Abstract
Accumulation of misfolded protein in the endoplasmic reticulum (ER) causes stress. The unfolded protein response (UPR), a transcriptional induction pathway, is activated to relieve ER stress. Although UPR is not essential for viability, UPR-deficient cells are more sensitive to ER stress; ire1Δ cells cannot grow when challenged with tunicamycin or by overexpression of misfolded CPY*. In these cells, multiple functions are defective, including translocation, ER-associated degradation (ERAD), and ER-to-Golgi transport. We tested whether heat shock response (HSR) can relieve ER stress. Using a constitutively active Hsf1 transcription factor to induce HSR without temperature shift, we find that HSR rescues growth of stressed ire1Δ cells, and partially relieves defects in translocation and ERAD. Cargo-specific effects of constitutively active Hsf1 on ER-to-Golgi transport are correlated with enhanced protein levels of the respective cargo receptors. In vivo, HSR is activated by ER stress, albeit to a lower level than that caused by heat. Genomic analysis of HSR targets reveals that >25% have function in common with UPR targets. We propose that HSR can relieve stress in UPR-deficient cells by affecting multiple ER activities.
Keywords: ER stress, heat shock response, unfolded protein response, vesicle transport
Introduction
In eukaryotic cells, there are sophisticated mechanisms to help polypeptides fold, distinguish misfolded proteins from those with native forms, and clear away conformationally aberrant and, in many cases, toxic proteins. Collectively, these mechanisms are called ‘quality control'. The importance of quality control is underscored by the many examples of disease resulting from misfolding and degradation of a critical protein that has an essential activity and localization, for example, cystic fibrosis. Accumulation of misfolded and/or aggregated protein can also lead to perturbation of cell function and stress-induced cell death, for instance, in neurodegenerative disease and diabetes.
In the secretory pathway, protein synthesis at the endoplasmic reticulum (ER) occurs concomitantly with translocation, modification, and folding with the assistance of molecular chaperones. When proteins are misfolded, there are two branches of ER quality control that address the situation. One is the ER-associated degradation (ERAD) pathway to dispose of misfolded proteins, which involves retro-translocation, polyubiquitination, and degradation in the cytosol through the 26S proteasome. A second major response is called the unfolded protein response (UPR) (Ron and Walter, 2007). Key components of the UPR include the luminal Hsp70 family member Kar2/BiP, the transmembrane signal transducer Ire1, and the transcription factor Hac1. Detection of misfolded protein by Kar2 and Ire1 results in transcriptional activation of ∼400 target genes (Cox et al, 1993; Mori et al, 1993; Travers et al, 2000). Although several of these encode factors involved in protein folding, several others are involved in ERAD and vesicular transport, which are proposed to work cooperatively to clear misfolded proteins. Indeed, degradation of some ERAD substrates requires transport between ER and Golgi (Caldwell et al, 2001; Taxis et al, 2002). These reports suggest that removal of defective ER proteins by either the ubiquitin–proteasome system or by delivery to lysosomal/vacuolar degradation is a rectifying response. Although UPR is not essential for viability, its importance in protecting ER function is revealed by analysis of ire1Δ cells stressed by overexpression of the misfolded luminal protein CPY*; the cells fail to grow and have defects in protein translocation and ER export (Ng et al, 2000; Spear and Ng, 2003).
In contrast to UPR, which serves the secretory pathway, heat shock response (HSR) is predominantly a response to stress conditions in the cytosol (Mager and Ferreira, 1993). Analogous to UPR, HSR causes transcriptional activation of molecular chaperones and elements of the ubiquitin–proteasome system (Parsell et al, 1993). Although originally discovered as a response to thermal stress, HSR is triggered by a variety of stress conditions that interfere with folding and result in accumulation of misfolded or aggregated proteins. HSR is mediated by Hsf1 transcription factor. In Saccharomyces cerevisiae, there is a single Hsf1 (Sorger et al, 1987), which binds to heat shock elements (HSE) in the promoters of a wide range of target genes (Hahn et al, 2004; Eastmond and Nelson, 2006). Interestingly, genomic analysis has revealed targets of Hsf1 in the secretory pathway that are also induced by UPR. For instance, the ER chaperone Kar2/BiP is dramatically increased by both HSR and UPR; accordingly, both HSR and UPR elements are found in the KAR2 promoter (Kohno et al, 1993). ERV29, encoding a COPII cargo receptor (Belden and Barlowe, 2001; Caldwell et al, 2001), is also induced by both heat (Hahn et al, 2004) and ER stress (Caldwell et al, 2001). Recently, we reported that HSR, but not UPR, is activated by a misfolded membrane substrate for ERAD, Pma1-D378S (Han et al, 2007). These findings suggest a role for HSR in ER quality control.
To address whether HSR can relieve ER stress, we used UPR-deficient ire1Δ cells overexpressing CPY* (Spear and Ng, 2003) or treated with tunicamycin to induce protein misfolding. We introduced an Hsf1 mutant, Hsf1-R206S, into these cells to constitutively activate HSR in the absence of temperature shift (Sewell et al, 1995). In stressed UPR-defective cells, constitutively active Hsf1 rescues the growth defect, corrects the protein translocation defect, and promotes ER export. Increased protein levels of Kar2 and specific cargo receptors appear to account for some of the ameliorative effects of HSR. ERAD is also increased by Hsf1-R206S. Our data indicate that HSR can relieve ER stress through multiple pathways.
Results
HSR helps UPR-deficient cells survive ER stress
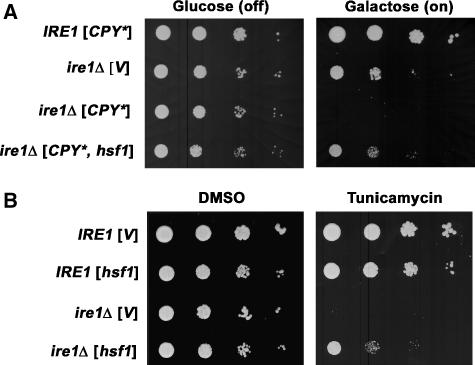
To determine whether HSR has a role in ER quality control, we used ire1Δ cells deficient in UPR. Previous work has shown that cells become more sensitive to ER stress without Ire1; in the presence of tunicamycin, which increases protein misfolding (Figure 1B; Cox et al, 1993), or upon overexpression of CPY* (under the control of the GAL1 promoter), ire1Δ cells cannot grow (Figure 1A; Spear and Ng, 2003). HSR was elicited in these cells in the absence of temperature shift by introducing a constitutively active Hsf1 mutant, Hsf1-R206S (Sewell et al, 1995). Figure 1A shows that ire1Δ cells overexpressing CPY* can grow in the presence of Hsf1-R206S. A similar effect of Hsf1-R206S on ire1Δ cells is seen with tunicamycin treatment in Figure 1B. Moreover, incubating ire1Δ cells at 37°C to induce a mild HSR rescues the cells from ER stress caused by CPY* overexpression (Supplementary Figure S1). These results suggest that HSR helps cells survive ER stress in the absence of UPR.
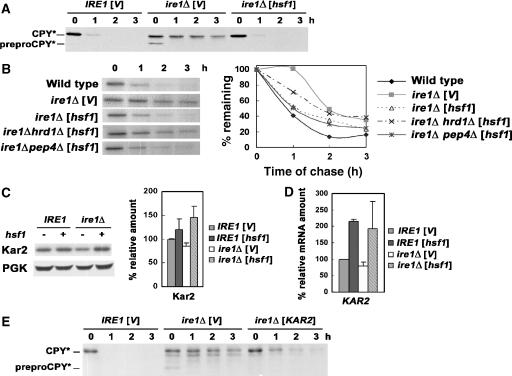
Figure 1.
Constitutively active Hsf1 restores the growth of ire1Δ cells under ER stress. (A) Wild-type (MATa W303) and ire1Δ (KKY100) cells bearing pGAL-CPY* (pES67) were co-transformed with constitutively active mutant hsf1-R206S (pYEP96) or vector (pRS314). Cells (normalized to OD600/ml) were spotted onto plates with synthetic complete (SC) medium with 2% glucose (GAL off) or 2% galactose (GAL on) and incubated at 30°C for 2 and 3 days, respectively. (B) Wild-type (MATa W303) and ire1Δ (KKY100) cells bearing hsf1-R206S (pYEP96) or vector (pRS314) were spotted on plates with SC medium with DMSO (solvent) or tunicamycin (0.05 μg/ml) and incubated at 30°C for 3 days.
Constitutively active Hsf1-R206S rescues translocation, degradation, and transport defects in ire1Δ cells overexpressing CPY*
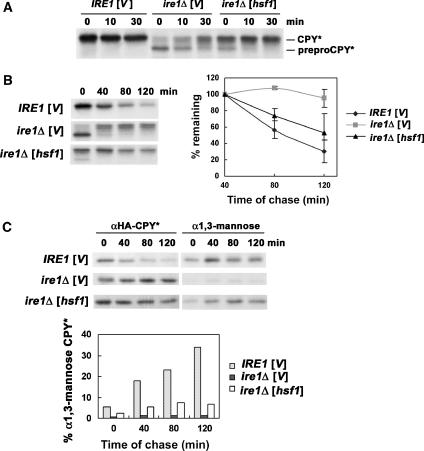
We next analysed the mechanism by which constitutively active Hsf1 rescues ire1Δ cells overexpressing CPY*. Previous work has shown that ire1Δ cells with overexpressed CPY* have multiple defects in ER function, including impaired protein translocation, ERAD, and ER export (Spear and Ng, 2003). Figure 2A shows a pulse–chase experiment with ire1Δ cells overexpressing HA-tagged CPY*. Immunoprecipitation (IP) of HA–CPY* immediately after pulse-labelling revealed that the major species of CPY* in these cells is an untranslocated unglycosylated prepro-form (Figure 2A, 0 min chase). A lesser amount of a higher molecular weight band corresponding to translocated glycosylated CPY* (pro-form) was also detected at 0 min chase, consistent with a previous report (Spear and Ng, 2003); at later times of chase, preproCPY* was almost completely converted to CPY*, possibly due to post-translational translocation. In the presence of constitutively active Hsf1, the preproCPY* form is decreased and, at the same time, a higher molecular weight band representing translocated glycosylated CPY* is increased (Figure 2A, 0 min chase), suggesting rapid conversion from the pre-form. Thus, HSR appears to facilitate ER translocation of newly synthesized polypeptide.
Figure 2.
Constitutively active Hsf1 facilitates CPY* translocation, degradation, and ER export. (A) Wild-type (MATa W303) and ire1Δ (KKY100) cells bearing pGAL-CPY* (pES67) were co-transformed with hsf1-R206S (pYEP96) or vector (pRS314). After pulse-labelling for 10 min at 25°C and chase, HA-tagged CPY* was immunoprecipitated with anti-HA and analysed by SDS–PAGE and fluorography. Mobilities of unglycosylated (preproCPY*) and glycosylated pro- (CPY*) forms are indicated on the right. (B) Same strains as (A). Cells were pulse-labelled as described in (A) and chased for longer times. HA-tagged CPY* was immunoprecipitated with anti-HA and analysed by SDS–PAGE and fluorography. CPY* quantification was performed by phosphorimager analysis, and the graph shows the mean±s.e. of three independent experiments. (C) Modification of CPY* by α1,3-mannose was monitored by pulse–chase analysis. After pulse-labelling and chase as in (B), HA-tagged CPY* was immunoprecipitated from lysate with anti-HA. A second IP was performed with anti-HA (left panel) or anti-α1,3-mannose antibody (right panel). IPs were treated with Endo H and analysed by SDS–PAGE and fluorography. Quantification was performed by phosphorimager analysis; modification by α1,3-mannose is expressed as a per cent of HA–CPY* recovered at each time point of chase. Exposure of the autoradiogram with α1,3-mannose IPs is increased compared with that of HA IPs.
To test whether suppression of the growth defect of ireΔ cells by constitutively active Hsf1 is related to degradation of CPY*, pulse–chase experiments were performed after inducing CPY* expression for 6 h. To discount the translocation defect of ire1Δ cells, newly synthesized CPY* was quantified at 40 min chase when the low-molecular-weight preproCPY* band had almost disappeared (Figure 2B). Nearly half of newly synthesized CPY* was degraded by 120 min in ire1Δ cells in the presence of constitutively active Hsf1, compared with >90% CPY* remaining in the absence of HSR (Figure 2B). Nevertheless, the degradation rate of CPY* in ire1Δ cells with constitutively active Hsf1 is slower than in wild-type IRE1 cells with a fully functional UPR.
Degradation of overexpressed CPY* is dependent on UPR and transport out of the ER (Spear and Ng, 2003). To test whether constitutively active Hsf1 increases CPY* degradation by promoting ER-to-Golgi transport, CPY* was examined for modification by α1,3-mannose, which is catalysed by Mnn1 in the medial Golgi (Raschke et al, 1973). Cells were pulse-labelled and chased for various times. Sequential IPs were then performed first with anti-HA to immunoprecipitate HA-tagged CPY* and then with either anti-HA or anti-α1,3-mannose. Compared with IRE1 cells, α1,3-mannose modification of CPY* was virtually undetectable in ire1Δ cells (Figure 2C). However, constitutively active Hsf1 restores modification of CPY* by α1,3-mannose. Thus, HSR promotes ER export of CPY* in the absence of UPR.
Constitutively active Hsf1 selectively releases the ER-to-Golgi transport block in ire1Δ cells overexpressing CPY*
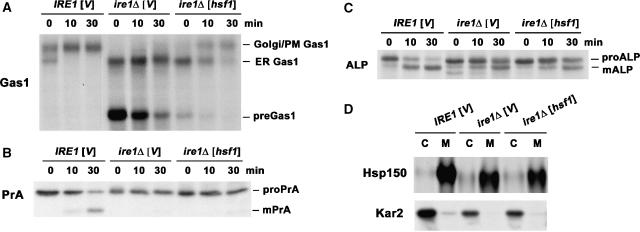
Because a general defect in ER export has previously been reported for ire1Δ cells overexpressing CPY* (Spear and Ng, 2003), we examined whether the effect of constitutively active Hsf1 is specific for CPY* export or has an effect on other cargoes also. Figure 3 shows pulse–chase experiments to follow the transport of several well-characterized cargoes. In Figure 3A, Gas1, a GPI-anchored protein of the plasma membrane (PM), was analysed (Nuoffer et al, 1991; Doering and Schekman, 1996). Newly synthesized Gas1 acquires its GPI anchor and core-glycosylation at the ER; upon delivery to the Golgi, it acquires Golgi glycosylation and its apparent molecular mass is increased as it is converted from the 105 kDa form to the mature 125 kDa form (Nuoffer et al, 1991; Doering and Schekman, 1996). In wild-type IRE1 cells overexpressing CPY*, Gas1 transport from the ER is not impaired, with the mature form detectable immediately after pulse (Figure 3A; Spear and Ng, 2003). By contrast, in ire1Δ cells, virtually no mature Gas1 was apparent, indicating an ER-to-Golgi transport defect (Figure 3A; Spear and Ng, 2003). Moreover, accumulation of untranslocated pre-Gas1 (∼60 kDa) was observed in these cells, consistent with the severe translocation defect (Figure 3A; Spear and Ng, 2003). In the presence of constitutively active Hsf1, pre-Gas1 is dramatically decreased, indicating reversal of the translocation defect. More importantly, the ER form of Gas1 is converted to the mature Golgi/PM form in the presence of constitutively active Hsf1, indicating that the transport block is relieved (Figure 3A). Even so, Hsf1-promoted conversion of Gas1 to its mature form is slower than that seen in UPR-competent IRE1 cells. Together, these data suggest that HSR relieves defects in Gas1 translocation and ER export in ire1Δ cells overexpressing CPY*.
Figure 3.
Constitutively active Hsf1 selectively relieves the ER-to-Golgi trafficking defect in ire1Δ cells overexpressing CPY*. (A–C) Wild-type (MATa W303) and ire1Δ (KKY100) cells bearing vector (pRS314) or hsf1 mutant (pYEP96) and pGAL-CPY* (pES67) were subjected to pulse–chase analysis as described in Figure 2A. Gas1 (A), PrA (B), and ALP (C) were immunoprecipitated and analysed by SDS–PAGE and fluorography. Untranslocated (pre-), ER, and mature forms (m-) of each protein are indicated. (D) Same strains as (A) were subjected to pulse–chase analysis and secretion assay as described in Materials and methods. Hsp150 and Kar2 were immunoprecipitated from cell lysate or medium and analysed by SDS–PAGE and fluorography.
Intracellular transport of vacuolar proteinase A (PrA) was also examined by monitoring its processing. PrA is synthesized at the ER as a pro-form of 48 kDa and undergoes proteolytic processing in the vacuole to generate a mature (m-) form of 42 kDa (Klionsky et al, 1988). In pulse–chase experiments in IRE1 cells overexpressing CPY*, m-PrA was observed after 10 min chase. In ire1Δ cells overexpressing CPY*, no processed m-PrA was detected even after 30 min, indicating a transport defect similar to that of Gas1 (Figure 3B; Spear and Ng, 2003). In contrast to Gas1, however, the transport block of PrA is not released by constitutively active Hsf1 (Figure 3B). The different fates of PrA and Gas1 suggest that the role of Hsf1 in promoting intracellular transport is cargo specific.
To examine possible cargo-specific effects of Hsf1, another vacuolar cargo protein, alkaline phosphatase (ALP), was examined (Klionsky and Emr, 1989). Similar to PrA, ALP is cleaved in the vacuole to yield the mature form. Surprisingly, ALP conversion to the mature form was observed in ire1Δ cells overexpressing CPY*, albeit the rate of processing is slower than that in wild-type IRE1 cells (Figure 3C). This observation suggests that in UPR-deficient cells, ER stress results in a selective, not general, block in vesicle transport. With constitutively active Hsf1, no significant change was observed in the rate of ALP processing (Figure 3C), indicating no stimulation of intracellular transport. These results are consistent with the idea that HSR selectively promotes ER-to-Golgi transport of specific cargo proteins in cells stressed by misfolded protein and loss of UPR.
Analysis of a secreted protein Hsp150 supports the idea of a cargo-specific transport defect in ire1Δ cells overexpressing CPY*. Extensive O-glycosylation of Hsp150 occurs during intracellular transport so that its apparent molecular mass shifts from 60 to 150 kDa (Russo et al, 1992). In Figure 3D, newly synthesized Hsp150 was immunoprecipitated from cell lysate or medium after pulse–chase. As in wild-type IRE1 cells, Hsp150 is present as the fully glycosylated 150 kDa form in the medium after 30 min chase in ire1Δ cells overexpressing CPY* (Figure 3D). A similar result was seen in the presence of constitutively active Hsf1. The ER luminal protein Kar2 serves as a control non-secreted protein and was detected only in cell lysate (Figure 3D, bottom panel).
Constitutively active Hsf1 selectively induces COPII cargo receptors
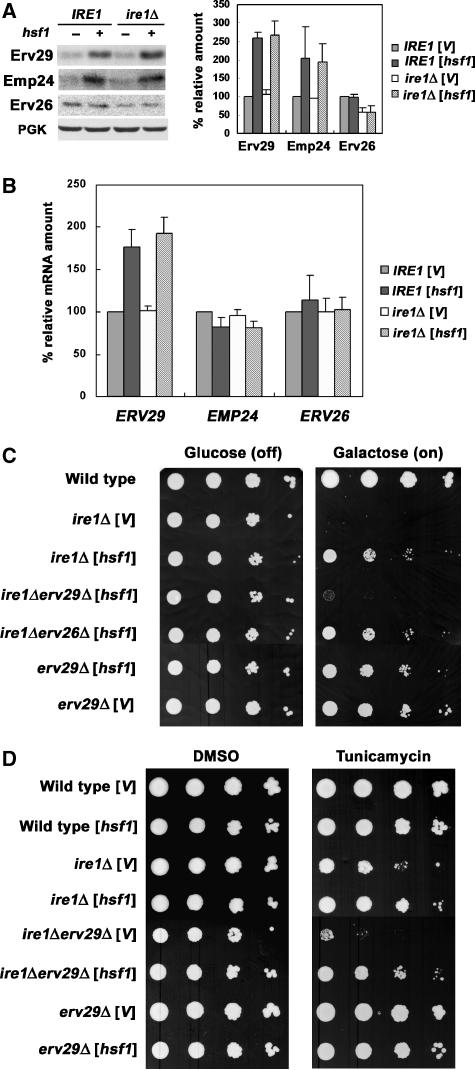
We next tested whether specific cargo receptors are targets of Hsf1. The COPII vesicle-associated proteins Erv29, Emp24, and Erv26 serve as cargo receptors to recruit CPY*, Gas1, and ALP, respectively, into transport vesicles (Muniz et al, 2000; Caldwell et al, 2001; Bue et al, 2006). In Figure 4A, the protein level of cargo receptors was measured by quantitative western blot. In the presence of constitutively active Hsf1, the protein level of Erv29 is dramatically increased (2.5-fold), consistent with the presence of several short consensus HSE, 5′-nGAAn-3′, in the ERV29 promoter; there is also an increase in Emp24 (two-fold), but the protein level of Erv26 remains the same with or without Hsf1 (Figure 4A). These results are in agreement with our observation that constitutively active Hsf1 facilitates ER exit of CPY* and Gas1 but not ALP (Figure 3). PrA transport is not enhanced by HSR in ire1Δ cells overexpressing CPY* (Figure 3B) although PrA export is partially dependent on Erv29 (Caldwell et al, 2001). It is possible that competition of PrA with overexpressed CPY* for the same receptor leaves PrA little chance to enter COPII vesicles.
Figure 4.
Induction of the cargo receptor Erv29 by HSR and its role in HSR-mediated suppression of ER stress. (A) Wild-type (MATa W303) and ire1Δ (KKY100) cells were transformed with hsf1-R206S (pYEP96) or vector (pRS314). Levels of cargo receptors Erv29, Emp24, and Erv26 were analysed by western blot with 125I-protein A. Scanned blots from two independent experiments were quantified. The loading control, PGK, was analysed by western blot visualized by ECL. (B) RT–PCR analysis of strains as in (A). These data are the mean±s.e. of three independent experiments. (C) Wild-type (MATa W303), ire1Δ (KKY100), ire1Δ erv29Δ (KKY110), ire1Δ erv26Δ (KKY112), and erv29Δ (KKY111) cells bearing pGAL-CPY* (pES67) were co-transformed with hsf1-R206S (pYEP96) or vector (pRS314). Cells were grown overnight in medium with raffinose and then spotted onto plates with SC medium with glucose (GAL off) or galactose (GAL on) for 2 and 3 days, respectively. (D) Wild-type (MATa W303), ire1Δ (KKY100), ire1Δ erv29Δ (KKY110), and erv29Δ (KKY111) cells were transformed with vector (pRS314) or hsf1-R206S (pYEP96). Cells were spotted on plates with SC medium and DMSO (solvent) or tunicamycin (0.05 μg/ml) and incubated at 30°C for 4 days.
RT–PCR was also used to measure mRNA levels of Erv29, Emp24, and Erv26. Consistent with the changes in protein level in the presence of constitutively active Hsf1, the mRNA level of ERV29 increased (1.8-fold) (Figure 4B), and a similar increase was observed when HSR was induced by mild heat stress, 37°C for 1 h (unpublished data). ERV26 mRNA was not increased by constitutively active Hsf1 (Figure 4B). These results indicate that ERV29, but not ERV26, is an Hsf1 target, consistent with a previous report using microarray and chromatin IP to identify Hsf1 targets (Hahn et al, 2004). Interestingly, the mRNA level of EMP24 is not increased by constitutively active Hsf1 (Figure 4B) or by shifting cells to 37°C (unpublished data). It appears that EMP24 transcription is not upregulated by Hsf1 although Emp24 protein level is increased (Figure 4A), suggesting the possibility that HSR may have a selective effect to enhance EMP24 translation. A similar observation has been made previously that HSR enhances translation without affecting transcription of the cargo receptor ERGIC53 (Spatuzza et al, 2004).
As impaired growth in ire1Δ cells is triggered by CPY* overexpression, we tested whether rescue by HSR is dependent on Erv29. Although constitutively active Hsf1 suppresses the growth defect of ire1Δ cells overexpressing CPY* (Figures 1A and 4C), suppression was no longer observed in ire1Δ erv29Δ cells (Figure 4C). Hsf1 action in this case is specifically dependent on Erv29; suppression by constitutively active Hsf1 is not affected by loss of ERV26 (Figure 4C). Therefore, HSR relieves defects associated with loss of UPR and CPY* overexpression through Erv29. Nevertheless, suppression of the growth defect of ire1Δ cells on tunicamycin by constitutively active Hsf1 is not affected by loss of ERV29 (Figure 4D), suggesting additional ameliorative effects of Hsf1.
Activated HSR stimulates ERAD
To test whether HSR can affect additional ER quality control mechanisms in the absence of UPR, we examined CPY* expressed at low copy number from its native promoter. Low-copy CPY* is cleared entirely through ERAD (Ng et al, 2000), in contrast to overexpressed CPY*, which appears to overflow to the vacuole for degradation (Spear and Ng, 2003). However, low-copy CPY* also requires UPR for efficient degradation, as it is partially stabilized in ire1Δ cells, as detected by western blot at various time points after cycloheximide addition (Figure 5A; Ng et al, 2000) and metabolic pulse–chase (Figure 5B). In wild-type IRE1 cells, low-copy CPY* is rapidly degraded (Figure 5A and B). In ire1Δ cells, a lower apparent molecular weight band corresponding to untranslocated preproCPY* indicates a translocation defect (Figure 5A; Ng et al, 2000). In the presence of constitutively active Hsf1, the prepro-form was no longer detected, and degradation of CPY* is as rapid as in wild-type IRE1 cells (Figure 5A and B).
Figure 5.
Constitutively active Hsf1 stimulates ERAD in ire1Δ cells. (A) Wild-type (MATa W303) and ire1Δ (KKY100) cells bearing vector (pRS314) or hsf1-R206S (pYEP96) and low-copy CPY* (pDN436) were analysed by western blot following cycloheximide chase. Untranslocated unglycosylated preproCPY* and translocated CPY* are indicated. (B) Wild-type (MATa W303), ire1Δ (KKY100), ire1Δ hrd1Δ (KKX29-1C), and ire1Δ pep4Δ (KKX30-1D) cells bearing vector (pRS314) or hsf1R206S (pYEP96) and low-copy CPY* (pDN436) were pulse-labelled and chased for various times. HA-tagged CPY* was immunoprecipitated, treated with Endo H, and then analysed by SDS–PAGE and fluorography. CPY* bands were quantified by phosphorimager analysis. (C) The steady-state protein level of Kar2 was examined by western blot in wild-type (MATa W303) and ire1Δ (KKY100) cells bearing constitutively active hsf1R206S (pYEP96) or vector (pRS314). The secondary antibody is 125I-protein A. Scanned films were quantified, and the graph shows the mean±s.e. of three independent experiments. (D) RT–PCR analysis of Kar2 on the same strains as in (D). The graph shows the mean±s.e. of three independent experiments. (E) Cycloheximide chase analysis as in (A) was carried out in wild-type (MATa W303) and ire1Δ (KKY100) cells bearing pGAL-KAR2 (pMR1341) or vector (pRS316) and low-copy CPY* (pDN436). To induce GAL-promoted KAR2, cells were grown overnight in medium with 2% galactose. Cells were harvested at various times after cycloheximide addition (10 μg/ml).
To show that constitutively active Hsf1 promotes degradation of low-copy CPY* through the ERAD pathway, dependence on the ubiquitin ligase Hrd1 was tested. Previous work has shown that Hrd1 is required for CPY* ubiquitination and its disposal by means of ERAD (Bordallo et al, 1998). Pulse–chase experiments in Figure 5B show that degradation of low-copy CPY* is not increased by constitutively active Hsf1 in ire1Δ hrd1Δ cells; by contrast, increased CPY* degradation promoted by HSR is not affected in ire1Δ pep4Δ cells. Because Kar2 is a known target of Hsf1 (Kohno et al, 1993), and works together with Hrd1 in ERAD (Plemper et al, 1997; Ismail and Ng, 2006), we tested whether the action of constitutively active Hsf1 on low-copy CPY* is mediated by Kar2. Figure 5C shows a quantitative western blot confirming that Kar2 is induced by constitutively active Hsf1, as reported previously (Kohno et al, 1993); Kar2 mRNA level is also increased (Figure 5D). Kar2 overexpression mimics some of the effects of constitutively active Hsf1: it rescues the translocation defect in ire1Δ cells, as virtually no preproCPY* was detectable (Figure 5E, 0 min chase). Moreover, Kar2 overexpression facilitates degradation of low-copy CPY*, as shown by metabolic pulse–chase analysis (Figure 5E; Plemper et al, 1997; Ng et al, 2000). We propose that HSR enhances ERAD by inducing KAR2.
Kar2 overexpression also suppresses the translocation defect of Gas1 in ire1Δ cells challenged by CPY* overexpression (Supplementary Figure S1A). However, only ∼20% of newly synthesized Gas1 leaves the ER after 30 min chase with overexpressed Kar2 compared with half (52%) arriving at the Golgi/PM with constitutively active Hsf1. Moreover, Kar2 overexpression (driven by the GAL1 promoter) cannot reverse the growth defect of ire1Δ cells challenged by CPY* overexpression (Supplementary Figure S1B; Spear and Ng, 2003). These results suggest that Kar2 has a major role in facilitating translocation but not vesicular transport, and support the idea that Hsf1 exerts an effect on multiple targets to relieve ER stress.
ER stress can induce HSR
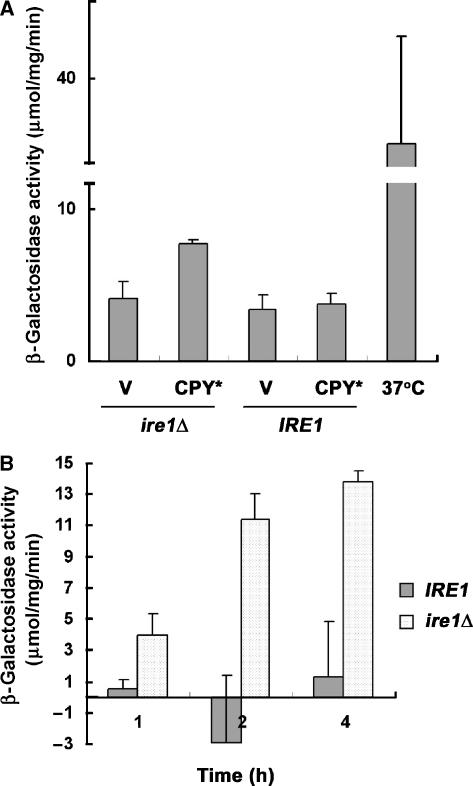
HSR induced artificially by introduction of constitutively active Hsf1 is able to relieve ER stress in the absence of UPR. We tested whether HSR is induced in vivo by ER stress, using a reporter in which the HSE is fused to lacZ (Liu et al, 1999). No increase in β-galactosidase activity was detected upon CPY* overexpression in wild-type IRE1 cells, indicating that HSR is not induced if UPR is functional. However, in ire1Δ cells, HSR is induced upon CPY* overexpression (Figure 6A) or in the presence of tunicamycin (Figure 6B). However, HSR induced by ER stress is significantly less (∼4 times) compared with HSR induced by mild heat stress (Figure 6A). These results are consistent with the inability of ire1Δ cells to grow in the presence of protein misfolding caused by tunicamycin or CPY* (Figure 1A and B), and suggest that HSR generated by these cells is insufficient to rescue their impaired growth.
Figure 6.
ER stress induces HSR in vivo. (A) IRE1 (MATa W303) and ire1Δ (KKY100) cells bearing an HSE-LacZ reporter (pCM64-SSA3-lacZ) and vector (pRS315) or pGAL-CPY* (pES67) were grown in SC medium with 2% raffinose at 30°C. CPY* was then induced in 2% galactose medium for 2 h. β-Galactosidase activity was measured in cell lysates. The graph shows the mean±s.e. of three independent experiments. (B) IRE1 (MATa W303) and ire1Δ (KKY100) cells transformed with an HSE-LacZ reporter (pCM64-SSA3-lacZ) were treated with tunicamycin (5 μg/ml) at the indicated times. β-Galactosidase activity was measured in cell lysates; activity from cells incubated in DMSO was subtracted. The graph shows the mean±s.e. of three independent experiments.
HSR targets in the secretory pathway
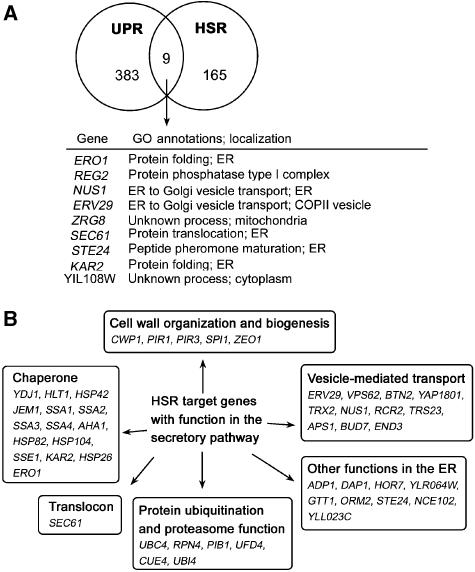
Because both Kar2 and Erv29 are involved in alleviating ER stress, we tested whether overexpression of Kar2 together with Erv29 can mimic the effect of constitutively active Hsf1. Supplementary Figure S1 shows that overexpression of both Kar2 and Erv29 cannot rescue growth of ire1Δ cells challenged by misfolded CPY*. These results suggest that other or additional target genes are involved in rescue by constitutively active Hsf1. It is possible that these targets are induced by both UPR and HSR, similar to KAR2 and ERV29. To compare HSR- and UPR-regulated genes, we used data generated from genome-wide studies (Travers et al, 2000; Hahn et al, 2004). We considered 165 HSR-regulated genes identified by Hahn et al and 383 UPR-regulated genes identified by Travers et al (including the well-known UPR target genes KAR2 and INO1). Nine genes found in both data sets are regulated by HSR and UPR (Figure 7A). Of these, five encode products localized to the ER: Kar2, Ero1, Sec61, Nus1, and Erv29. Based on their GO annotations in the Saccharomyces Genome Database (SGD), Kar2 and Ero1 are involved in protein folding; Kar2 also has a role in protein translocation together with Sec61, consistent with the idea that HSR enhances translocation; Nus1 and Erv29 function in vesicle transport.
Figure 7.
HSR target genes in the secretory pathway. (A) Nine of 383 UPR target genes are also induced by HSR. GO annotations and localization of these genes are from SGD (http://www.yeastgenome.org). (B) Forty-seven of 165 HSR target genes have functions in the secretory pathway. Genes are categorized based on known function and/or GO annotations. The functions of these genes are listed in Supplementary Table I.
As it is possible that rescue of UPR-deficient cells by constitutively active Hsf1 may occur by bypass of UPR-induced genes, we analysed HSR-dependent genes that function in the secretory pathway (Figure 7B and Supplementary Table I). Of the HSR-induced genes (Hahn et al, 2004), >25% (47 genes) appear to be important in the secretory pathway based on GO annotations in the SGD. Strikingly, these targets fall into the same functional categories as UPR targets (Figure 7; Travers et al, 2000). Among these, 15 have chaperone function, participating in protein folding and/or ERAD. Several genes upregulated by Hsf1 encode ERAD components, including ubiquitin and regulators of ubiquitination and proteasome activity. Other genes (PIB1, UBC4) involved in ubiquitination may participate in protein sorting in the distal secretory pathway. In addition to ERV29, 11 Hsf1-regulated genes function in vesicle transport, mediating transport at multiple steps of the secretory pathway. Cell wall biosynthesis genes are targets of both HSR and UPR. Finally, a group of HSR-regulated genes are proposed to encode products with ER function because they are localized to the ER (Huh et al, 2003). This extensive list supports a role for HSR in relieving stress in the secretory pathway.
Discussion
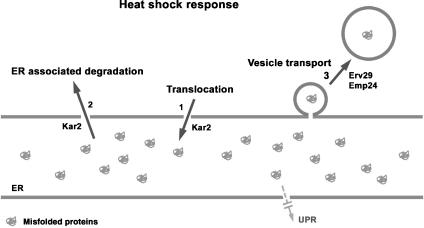
We have explored the role of HSR in ER quality control. Although UPR is the major response to ER stress, it is sometimes overwhelmed, for instance in ER stress–mediated diabetes as well as multiple conformational diseases characterized by protein misfolding and/or aggregation (Kaufman, 2002). We report that in the absence of UPR, activation of HSR can promote many pathways. Similar to UPR, HSR facilitates ER translocation of newly synthesized polypeptides, enhances the ERAD pathway, and promotes vesicular transport out of the ER (Figure 8). Our results point to a clear ameliorative effect of HSR on ER stress.
Figure 8.
HSR relieves ER stress through multiple pathways. The ER experiences stress when misfolded proteins are accumulated. In UPR-deficient cells challenged by misfolded proteins, HSR can relieve ER stress by at least three mechanisms. (1) HSR rescues defective protein translocation. (2) HSR promotes disposal of misfolded proteins through ERAD. (3) HSR promotes clearing of misfolded proteins by enhancing the export of some cargo proteins from the ER. We propose that the HSR target Kar2 has an important role in protein translocation and ERAD, and the cargo receptors Erv29 and Emp24 are upregulated during HSR to increase selective ER-to-Golgi transport.
A major finding of this study is that HSR enhances ER export of misfolded proteins. Vesicle transport from the ER with retrieval from the Golgi is one strategy for delivery of misfolded proteins for ERAD (Caldwell et al, 2001; Hermosilla et al, 2004). In both yeast and mammalian cells, transport of conformationally defective proteins to the vacuole/lysosome for degradation is another mechanism for relieving ER stress (Arvan et al, 2002). Furthermore, in mammalian cells, ER export is required during UPR, as transport to the Golgi results in proteolytic activation of the ATF6 transcription factor (Kaufman, 2002). These findings indicate that vesicle transport has an important role in response to ER stress. We find that HSR promotes ER export of the misfolded protein CPY* (Figure 2C). Although UPR upregulates multiple components essential for the formation of COPII vesicles (Travers et al, 2000), these targets are not affected by Hsf1 (Hahn et al, 2004; Yamamoto et al, 2005; our unpublished results). Instead, HSR promotes vesicular transport from the ER by upregulating other components, such as Erv29, to increase inclusion of specific cargo into the COPII vesicles.
Erv29 is required for Hsf1-mediated suppression of impaired growth of ire1Δ cells overexpressing CPY* (Figure 4). Erv29 has been shown to participate in the selection of a number of misfolded and native cargo proteins for ER export (Belden and Barlowe, 2001; Caldwell et al, 2001), and it is a high-copy suppressor of the ER-to-Golgi trafficking defect caused by expression in yeast of α-synuclein (Cooper et al, 2006). ERV29 is also one of a few genes upregulated by both UPR and HSR (Figure 7). Thus, Erv29 is an important participant in ER quality control. Nevertheless, ERV29 overexpression (or even in combination with KAR2 overexpression) is not sufficient to rescue the growth defect of ire1Δ cells challenged by CPY* (Supplementary Figure S1). Constitutively active Hsf1 can rescue sensitivity of ire1Δ cells to tunicamycin (Figure 1B), but Erv29 is not required for suppression in this case (Figure 4D). These observations support the idea that Hsf1 causes upregulation of additional factors that contribute to stress relief.
The ER-to-Golgi transport block of ire1Δ cells challenged by CPY* has been well characterized previously by Spear and Ng (2003). Even so, we were surprised to discover that the transport block is not a general one. We find that ALP and Hsp150 undergo normal intracellular transport (Figure 3C and D), confirming selective block in ER export. Our results imply that COPII vesicle formation and transport are not impaired but packaging of select cargo is affected by ER stress in these cells. It seems possible that different effects of ER stress on cargo packaging reflect different requirements of cargo for folding by engagement with distinct chaperones, as well as different requirements for export through bulk flow or association with specific cargo receptors.
Constitutively active Hsf1 also enhances ER translocation (Figures 2A and 5A). The translocon Sec61, many cytosolic chaperones, and Kar2 are under heat shock control (Figure 7), and both cytosolic and ER lumenal chaperones are required to facilitate translocation of newly synthesized proteins into the ER (Deshaies et al, 1988; Zimmermann, 1998). We find that overexpression of KAR2 corrects the translocation defect seen in ire1Δ cells with low-copy CPY* (Figure 5E) or overexpressed CPY* (Supplementary Figure S1A), suggesting that KAR2 upregulation has a critical role in facilitating protein translocation during HSR.
KAR2 overexpression also mimics the effect of constitutively active Hsf1 to increase ERAD (Figure 5E). This observation suggests that Kar2 mediates enhancement of ERAD by HSR, consistent with Kar2 participation in substrate recognition and delivery for polyubiquitination for ERAD (Ismail and Ng, 2006). In addition, HSR may promote ERAD through activation of the ubiquitin–proteasome system. The importance of the proteasome in HSR is indicated by cell sensitivity to heat stress when proteasome function is defective (Heinemeyer et al, 1991). Heat stress increases transcription and translation of polyubiquitin, encoded by UBI4 in yeast (Finley et al, 1987; Hahn et al, 2004), and Rpn4, which exerts transcriptional control over proteasome components, is also regulated by Hsf1 (Lee et al, 2002; Hahn et al, 2004). Involvement of HSR in ERAD is underscored by the observation that a misfolded membrane-bound ERAD(-C) substrate, Pma1-D378S, induces HSR rather than UPR, and degradation of this substrate is increased by constitutively active Hsf1 (Han et al, 2007).
In the absence of therapeutic intervention by constitutively active Hsf1, mild heat shock induced at 37°C is sufficient to rescue ire1Δ cells overexpressing CPY* (Supplementary Figure S1), suggesting that rescue is not dependent on a hyperactive Hsf1. In the absence of Hsf1-R206S or high temperature, a small HSR is detectable in UPR-deficient cells challenged with misfolded protein (Figure 6). It seems possible that HSR becomes involved in alleviating stress in the secretory pathway under specific physiologic conditions. For instance, during heat stress, UPR is activated (Gasch et al, 2000; Matsumoto et al, 2005) together with HSR to ameliorate protein misfolding in the secretory pathway. Consistent with this idea, ire1Δ cells are sensitive to heat stress (Supplementary Figure S3). An abundance of misfolded proteins in the cytosol is thought to activate HSR under various stress conditions, including heat (Morimoto, 1998). Initiation of HSR, specifically in response to ER stress, could occur by means of accumulation of misfolded proteins in the cytosol as a result of failed ER translocation and/or accumulation of ER membrane proteins with misfolded cytoplasmic domains that are recognized by cytoplasmic chaperones (Han et al, 2007).
Analysis of HSR-regulated genes supports the idea that quality control in the secretory pathway is within HSR purview (Figure 7). Although there are only a few genes that are under transcriptional regulation by both pathways (Figure 7A), >25% of Hsf1 targets have function in the secretory pathway (Figure 7B). Thus, it seems possible that rescue of UPR-deficient cells by constitutively active Hsf1 may occur by enhancement of common activities (translocation, vesicle transport, ERAD) albeit by distinct target genes. The effect of HSR on the endomembrane system is further emphasized by recent reports that Hsp90 and its cofactors have major roles in regulating protein folding at the ER, ER export, as well as multiple other vesicular transport steps (Wang et al, 2006; McClellan et al, 2007). As a subset of UPR targets function in the distal secretory pathway, several Hsf1 targets do so as well: enzymes involved in ubiquitination, such as Ubc4 (an E2) and Pib1 (an E3), and components of vesicle transport machinery, such as Rcr2 and Vps62 (Figure 7). These HSR targets may have a role in post-ER quality control.
Misfolded protein accumulation in cells is often toxic. Prolonged ER stress and UPR activation resulting in cell death is associated with numerous protein conformational diseases (Ron and Walter, 2007). Recent therapeutic efforts have been focused on chemical and pharmacological chaperones that promote protein folding and modulators of ER quality control (Romisch, 2004). Because of the importance of heat shock proteins in normal and disease states, modulators of HSR are also under investigation as therapeutic tools (Westerheide and Morimoto, 2005). Our results emphasize the potential of Hsf1 and other components of HSR as therapeutic targets for diseases involving ER stress and protein misfolding in the secretory pathway.
Materials and methods
Strains and plasmids
Yeast strains are listed in Table I. KKY100 is the same as MN5 (gift from J Warner, Albert Einstein College of Medicine) except that the LEU2 marker was swapped to HIS3 by transformation with pLH4 (Cross, 1997). KKY100 was crossed to WQY4 to generate KKX29-1C, an ire1∷HIS3 hrd1∷URA3 double knockout. KKY100 was crossed to WQY2 to generate KKX30-1D, an ire1∷HIS3 pep4 double mutant. KKY111 and KKY110 are W303 and KKY100 strains, respectively, transformed with PCR products to generate an ERV29 knockout marked by resistance to clonNAT (Werner BioAgents) (Goldstein and McCusker, 1999). The PCR products were amplified using pAG25 as template with primers 462 and 463 (Supplementary data). pAG25 was a gift from C Boone (University of Toronto, Canada). KKY112 is KKY100-derived with an ERV26 knockout marked by the resistance to clonNAT, which was also generated by transformation with PCR products. PCR products of ERV26 were amplified using primers 472 and 473 (Supplementary data).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| MN5 | MATa ire1∷LEU2 | J Warner (Albert Einstein College of Medicine) |
| WQY2 | MATa pep4∷URA3 (URA3 pop-out) | Wang and Chang |
| WQY4 | MATa hrd1∷URA3 | Wang and Chang |
| KKY100 | MATa ire1∷HIS3 | This study |
| KKX29-1C | MATα ire1∷HIS3 hrd1∷URA3 | This study |
| KKX30-1D | MAT? ire1∷HIS3 pep4 | This study |
| KKY110 | MATa ire1∷HIS3 erv29∷clonNAT | This study |
| KKY111 | MATα erv29∷clonNAT | This study |
| KKY112 | MATa ire1∷HIS3 erv26∷clonNAT | This study |
| All the strains were derived from W303 (MATa/α, leu2-3, 112, his3-11, trp1-1, ura3-1, can1-100, ade2-1). | ||
pES67 is a LEU2-marked centromeric plasmid bearing HA-tagged CPY* under the control of a GAL promoter; pDN436 is a LEU2-marked centromeric plasmid with HA-tagged CPY* under its own promoter (Spear and Ng, 2003); both were gifts from D Ng (National University of Singapore, Singapore). pCM64-SSA3-lacZ is a URA3-marked 2μ plasmid bearing the SSA3 HSE fused to lacZ (Liu et al, 1999), provided by D Thiele (Duke University Medical Center). pYEP96 is a TRP1-marked 2μ plasmid bearing hsf1-R206S, a constitutively active mutant of HSF1 (Sewell et al, 1995), and was a gift from D Winge (University of Utah Health Sciences Center). pGAL-KAR2 is a URA3-marked CEN plasmid bearing KAR2 under the control of the GAL promoter, and was a gift from M Rose (Princeton University). The vector pRS314 is a TRP1-marked centromeric plasmid (Sikorski and Hieter, 1989).
Quantitative real-time PCR
See Supplementary data.
Protein induction, metabolic pulse–chase, cycloheximide analysis, and western blot
Strains bearing pGAL-HA-CPY* were grown overnight at 30°C in SC medium with 2% raffinose. To induce protein expression, mid-log cells were shifted to medium with 2% galactose for 6 h.
Metabolic pulse–chase was performed as described previously (Luo and Chang, 2000; Liu et al, 2006). Briefly, cells were pulse-labelled with Expre35S35S (PerkinElmer) for 10 min and then chased in medium with 20 mM cysteine and methionine for various times. Cell lysate was prepared by vortexing with glass beads, as described before (Chang and Slayman, 1991). For IPs with anti-HA (Covance Inc.), anti-Gas1 (H Riezman, University of Geneva, Switzerland), anti-ALP (G Payne, University of California at Los Angeles), anti-PrA (T Stevens, University of Oregon), and anti-Hsp150 (M Makarow, University of Helsinki, Finland), the lysate was boiled in 1% SDS. Samples were then diluted to 0.1% SDS in SDS-free RIPA buffer and incubated with antibody and protein-A beads. To detect secretion of Hsp150, IP from cell lysate and medium was carried out as described before (Luo and Chang, 1997). For analysis of α1,3-mannose modification, anti-HA IPs were released from protein-A beads by boiling in 1% SDS. Samples were then diluted to 0.1% SDS in SDS-free RIPA buffer and anti-α1,3-mannose antibody (R Schekman, University of California at Berkeley) was added for a second round of IP. IPs were analysed by SDS–PAGE and fluorography. When indicated, N-linked glycosylation was removed by incubating IPs with 50 U endoglycosidase H (New England Biolabs). Quantification of pulse–chase experiments was performed by phosphorimager analysis using QuantityOne (Bio-Rad).
For cycloheximide chase analysis, cycloheximide was added to mid-log cells to a final concentration of 10 μg/ml. At various times after cycloheximide addition, aliquots were removed and added to 10 mM Na azide on ice. Lysates were normalized to protein content by Bradford assay and analysed by SDS–PAGE and western blotting.
For western blots, antibody binding was visualized by peroxidase-conjugated secondary antibody followed by a chemiluminescence detection system. Quantitative western blots were carried out using 125I-protein A (GE Healthcare). Rabbit anti-Kar2 was a gift from M Rose (Princeton University, Princeton, NJ). Rabbit anti-Erv29 and anti-Erv26 were gifts from C Barlowe (Dartmouth University, New Hampshire). Rabbit anti-Emp24 was a gift from H Riezman (University of Geneva, Switzerland).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary data
Acknowledgments
We thank Chong Chen for technical support and Jon Warner, Davis Ng, Dennis Thiele, Dennis Winge, Mark Rose, Charles Barlowe, Howard Riezman, Greg Payne, Tom Stevens, Randy Schekman, and Marja Makarow for strains, plasmids, and antibodies. This work was supported by NIH grant GM 58212.
References
- Arvan P, Zhao X, Ramos-Castaneda J, Chang A (2002) Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic 3: 771–780 [DOI] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C (2001) Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science 294: 1528–1531 [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH (1998) Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9: 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bue CA, Bentivoglio CM, Barlowe C (2006) Erv26p directs pro-alkaline phosphatase into endoplasmic reticulum-derived coat protein complex II transport vesicles. Mol Biol Cell 17: 4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell SR, Hill KJ, Cooper AA (2001) Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem 276: 23296–23303 [DOI] [PubMed] [Google Scholar]
- Chang A, Slayman CW (1991) Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J Cell Biol 115: 289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S (2006) Alpha-synuclein blocks ER–Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science 313: 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P (1993) Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73: 1197–1206 [DOI] [PubMed] [Google Scholar]
- Cross FR (1997) ‘Marker swap' plasmids: convenient tools for budding yeast molecular genetics. Yeast 13: 647–653 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R (1988) A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332: 800–805 [DOI] [PubMed] [Google Scholar]
- Doering TL, Schekman R (1996) GPI anchor attachment is required for Gas1p transport from the endoplasmic reticulum in COP II vesicles. EMBO J 15: 182–191 [PMC free article] [PubMed] [Google Scholar]
- Eastmond DL, Nelson HC (2006) Genome-wide analysis reveals new roles for the activation domains of the Saccharomyces cerevisiae heat shock transcription factor (Hsf1) during the transient heat shock response. J Biol Chem 281: 32909–32921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48: 1035–1046 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR (2004) Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24: 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Liu Y, Chang A (2007) Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem 282: 26140–26149 [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH (1991) Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J 10: 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermosilla R, Oueslati M, Donalies U, Schonenberger E, Krause E, Oksche A, Rosenthal W, Schulein R (2004) Disease-causing V(2) vasopressin receptors are retained in different compartments of the early secretory pathway. Traffic 5: 993–1005 [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ismail N, Ng DT (2006) Have you HRD? Understanding ERAD is DOAble!. Cell 126: 237–239 [DOI] [PubMed] [Google Scholar]
- Kaufman RJ (2002) Orchestrating the unfolded protein response in health and disease. J Clin Invest 110: 1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD (1988) Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol 8: 2105–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (1989) Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J 8: 2241–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K (1993) The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol 13: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ (1999) The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem 274: 26654–26660 [DOI] [PubMed] [Google Scholar]
- Liu Y, Sitaraman S, Chang A (2006) Multiple degradation pathways for misfolded mutants of the yeast plasma membrane ATPase, PMA1. J Biol Chem 281: 31457–31466 [DOI] [PubMed] [Google Scholar]
- Luo W, Chang A (1997) Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J Cell Biol 138: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Chang A (2000) An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast. Mol Biol Cell 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager WH, Ferreira PM (1993) Stress response of yeast. Biochem J 290 (Part 1): 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto R, Akama K, Rakwal R, Iwahashi H (2005) The stress response against denatured proteins in the deletion of cytosolic chaperones SSA1/2 is different from heat-shock response in Saccharomyces cerevisiae. BMC Genomics 6: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J (2007) Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131: 121–135 [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J (1993) A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74: 743–756 [DOI] [PubMed] [Google Scholar]
- Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Muniz M, Nuoffer C, Hauri HP, Riezman H (2000) The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J Cell Biol 148: 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Spear ED, Walter P (2000) The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol 150: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C, Jeno P, Conzelmann A, Riezman H (1991) Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol 11: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Taulien J, Lindquist S (1993) The role of heat-shock proteins in thermotolerance. Philos Trans R Soc London B 339: 279–285; discussion 285–276 [DOI] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH (1997) Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388: 891–895 [DOI] [PubMed] [Google Scholar]
- Raschke WC, Kern KA, Antalis C, Ballou CE (1973) Genetic control of yeast mannan structure. Isolation and characterization of mannan mutants. J Biol Chem 248: 4660–4666 [PubMed] [Google Scholar]
- Romisch K (2004) A cure for traffic jams: small molecule chaperones in the endoplasmic reticulum. Traffic 5: 815–820 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Russo P, Kalkkinen N, Sareneva H, Paakkola J, Makarow M (1992) A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc Natl Acad Sci USA 89: 8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell AK, Yokoya F, Yu W, Miyagawa T, Murayama T, Winge DR (1995) Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J Biol Chem 270: 25079. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Lewis MJ, Pelham HR (1987) Heat shock factor is regulated differently in yeast and HeLa cells. Nature 329: 81–84 [DOI] [PubMed] [Google Scholar]
- Spatuzza C, Renna M, Faraonio R, Cardinali G, Martire G, Bonatti S, Remondelli P (2004) Heat shock induces preferential translation of ERGIC-53 and affects its recycling pathway. J Biol Chem 279: 42535–42544 [DOI] [PubMed] [Google Scholar]
- Spear ED, Ng DT (2003) Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol Biol Cell 14: 2756–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Vogel F, Wolf DH (2002) ER–Golgi traffic is a prerequisite for efficient ER degradation. Mol Biol Cell 13: 1806–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, Riordan JR, Kelly JW, Yates JR III, Balch WE (2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127: 803–815 [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Morimoto RI (2005) Heat shock response modulators as therapeutic tools for disease of protein conformation. J Biol Chem 280: 33097–33100 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Mizukami Y, Sakurai H (2005) Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J Biol Chem 280: 11911–11919 [DOI] [PubMed] [Google Scholar]
- Zimmermann R (1998) The role of molecular chaperones in protein transport into the mammalian endoplasmic reticulum. Biol Chem 379: 275–282 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary data