthologues of BTG2 were initially reported to be primary response genes induced by growth factors and tumour promoters, as the corresponding transcripts and proteins are detected very rapidly after phorbol ester treatment of NIH3T3 cells or NGF induction of PC12 cells (Bradbury et al, 1991; Fletcher et al, 1991; Varnum et al, 1994). Their expression is transient, as BTG2 transcripts and proteins almost completely disappeared 3 h after treatment. The BTG2 protein is remarkably labile, exhibiting a half-life of less than 15 min (Varnum et al, 1994), potentially because it is a ubiquitin–proteasome target (Sasajima et al, 2002). The BTG2 gene is also induced upon several cellular stresses by p53-dependent and p53-independent mechanisms and shows reduced expression in a large number of tumour tissues (Boiko et al, 2006; Lim, 2006). Overexpression of BTG2 in diverse cell lines induces a partial inhibition of cell proliferation (Montagnoli et al, 1996; Lim et al, 1998). The detection of an interaction of BTG2 with Pop2/Caf1 prompted us to test whether it could affect mRNA decay. Here, we demonstrate that overexpression of BTG2 causes accelerated deadenylation of reporters and of endogenous transcripts. This BTG2 function requires a direct interaction between BTG2 and Pop2/Caf1 as well as active Pop2/Caf1 and Ccr4 deadenylases. These observations implicate BTG2 in the general control of mRNA decay.
Results
BTG2 increases the turnover rates of reporter transcripts
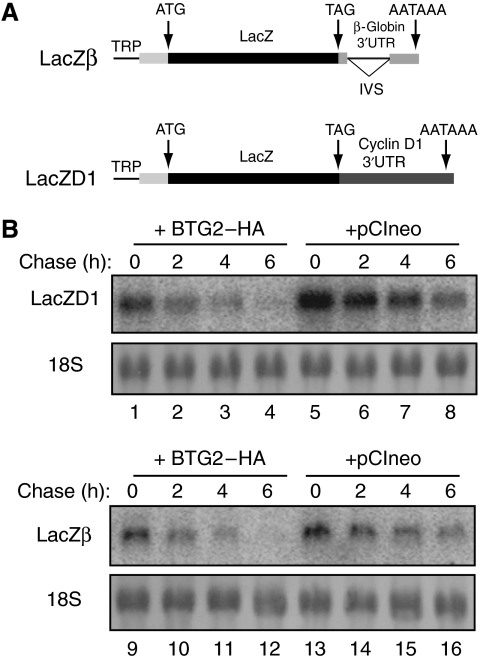
Several studies have reported that the 3′ UTR of the cyclin D1 mRNA that contains AU-rich elements renders this transcript unstable (Dufourny et al, 2000; Lin et al, 2000; Guo et al, 2005). Because BTG2 expression was shown to downregulate cyclin D1 mRNA level (Guardavaccaro et al, 2000; Kwon et al, 2005), we tested whether BTG2 could modulate the degradation of a reporter transcript carrying the cyclin D1 3′UTR. For this purpose, we constructed two reporter plasmids that contain the LacZ coding sequence fused either to the β-globin 3′UTR (LacZβ) or to the cyclin D1 3′UTR (LacZD1). Both reporters are under the control of a Tet-regulated promoter (Figure 1A). Reporters LacZβ and lacZD1 were transfected in HEK293-TOF cells, stably expressing the Tet-Off transcriptional activator, together with either a plasmid expressing HA-tagged human BTG2 or as a control the corresponding empty vector. Two days after transfection, transcriptional chase experiments were performed by adding doxycyclin in the cell medium to block further transcription of the reporter. RNA analyses revealed that the turnover of the LacZD1 reporter was increased in cells coexpressing BTG2–HA (Figure 1B, compare lanes 1–4 to lanes 5–8). Unexpectedly, similar results were also observed with the LacZβ reporter (Figure 1B, compare lanes 9–12 to lanes 13–16). Quantitative RT–PCR analyses allowed the precise determination of the half-lives of the two reporters, confirming the northern blot results (Table I). We conclude that BTG2 expression shortened the half-lives of the two reporter mRNAs tested, independently of their 3′UTR sequence.
Figure 1.
Cotransfection with BTG2 increases turnover of two reporter transcripts. (A) Schematic representation of the LacZβ and LacZD1 reporters. Black bars, coding sequences; grey bars, 5′ and 3′ UTR. TRP: tetracyclin responsive promoter; IVS: intervening sequence. (B) Transcriptional chase experiments showing mRNA decay of the LacZβ and LacZD1 reporters in the presence (+BTG2–HA) or absence (+pCIneo) of ectopically expressed BTG2 protein. HEK293-TOF cells were cotransfected with 0.2 μg of reporter plasmids and 0.4 μg of empty or BTG2–HA-expressing pCIneo plasmids. Chase time indications correspond to hours after doxycyclin addition. 18S ribosomal RNA (rRNA) staining with methylene blue is shown to demonstrate equal loading.
Table 1.
Effect of BTG2 expression on reporter mRNA half-lives
| Reporters | mRNA half-lives (h) |
|---|---|
| LacZβ+pCIneo | 3.80±0.26 |
| LacZβ+BTG2-HA | 1.80±0.3 |
| LacZD1+pCIneo | 2.80±0.3 |
| LacZD1+BTG2-HA | 1.95±0.05 |
| Half-lives of the LacZβ and LacZD1 reporters in cells cotransfected with empty pCIneo vector or BTG2-HA-expressing vector were quantified by quantitative RT–PCR. Values are the average of three independent biological replicates. | |
BTG2 stimulates mRNA decay by increasing the deadenylation rate
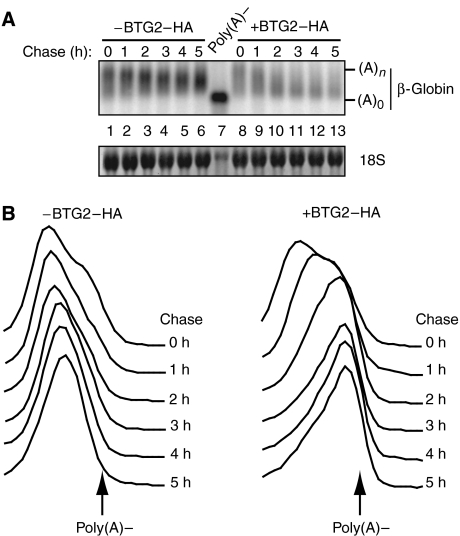
The observation that BTG2–HA affected the LacZβ reporter prompted us to test whether the small β-globin reporter (pTet-β-globin; Xu et al, 1998; Couttet and Grange, 2004) was also affected. Pulse–chase experiments were performed after cotransfection of the β-globin reporter in HEK293-TOF cells with either a control plasmid or a plasmid expressing BTG2–HA. Northern blot analysis of RNA extracted at various time points after doxycyclin addition revealed an increased rate of deadenylation for the β-globin reporter cotransfected with BTG2–HA as compared to the control (Figure 2A). This can be more easily visualized by comparing the evolution of migration profile of reporter mRNAs (Figure 2B). When the reporter was expressed alone, a peak shifting slowly as a function of time towards short poly(A) tail length was observed, reflecting a slow deadenylation. In contrast, for the reporter coexpressed with BTG2–HA, a rapid shift of the peak was observed, indicating a rapid deadenylation. In the latter case, deadenylation was essentially complete after 3 h of chase (Figure 2B), whereas in the former case deadenylation was still incomplete at the 5 h time point (note that deadenylation did not totally remove the poly(A) tail but left a few A residues at the end of the mRNA (compare lanes 13 and 7 in Figure 2A), as is usually observed during mRNA degradation by the general decay pathways (Beelman and Parker, 1995).) In conclusion, these experiments demonstrate that BTG2 expression increases drastically deadenylation of the β-globin reporter.
Figure 2.
BTG2 stimulates poly(A) shortening of the β-globin reporter. (A) Transcriptional pulse–chase experiments showing deadenylation of the β-globin reporter. HEK293-TOF cells were transfected with 0.8 μg of pTet-β-globin plasmid and 1.6 μg of empty (−BTG2–HA) or BTG2–HA-expressing (+BTG2–HA) pCIneo plasmids. Immediately after transfection, doxycyclin (1 ng/ml) was added to the medium to block transcription of the reporter. Two days after transfection, cells were washed and a 3-h transcriptional pulse was performed before re-addition of doxycyclin (2 μg/ml). Chase times indicate hours after doxycyclin addition. An RNA sample treated with oligo(dT) and RNase H was used as a marker for the migration of the fully deadenylated β-globin mRNA (poly(A)−). 18S rRNA staining demonstrates equal loading. Five independent experiments gave essentially identical results. Variations in total intensities between lanes reflect mainly differences in transfection efficiencies. (B) Profiles of mRNA migration. Northern blot image shown in (A) was quantified with the ImageQuant software (Molecular Dynamics). Briefly, 30 adjacent rectangles encompassing the mRNA migration region from fully adenylated to deadenylated mRNAs were drawn for each time point to quantify signal intensity. These intensities for each chase point were then plotted as a function of the poly(A) tail length (i.e., rectangle position) as previously described (Yamashita et al, 2005).
Interaction between BTG2 and Caf1/Pop2 is required for BTG2 activation of deadenylation
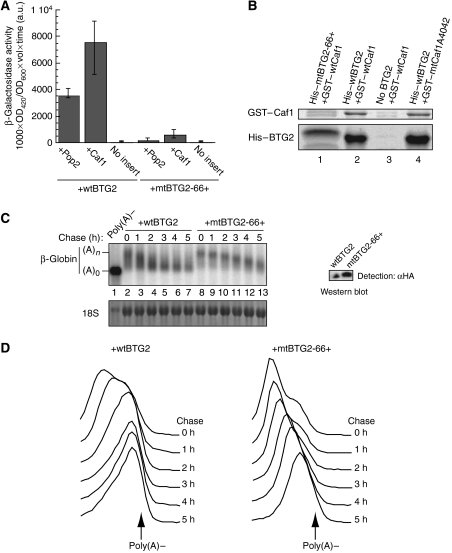
BTG2 has been shown to interact directly with the mammalian Pop2 and Caf1 deadenylase subunits. It was thus tempting to propose that this interaction was essential to mediate the BTG2 effect on mRNA decay. To test this hypothesis, we first screened for BTG2 mutants defective in their interaction with Caf1 and Pop2 using a two-hybrid assay. For this purpose, the human BTG2, mouse Caf1 and human Pop2 coding sequences were fused in appropriate yeast expression vectors with either the GAL4 activation domain or the GAL4-binding domain (GAL4BD). After transformation in yeast, we verified that the interaction of wild-type (wt) BTG2 with Caf1 or Pop2 was detectable by both colony growth on uracil-free plates and β-galactosidase production (data not shown; see also Rouault et al, 1998; Prevot et al, 2001). The BTG2-encoding plasmid was then mutagenized using a transposon-based strategy (Mutation Generation System, Finnzymes) to generate a library of insertion mutants. The mutagenized library was transformed in yeast, together with the plasmid encoding human Pop2 fused to the GAL4BD, and colonies not able to grow further on uracil-free plates were recovered. These candidates were then tested for interactions with Pop2 or Caf1 using a quantitative β-galactosidase assay to ascertain that interactions with both factors were abolished. We finally selected a mutant, mtBTG2-66+, displaying barely detectable interactions with Caf1 and Pop2 in the β-galactosidase assay (Figure 3A). Interestingly, mtBTG2-66+ contains a 5-amino-acid insertion at position 66 of the BTG2 factor (Supplementary Figure 1), in the conserved box A motif of the BTG/Tob family (Matsuda et al, 2001).
Figure 3.
A direct interaction between BTG2 and Pop2/Caf1 is required for BTG2 activation of deadenylation. (A) β-Galactosidase assays in a yeast two-hybrid system. MAV203 yeast strain was cotransformed with plasmids encoding the GAL4 activation domain fused to wtBTG2 or mtBTG2-66+ proteins as indicated together with empty GAL4-binding-domain plasmid (no insert) or plasmids encoding the GAL4BD fused to Pop2 or Caf1 as indicated. β-Galactosidase activity was expressed in arbitrary units. (B) Pull-down experiments characterizing the interaction between wtBTG2 or mtBTG2 and wtCaf1 or mtCaf1 proteins. GST-tagged wtCaf1 or GST-tagged mtCaf1A4042 was expressed in bacteria either alone or with 6His-tagged wtBTG2 or 6His-tagged mtBTG2-66+ proteins as indicated. After purification on nickel columns, eluates were analysed by SDS–PAGE and Coomassie staining. The two bands visible in the top part of the figure in lanes 1 and 3 are contaminants migrating just above and below GST–Caf1. The difference in migration between His–wtBTG2 and His–mtBTG2-66+ is due to the 5-amino-acid insertion that increases the molecular weight of BTG2 (18 kDa). (C) Northern blot showing deadenylation of the β-globin mRNA reporter in cells transiently expressing HA-tagged wtBTG2 or HA-tagged mtBTG2-66+ proteins. Transfection and transcriptional pulse–chase conditions were as described in the legend of Figure 2A. Chase time corresponds to hours after doxycyclin addition. Poly(A)− control is as in Figure 2 and lanes 1–7 correspond to lanes 7–13 of Figure 2A. 18S rRNA staining is shown to demonstrate equal loading. Three biological replicates gave essentially identical results. Inset: western blot analysis with anti-HA antibody to demonstrate expression of wtBTG2–HA and mtBTG2–HA proteins. Duplicate wells were transfected under the same conditions and used for western blot analysis. (D) Profiles of mRNA migration. Northern blot image shown in (C) was processed as described in the legend of Figure 2B.
Lack of interaction of mtBTG2-66+ protein with Caf1 was further confirmed in pull-down experiments using recombinant proteins. In this assay, a wt mouse Caf1 protein fused to GST was expressed in bacteria, either alone or together with 6His-tagged wtBTG2 or mtBTG2-66+ proteins. After coexpression, GST–wtCaf1 copurified on nickel column with 6His–wtBTG2 (Figure 3B, lane 2), whereas no band corresponding to GST–wtCaf1 was detectable when it was expressed alone (Figure 3B, lane 3) or with the His–mtBTG2-66+ protein (Figure 3B, lane 1), thus confirming absence of interaction between mtBTG2-66+ and Caf1.
The β-globin reporter was cotransfected in HEK293-TOF cells with plasmids expressing wtBTG2–HA or mtBTG2-66+–HA. Transcriptional pulse–chase experiments were performed and deadenylation kinetics of the reporter transcripts analysed by northern blot analysis. Deadenylation of the transcript transfected with wtBTG2 was rapid and reached a maximum after 3 h of chase, whereas deadenylation of the transcript cotransfected with mtBTG2 was slower and did not reach a maximum before 5 h (Figure 3C). This effect is clearly seen on the profiles of mRNA migration: the peaks corresponding to the mRNA reporter from cells expressing mtBTG2-66+ shifted more slowly towards short poly(A) tails than those from cells expressing wtBTG2 (Figure 3D). Western blot analysis indicates that this result cannot be attributed to a lower expression of the mutant factor (Figure 3C, inset). Thus, we concluded that mtBTG2 protein unable to interact with Caf1 or Pop2 was unable to activate deadenylation of a reporter transcript.
Caf1 is an active deadenylase in vivo whose activity is required, together with Ccr4's, for BTG2-induced deadenylation
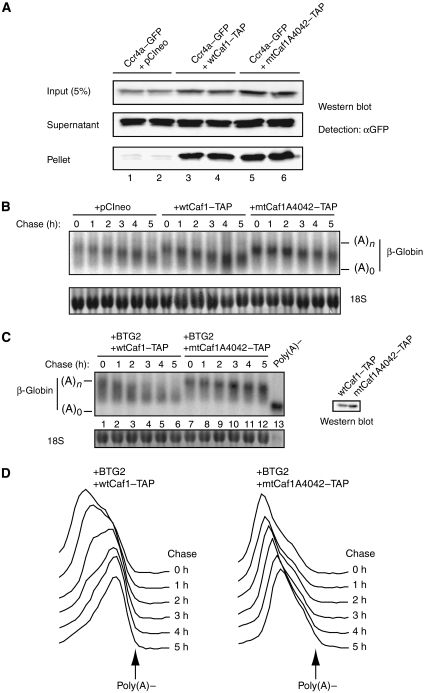
In the previous section, we have shown that BTG2-induced deadenylation requires an interaction between BTG2 and Caf1/Pop2. The Caf1/Pop2 subunit associates with Ccr4a/Ccr4b, which has been shown to mediate the final phase of deadenylation (Yamashita et al, 2005). Interestingly, both mammalian Ccr4 and Caf1/Pop2 subunits are endowed with deadenylase activity in vitro (Chen et al, 2002b; Viswanathan et al, 2004; Bianchin et al, 2005; Wagner et al, 2007) but their respective contribution to deadenylation in vivo is unclear. To test whether the deadenylase activity of Caf1/Pop2 was required for BTG2 function, an inactive Caf1 mutant (Caf1A4042) that displays no in vitro nuclease activity was used (Bianchin et al, 2005). We first checked whether the Caf1A4042 mutant retained its ability to interact with the Ccr4a/Ccr4b subunit. Plasmids encoding TAP-tagged versions of wt Caf1 or Caf1A4042 were cotransfected together with an empty pCIneo vector or a plasmid encoding human Ccr4a fused to GFP. Two days after transfection, cells were lysed and interactions between the proteins were tested by monitoring the co-precipitation of Ccr4a—GFP with TAP-tagged Caf1. Western blotting using an anti-GFP antibody demonstrated that Ccr4a–GFP associated similarly with wtCaf1 or mtCaf1 proteins (Figure 4A, lanes 3, 4 and 5, 6, respectively) well above background (Figure 4A, lanes 1 and 2). The Caf1A4042 mutant was thus used for activity test in vivo. Transcriptional pulse–chase experiments revealed that deadenylation kinetics of the β-globin transcript were similar in cells expressing wtCaf1 and in cells transfected with control plasmid, whereas they were slightly slower in cells expressing mtCaf1A4042–TAP (Figure 4B and data not shown). This indicates that the mutant protein slowed down deadenylation through a dominant-negative action. To test whether the deadenylase activity of Caf1/Pop2 was necessary for BTG2 function, the β-globin reporter was cotransfected with plasmids expressing HA-tagged wtBTG2 and either TAP-tagged wtCaf1 or dominant-negative Caf1 mutant. This mutant still interacts with wtBTG2 (Figure 3B, lane 4). Transcriptional pulse–chase experiments revealed that expression of wtCaf1 did not change the BTG2-activated deadenylation kinetics of the β-globin reporter (compare Figure 4C, lanes 1–6 to Figure 2A, lanes 7–12 and Figure 4D to Figure 2B). In contrast, when the catalytically inactive Caf1A4042 mutant was overexpressed, deadenylation of the β-globin reporter was drastically slowed down even in the presence of BTG2 (Figure 4C, lanes 7–12, and Figure 4D). Because levels of wtCaf1 and mtCaf1 proteins were similar, the reduced deadenylation cannot be attributed to a difference in the expression of the transfected constructs (Figure 4C, inset). We also verified that the BTG2–HA transgene was correctly expressed when cotransfected with Caf1-expressing plasmids (data not shown). These results demonstrate that the deadenylase activity of the Caf1/Pop2 subunit is necessary for BTG2 activation of mRNA deadenylation.
Figure 4.
The catalytic site mutant Caf1A4042 has a dominant-negative effect on BTG2-activated deadenylation. (A) Copurification assays performed with HEK293 cell lysates transiently transfected with a plasmid expressing Ccr4a–GFP fusion protein and empty pCIneo plasmid (lanes 1 and 2) or pCIneo plasmid expressing TAP-tagged wtCaf1 (lanes 3 and 4) or TAP-tagged mtCaf1A4042 (lanes 5 and 6). Affinity purifications were performed with IgG sepharose beads. Eluates were resolved by SDS–PAGE and revealed by western blotting with an anti-GFP antibody. Input, supernatants (showing the absence of protein degradation) and pellets of biological duplicates are shown. (B) Northern blot showing deadenylation of the β-globin reporter in cells cotransfected with empty pCIneo plasmid or pCIneo plasmids expressing wtCaf1–TAP or mtCaf1A4042–TAP. Transfection and transcriptional pulse–chase conditions are as described in Figure 2. Three biological replicates gave essentially identical results. (C) Northern blot showing deadenylation of the β-globin reporter in cells coexpressing wtBTG2 and wtCaf1 deadenylase or the catalytic site mutant Caf1A4042. HEK293-TOF cells were transfected with 0.8 μg of pTet-β-globin plasmid, 0.8 μg of plasmid encoding BTG2 and 0.8 μg of plasmids encoding wtCaf1 or mtCaf1A4042. Transcriptional pulse–chase experiments were as described in the legend of Figure 2A. Chase time corresponds to hours after doxycyclin addition. Poly(A)− control is as in Figure 2. 18S rRNA staining is shown to demonstrate equal loading. Two biological replicates gave essentially identical results. Inset: western blot analysis of cells transfected under the same conditions to verify expression of wtCaf1 and mtCaf1 proteins. (D) Profile of mRNA migration. Processing of the northern blot image was as described in the legend of Figure 2.
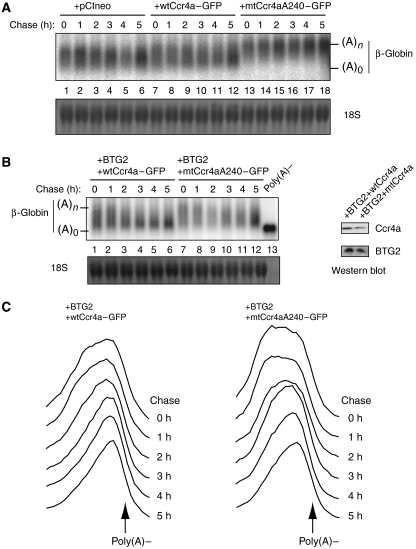
To assess the contribution of Ccr4, we similarly mutated the catalytic site of the Ccr4a factor (position 240 changed to alanine). Consistent with previous observations (Chang et al, 2004; Yamashita et al, 2005), transfection of a catalytically inactive mtCcr4a fused to a GFP tag resulted in a strong dominant-negative effect on deadenylation of the reporter globin mRNA (Figure 5A). Cotransfection of the mtCcr4a construct with BTG2 resulted in a reduced rate of deadenylation of the reporter transcript (Figure 5B and C). However, in contrast to constitutive deadenylation, the effect of the catalytically inactive Ccr4a mutant was not as strong as the one observed with the Caf1 mutant (compare Figures 4 and 5). Overall, our results demonstrate that, in contrast to yeast (Chen et al, 2002b; Tucker et al, 2002; Viswanathan et al, 2004), efficient constitutive deadenylation in mammalian cells requires both Pop2/Caf1 and Ccr4 activities. Moreover, our data indicate that, for BTG2-induced deadenylation, Caf1 plays a preponderant role over Ccr4a.
Figure 5.
The catalytic site mutant Ccr4aA240 impairs, in a dominant-negative manner, BTG2-activated deadenylation. (A) Northern blot showing deadenylation of the β-globin reporter in cells cotransfected with empty pCIneo plasmid or pCIneo plasmids expressing wtCcr4a–GFP or mtCcr4aA240–GFP. Transfection and transcriptional pulse–chase conditions were as described in the legend of Figure 2 except that the transcriptional pulse was for 4 h. Two biological replicates gave essentially identical results. (B) Northern blot showing deadenylation of the β-globin reporter in cells coexpressing wtBTG2 and wtCcr4a deadenylase or the catalytic site mutant Ccr4aA240. HEK293-TOF cells were transfected with 0.8 μg of pTet-β-globin plasmid, 0.8 μg of plasmid encoding BTG2 and 0.8 μg of plasmid encoding wtCcr4a or 1 μg of plasmid encoding mtCcr4aA240. Transcriptional pulse–chase experiments were as described in the legend of Figure 2A except that the transcriptional pulse was for 4 h. Chase time corresponds to hours after doxycyclin addition. Poly(A)− control is as in Figure 2. 18S rRNA staining is shown to demonstrate equal loading. Three biological replicates gave essentially identical results. Inset: western blot analysis of cells transfected under the same conditions to verify the expression of BTG2 and Ccr4a constructs. (C) Profile of mRNA migration. Processing of the northern blot image was as described in the legend of Figure 2.
BTG2 is a general activator of mRNA decay
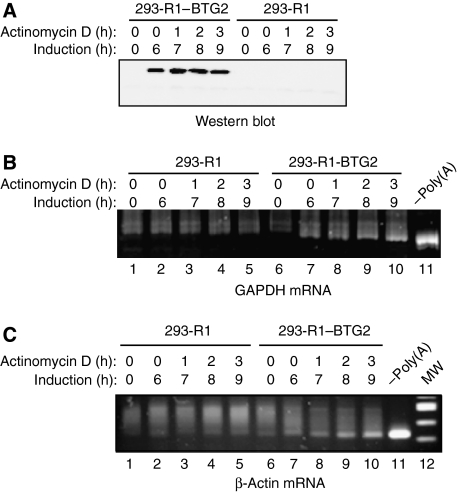
BTG2 expression activated the degradation of the three reporter transcripts that we tested in transcriptional chase experiments, suggesting that BTG2 activity was not limited to a restricted number of mRNAs but displayed rather a large spectrum of specificity. To test whether BTG2 affects cellular encoded mRNAs, we generated cell lines in which stable expression of BTG2 can be induced using the RheoSwitch Mammalian Inducible Expression System (New England Biolabs). Western blot analysis confirmed that, after a 6 h treatment with the RSL1 inducer, BTG2–HA was expressed in the 293-R1-BTG2 cell line but not in 293-R1 parental cell line (Figure 6A). Cells were then treated by the addition of actinomycin D to shut off transcription, hence allowing the analysis of endogenous mRNA decay. RNA was extracted at the indicated times and the poly(A) tail length distribution of two endogenous transcripts was assayed using a modified RACE-PAT assay (Salles et al, 1999). Briefly, a synthetic RNA oligonucleotide blocked at its 3′ end and 5′ phosphorylated was ligated to total cellular RNA. These molecules were converted into DNA by reverse transcription and the region covering the poly(A) tail and the extremity of the 3′UTR was amplified by PCR. This assay revealed that treatment of parental 293-R1 cells with RSL1 and actinomycin D induced little changes in the length of the poly(A) tail of GAPDH and β-actin transcripts (Figure 6B and C, lanes 1–5). By contrast, the length of the poly(A) tail of these transcripts shortened noticeably in 293-R1-BTG2 cells after BTG2 induction (Figure 6B and C, lanes 6–10). During the treatment with actinomycin D, the poly(A) tail shortening was so accentuated that after 3 h of transcriptional shut-off, poly(A) tails of a large fraction of the test mRNAs appeared to have no or little poly(A) left (compare to the signal observed with the sample treated in vitro with oligo(dT) and RNase H; Figure 6B and C). These results demonstrate that inducing BTG2 expression in stable cell lines activates deadenylation of two endogenous transcripts that are involved in different cellular processes. Altogether, our results show that BTG2 activation of deadenylation is not transcript specific and could rather reflect a general mechanism to modify gene expression.
Figure 6.
BTG2 activation of deadenylation is not transcript specific. (A) Western blot analysis showing induction of BTG2 expression in stable cell lines. Parental 293-R1 cells and 293-R1-BTG2 cells were treated for 6 h with RSL1 (500 nM) to induce BTG2 expression. Actinomycin D (5 μg/ml) was then added to the cell medium. Time indication corresponds to hours after RSL1 addition (induction) or after actinomycin D addition. Western blot was revealed with an anti-HA antibody. (B, C) Poly(A) shortening of endogenous GAPDH transcript (B) or endogenous β-actin transcript (C) after induction of BTG2 expression in stable cell lines. RNA extracted from cells treated with RSL1 and actinomycin D as in panel A was used in RACE-PAT assays (see Materials and methods). Time indications correspond to hours after RSL1 addition (induction) or actinomycin D addition. An RNA sample treated with oligo(dT) and RNase H was used as a control for fully deadenylated mRNAs (−poly(A)). PCR products specific for the GAPDH transcript were resolved on a 6% non-denaturing acrylamide gel, whereas PCR products specific for the β-actin transcript were resolved on a 2% Nusieve agarose gel.
Discussion
Two decades ago, the rat and mouse homologues of BTG2 were identified and reported to belong to the primary response genes induced by growth factors and tumour promoters (Bradbury et al, 1991; Fletcher et al, 1991). Since then, the precise molecular functions of BTG2 have been under study. Here, we demonstrate that BTG2 is a general activator of mRNA decay affecting reporter transcripts and endogenous mRNAs. BTG2 acts by enhancing deadenylation through a direct interaction with Pop2/Caf1 factors. The requirement of active Ccr4 and Caf1 deadenylases for mediating the BTG2 effect demonstrates a specific implication of BTG2 in the deadenylation process.
Interestingly, our experiments reveal that BTG2 activates the deadenylation of all transcripts tested. This observation leads us to propose that one of the main functions of BTG2 is to generally activate degradation of pre-existing transcripts. The existence of general activators of deadenylation has, to the best of our knowledge, not been reported. Indeed, previous published examples of activation of deadenylation involved the recruitment of deadenylases on specific transcripts by factors that interact with specific mRNA sequences located primarily in the 3′UTR, such as AU-rich elements (Lykke-Andersen and Wagner, 2005). Similarly, Smaug (Semotok et al, 2005; Zaessinger et al, 2006) or PUF (Goldstrohm et al, 2006) proteins recruit the Ccr4–Pop2 deadenylase complex on specific transcripts to regulate their deadenylation as well as their localization and translation. The existence of general activators of deadenylation, such as BTG2, suggests that they could act by new, so far undescribed, mechanisms. It is also possible that other factors interacting with Pop2–Ccr4, such as some Not proteins (Tucker et al, 2002; Denis and Chen, 2003; Temme et al, 2004), could similarly act as global regulators of mRNA decay.
Consistent with a general function, proteins of the BTG/Tob family all interact with Caf1/Pop2 (Bogdan et al, 1998; Rouault et al, 1998; Ikematsu et al, 1999; Prevot et al, 2001) but lack known RNA-binding domains. This observation supports the idea that their function does not involve direct binding to specific mRNA sequences. One possibility would be that BTG/Tob factors activate mRNA deadenylation by stimulating directly the catalytic activity of the Ccr4–Caf1 complexes. However, preliminary in vitro deadenylation assays performed with recombinant Caf1 and BTG2 proteins indicated that the latter was not sufficient by itself to activate in vitro the catalytic deadenylase activity of Caf1 (data not shown). For the Tob subfamily, the presence of two PAM2 motifs (PABP-interacting motif 2; Albrecht and Lengauer, 2004; Okochi et al, 2005; Lim et al, 2006) suggests that Tob factors may recruit the main deadenylase to all poly(A)-containing transcripts by interacting with the C-terminal domain of the poly(A)-binding protein (PABP) and supports further the role of these proteins in mRNA metabolism. Even though BTG factors lack PAM2 motifs and have not been shown to interact directly with PABP, it is tempting to speculate that they may also recruit the main deadenylase to all transcripts with the help of an unknown protein bridging them to PABP. Alternatively, BTG factors may interact directly or indirectly with other conserved features of mRNAs such as the cap, bound ribosomes or translation factors. In this vein, it is interesting to note that BTG2 has been reported to interact with diverse partners (Lin et al, 1996; Prevot et al, 2000; Berthet et al, 2002), which may be involved in activation of deadenylation. Clearly, more experiments are required to elucidate the exact mechanism by which BTG2 and other BTG family members activate mRNA deadenylation.
Our data also demonstrate that overexpression of a catalytically inactive Caf1 deadenylase is sufficient to inhibit general deadenylation. These results indicate that, in contrast to the situation in yeast (Chen et al, 2002a; Tucker et al, 2002; Viswanathan et al, 2004), the deadenylase activity of the Caf1/Pop2 subunit of the mammalian Ccr4–Pop2 complex plays a role in cytoplasmic deadenylation in cultured human cells. Because residual deadenylation is detectable in the mutated Caf1 context but not with the mtCcr4 (compare Figures 4B and 5A), this suggests that Ccr4 plays a preponderant role in general deadenylation. Interestingly, inactivating Caf1/Pop2 provokes a strong block of BTG2-induced deadenylation, whereas inactivation of Ccr4 has a milder effect. This observation could suggest that BTG2 recruits the Caf1/Pop2 subunit independently of Ccr4a/Ccr4b. An alternative possibility is that both Ccr4 and Caf1 contribute to all events of deadenylation in mammalian cells, with each subunit contributing a different level of degradation depending on the conditions. The use of a dimeric deadenylase may thus offer a wider range of regulation potential and allow a fine-tuning of the deadenylation kinetics in mammalian cells. In such a case, dominant-negative effect resulting from the presence of one inactive subunit may be explained by the sequestering of the 3′ end of the substrate poly(A) tail blocking access to the remaining active subunit. Although the exact contributions of human Caf1 and Ccr4 to deadenylation remain to be precisely established, our data demonstrate that both enzymes contribute catalytically to this process and indicate that their contribution may vary according to the conditions, as observed in the case of BTG2-induced deadenylation.
Since BTG2 was identified, its expression has been reported to induce pleiotropic effects. Thus, BTG2 has been implicated in the regulation of cell-cycle progression, differentiation of cell lines as well as the control of cellular apoptosis (Matsuda et al, 2001; Tirone, 2001; Duriez et al, 2004; Lim et al, 2006). More recently, BTG2 was also shown to be a downstream mediator of tumour suppression by p53 (Boiko et al, 2006). The function of BTG2 in cytoplasmic deadenylation is compatible with the previously known effects of BTG2. Indeed, our results indicate that it may affect the expression of numerous proteins through a general destabilization of cellular mRNAs. It is interesting to note that many biological consequences of the expression of BTG/Tob proteins may involve rapid and dramatic changes in the expression profile of cells. This is indeed the case during differentiation or cell-cycle progression. BTG/Tob factors may contribute to these events by speeding up the degradation of previously made mRNAs, thus facilitating the rapid installation of a new gene expression programme. It will be of interest to test whether defective reprogramming due to the expression of BTG/Tob mutants prevents the establishment of a normal growth control programme and thus explains how BTG/Tob factors contribute to cancer.
Note: During the revision of this manuscript, two articles reporting the implication of Tob factors in deadenylation were published (Ezzeddine et al, 2007; Funakoshi et al, 2007).
Materials and methods
Plasmid construction
Reporter plasmids were constructed following standard cloning strategies. Construction details of the LacZβ and LacZD1 reporters, the BTG2 expression plasmids and the vectors encoding tagged Caf1 are provided in Supplementary data. The pTet-β-globin plasmid (Couttet and Grange, 2004) was kindly provided by T Grange. pBS2560 encoding GFP–hCcr4a has been described (Cougot et al, 2004). Mutagenesis of Ccr4a was performed with the QuickChange mutagenesis kit (Stratagene) using the OBS2818 and OBS2819 oligonucleotides. Oligonucleotides used in this study are presented in Supplementary Table SI. All constructions involving PCR amplification were verified by sequencing.
Two-hybrid and β-galactosidase assays
For two-hybrid assays, the human BTG2 ORF was inserted in pDEST22 vector (generating pBS2669), whereas ORFs encoding human Pop2 and mouse Caf1 were inserted in pDEST32 vector (yielding pBS2676 and pBS2789, respectively) using the Gateway Technology. Mutagenesis of plasmid pBS2669 was performed with the Mutation Generation System (Finnzymes). Yeast strain MAV203 (Invitrogen) was transformed simultaneously with plasmid pBS2676 and a library of mutagenized plasmid as previously described (Ito et al, 1983). Transformants growing on −Leu −Trp plates but unable to grow on −Ura plates were identified. DNA was recovered from such clones and the sequence encompassing the BTG2 ORF of some interaction-defective pBS2669 derivatives was determined. After retransformation in the tester yeast strain with plasmids encoding either wt Caf1 or Pop2 proteins, the interaction ability of BTG2 mutants of interest was further confirmed by testing growth on −Ura media and performing β-galactosidase assays (Seraphin and Kandels-Lewis, 1993).
Cell culture and transfection
HEK293 cells, maintained in DMEM medium supplemented with 10% FCS, were stably transfected with plasmid pTet-off (Clontech) by the Effectene Transfection Reagent (Qiagen) and selected with 400 μg/ml geneticin (Invitrogen). The cell line, HEK293-TOF, giving a high inducible expression level by the Tet-off Gene Expression System was selected. HEK293-TOF cells, grown in 60-mm dishes to approximately 60% confluence, were transiently transfected using the Effectene Transfection Reagent (Qiagen).
HEK293 cells allowing the regulated expression of BTG2 were constructed in two steps: first, HEK293 cells were stably transfected with plasmid pNEBR-R1 to generate a cell clone, 293-R1, allowing inducible expression of target genes by the RheoSwitch Inducible Expression System (New England Biolabs). This cell line was further transfected with plasmid pBS2943 encoding BTG2–HA and plasmid pcDNA3.1/Hygro(−) (Invitrogen) and the 293-R1-BTG2 resistant cells, selected with 300 μg/ml hygromycin B (Invitrogen), were tested for inducible expression of BTG2–HA after treatment with the RheoSwitch ligand RSL1. Addition of RSL1 to the 293-R1-BTG2 cell growth medium induced BTG2–HA expression, which reached a plateau after 8 h of induction (data not shown).
RNA extraction, northern blotting analysis and quantitative RT–PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen). For northern blot analysis, 10 μg of total RNA was electrophoresed onto 1.4% agarose/6% formaldehyde gels and transferred to Hybond-N+ membranes (GE Healthcare). Blots were hybridized to probes synthesized by the Megaprime DNA Labelling System (GE Healthcare) or by in vitro transcription with the T7 RNA polymerase (Promega). Hybridization signals were detected and visualized with a PhosphorImager (Molecular Dynamics) and analysed with ImageQuant and Kaleidagraph softwares. For quantitative RT–PCR, 1 μg of total RNA was reverse-transcribed with oligo(dT) and RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas) and RT products were analysed by real-time PCR using the LightCycler FastStart DNA Master SYBR Green I kit in a LightCycler apparatus (Roche Applied Science) according to the manufacturer's protocol.
RACE-PAT assay
A modified RACE-PAT assay (Salles et al, 1999) was used to monitor the length of the poly(A) tail of specific endogenous mRNAs. Briefly, 3 nmol of the synthetic RNA oligonucleotide T7-rev (for sequence, see Supplementary Table SI) was oxidized by reaction with 5 μl of 10 mg/ml NaIO4 in a volume of 100 μl for 30 min in the dark on ice. The reaction was stopped by the addition of 100 μl of 50% ethylene glycol and precipitated with ethanol. After resuspension, 1 nmol of oxidized T7-rev oligonucleotide was phosphorylated with T4 Polynucleotide Kinase (New England Biolabs) as recommended by the manufacturer. A 50 pmol portion of oxidized and kinased T7-rev oligonucleotide was ligated to 1 μg of total cellular RNA with T4 RNA ligase (New England Biolabs) under the conditions recommended by the manufacturer. One-half of the ligation reaction was then reverse-transcribed with oligonucleotide OBS2138 and RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas) as recommended by the manufacturer. PCR amplification was performed with 1 μl of the reverse transcription reaction and oligonucleotide OBS2313 and either oligonucleotide OBS2140 or oligonucleotide OBS2141 depending on the target mRNA.
Protein purification and western blotting
Transfected cells were lysed in IPP150 buffer (10 mM Tris–HCl pH 8, 150 mM NaCl, 1% Igepal CA-630 (Sigma) and proteases inhibitors) by standard procedures. For TAP purification, cell lysates were incubated with IgG Sepharose 6 Fast Flow beads (GE Healthcare), beads were washed four times with IPP150 buffer and eluted with IPP150 buffer supplemented with 1% sodium dodecyl sulphate. Western blotting was performed by standard procedures. The HA tag was revealed with the monoclonal antibody HA-11 (Covance) using the SuperSignal West Femto Maximum Sensitivity Substrate kit (Pierce), the TAP tag was revealed with the immunocomplex PAP (Dako) and the ECL Western Blotting Detection Reagents (GE Healthcare) and the GFP tag was revealed with the monoclonal antibody JL8 (BD Biosciences), a secondary horseradish peroxidase-conjugated anti-mouse antibody (Pierce) and the ECL Western Blotting Detection Reagents (GE Healthcare). Chemiluminescent signals were visualized with the LAS-3000 apparatus (Fujifilm). Recombinant His-tagged proteins were expressed and purified on Ni-agarose (Qiagen) essentially as recommended by the manufacturer.
Supplementary Material
Supplementary data
Acknowledgments
We thank L Corbo and T Grange for providing plasmids and S De Carvalho and S Ait Abdellah for technical assistance. We are grateful to group members for useful discussions and to S Camier, A Lebreton and J Marie for manuscript corrections. This work was supported by La Ligue contre le Cancer (Equipe Labellisée 2005), HSFP, the Ministry for Research (ACI-BCMS 0233), Agence Nationale de la Recherche and the CNRS.
References
- Albrecht M, Lengauer T (2004) Survey on the PABC recognition motif PAM2. Biochem Biophys Res Commun 316: 129–138 [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB (2005) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33: 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Parker R (1995) Degradation of mRNA in eukaryotes. Cell 81: 179–183 [DOI] [PubMed] [Google Scholar]
- Berthet C, Guehenneux F, Revol V, Samarut C, Lukaszewicz A, Dehay C, Dumontet C, Magaud JP, Rouault JP (2002) Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells 7: 29–39 [DOI] [PubMed] [Google Scholar]
- Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L (2005) Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA 11: 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan JA, Adams-Burton C, Pedicord DL, Sukovich DA, Benfield PA, Corjay MH, Stoltenborg JK, Dicker IB (1998) Human carbon catabolite repressor protein (CCR4)-associative factor 1: cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem J 336 (Part 2): 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko AD, Porteous S, Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV (2006) A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev 20: 236–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury A, Possenti R, Shooter EM, Tirone F (1991) Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc Natl Acad Sci USA 88: 3353–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Sachs AB (1998) Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol 18: 6548–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yamashita A, Chen CY, Yamashita Y, Zhu W, Durdan S, Kahvejian A, Sonenberg N, Shyu AB (2004) UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev 18: 2010–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465–470 [DOI] [PubMed] [Google Scholar]
- Chen CY, Xu N, Shyu AB (2002a) Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol 22: 7268–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiang YC, Denis CL (2002b) CCR4, a 3′–5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B (2004) Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Grange T (2004) Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res 32: 488–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron MC, Mauxion F, Seraphin B (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res 29: 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis CL, Chen J (2003) The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol 73: 221–250 [DOI] [PubMed] [Google Scholar]
- Dufourny B, van Teeffelen HA, Hamelers IH, Sussenbach JS, Steenbergh PH (2000) Stabilization of cyclin D1 mRNA via the phosphatidylinositol 3-kinase pathway in MCF-7 human breast cancer cells. J Endocrinol 166: 329–338 [DOI] [PubMed] [Google Scholar]
- Duriez C, Moyret-Lalle C, Falette N, El-Ghissassi F, Puisieux A (2004) BTG2, its family and its tutor. Bull Cancer 91: E242–E253 [PubMed] [Google Scholar]
- Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB (2007) Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 27: 7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yang X, Wang W, Wood WH III, Becker KG, Gorospe M (2002) Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci USA 99: 10611–10616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken MB, Corbett AH (2005) Process or perish: quality control in mRNA biogenesis. Nat Struct Mol Biol 12: 482–488 [DOI] [PubMed] [Google Scholar]
- Fletcher BS, Lim RW, Varnum BC, Kujubu DA, Koski RA, Herschman HR (1991) Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J Biol Chem 266: 14511–14518 [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S (2007) Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 21: 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M (2006) PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D, Corrente G, Covone F, Micheli L, D'Agnano I, Starace G, Caruso M, Tirone F (2000) Arrest of G(1)–S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol Cell Biol 20: 1797–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Harwalkar J, Stacey DW, Hitomi M (2005) Destabilization of cyclin D1 message plays a critical role in cell cycle exit upon mitogen withdrawal. Oncogene 24: 1032–1042 [DOI] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A (1990) Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 10: 2269–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, Ohsugi M, Onda M, Hirai M, Fujimoto J, Yamamoto T (1999) Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene 18: 7432–7441 [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Jun JM, Shin SW, Cho JW, Suh SI (2005) Curcumin decreases cell proliferation rates through BTG2-mediated cyclin D1 down-regulation in U937 cells. Int J Oncol 26: 1597–1603 [PubMed] [Google Scholar]
- Lim IK (2006) TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule. J Cancer Res Clin Oncol 132: 417–426 [DOI] [PubMed] [Google Scholar]
- Lim IK, Lee MS, Ryu MS, Park TJ, Fujiki H, Eguchi H, Paik WK (1998) Induction of growth inhibition of 293 cells by downregulation of the cyclin E and cyclin-dependent kinase 4 proteins due to overexpression of TIS21. Mol Carcinog 23: 25–35 [DOI] [PubMed] [Google Scholar]
- Lim NS, Kozlov G, Chang TC, Groover O, Siddiqui N, Volpon L, De Crescenzo G, Shyu AB, Gehring K (2006) Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J Biol Chem 281: 14376–14382 [DOI] [PubMed] [Google Scholar]
- Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M (2000) Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol 20: 7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR (1996) The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem 271: 15034–15044 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E (2005) Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Rouault J, Magaud J, Berthet C (2001) In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett 497: 67–72 [DOI] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E (2004) Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 39: 197–216 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Guardavaccaro D, Starace G, Tirone F (1996) Overexpression of the nerve growth factor-inducible PC3 immediate early gene is associated with growth inhibition. Cell Growth Differ 7: 1327–1336 [PubMed] [Google Scholar]
- Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T (2005) Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells 10: 151–163 [DOI] [PubMed] [Google Scholar]
- Prevot D, Morel AP, Voeltzel T, Rostan MC, Rimokh R, Magaud JP, Corbo L (2001) Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J Biol Chem 276: 9640–9648 [DOI] [PubMed] [Google Scholar]
- Prevot D, Voeltzel T, Birot AM, Morel AP, Rostan MC, Magaud JP, Corbo L (2000) The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem 275: 147–153 [DOI] [PubMed] [Google Scholar]
- Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR (2002) Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res 30: 5529–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault JP, Prevot D, Berthet C, Birot AM, Billaud M, Magaud JP, Corbo L (1998) Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J Biol Chem 273: 22563–22569 [DOI] [PubMed] [Google Scholar]
- Salles FJ, Richards WG, Strickland S (1999) Assaying the polyadenylation state of mRNAs. Methods 17: 38–45 [DOI] [PubMed] [Google Scholar]
- Sasajima H, Nakagawa K, Yokosawa H (2002) Antiproliferative proteins of the BTG/Tob family are degraded by the ubiquitin–proteasome system. Eur J Biochem 269: 3596–3604 [DOI] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA (2005) Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol 15: 284–294 [DOI] [PubMed] [Google Scholar]
- Seraphin B, Kandels-Lewis S (1993) 3′ splice site recognition in S.cerevisiae does not require base pairing with U1 snRNA. Cell 73: 803–812 [DOI] [PubMed] [Google Scholar]
- Tang G (2005) siRNA and miRNA: an insight into RISCs. Trends Biochem Sci 30: 106–114 [DOI] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E (2004) A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 23: 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirone F (2001) The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol 187: 155–165 [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R (2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Varnum BC, Reddy ST, Koski RA, Herschman HR (1994) Synthesis, degradation, and subcellular localization of proteins encoded by the primary response genes TIS7/PC4 and TIS21/PC3. J Cell Physiol 158: 205–213 [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL (2004) Mouse CAF1 can function as a processive deadenylase/3′–5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem 279: 23988–23995 [DOI] [PubMed] [Google Scholar]
- Wagner E, Clement SL, Lykke-Andersen J (2007) An unconventional human Ccr4–Caf1 deadenylase complex in nuclear cajal bodies. Mol Cell Biol 27: 1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO (2002) Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 99: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Loflin P, Chen CY, Shyu AB (1998) A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res 26: 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB (2005) Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M (2006) Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133: 4573–4583 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data