Summary
Complex organisms require tissue-specific transcriptional programs, yet little is known about how these are established. The transcription factor FoxA1 is thought to contribute to gene regulation though its ability to act as a pioneer factor binding to nucleosomal DNA. Through genome-wide positional analyses, we demonstrate that FoxA1 cell type-specific functions rely primarily on differential recruitment to chromatin predominantly at distant enhancers rather than proximal promoters. This differential recruitment leads to cell-type specific changes in chromatin structure and functional collaboration with lineage-specific transcription factors. Despite the ability of FoxA1 to bind nucleosomes, its differential binding to chromatin sites is dependent on the distribution of histone H3 lysine 4 dimethylation. Together, our results suggest that methylation of histone H3 lysine 4 is part of the epigenetic signature that defines lineage-specific FoxA1 recruitment sites in chromatin. FoxA1 translates this epigenetic signature into changes in chromatin structure thereby establishing lineage-specific transcriptional enhancers and programs.
Introduction
Over the course of development, cells transit from a pluripotent state to one of many committed cell lineages. During this process, transcription factor networks are activated in order to establish cell type-specific transcriptional programs (Son et al., 2005). FoxA1 (Hepatocyte Nuclear Factor 3α), a member of the Forkhead family of winged-helix transcription factors, is involved in the development and differentiation of several organs including liver, kidney, pancreas, lung, prostate and mammary gland (Friedman and Kaestner, 2006; Kouros-Mehr et al., 2006; Spear et al., 2006). In addition, high expression of FoxA1 is commonly observed in tumors arising from these organs, including prostate and estrogen receptor α (ERα)-positive breast tumors (Lacroix and Leclercq, 2004; Lin et al., 2002; Mirosevich et al., 2006). Interestingly, FoxA1 expression is a positive prognostic factor among patients with ERα-positive breast tumors and correlates with sensitivity to endocrine therapy (Badve S, 2007). Consistent with its originally reported role as a pioneer factor involved in liver-specific gene expression (Bossard and Zaret, 2000; Cirillo et al., 1998; Gualdi et al., 1996), FoxA1 acts as a pioneer factor in the recruitment of ERα to several cis-regulatory elements in the genome and subsequent transcriptional induction of target genes such as Cyclin D1 (CCND1) in breast cancer cells (Carroll et al., 2005; Eeckhoute et al., 2006; Laganiere et al., 2005). This is mediated in part through the chromatin remodeling activity of FoxA1 (Cirillo et al., 2002; Eeckhoute et al., 2006), reminiscent of its role in the induction of liver specific gene expression (Friedman and Kaestner, 2006). FoxA1 also interacts with the androgen receptor (AR) in prostate cancer cells where it is thought to impact the regulation of AR target genes (Gao et al., 2003). Hence, FoxA1 appears capable of regulating distinct transcriptional programs in cells of different lineages. However, the molecular bases for the differential transcriptional activities of FoxA1 remain to be established. In the present study, we have investigated FoxA1 differential transcriptional activities in breast and prostate cancer cells and their functional relation with the epigenome of these cells.
Results
Dual regulatory role of FoxA1 in E2-signaling revealed by genome-wide ChIP-chip
Estrogen stimulation leads to the establishment of specific transcriptional programs in ERα-positive breast cancer cells. To address how FoxA1 participates in this process we initially performed an unbiased genome-wide chromatin immunoprecipitation study using tiling-microarrays (ChIP-chip) to define the repertoire of FoxA1 binding sites, which we define as its “cistrome” (http://en.wikipedia.org/wiki/Cistrome), in the MCF7 breast cancer cell-line. A total of 12904 high-confidence FoxA1 recruitment sites were identified in these cells [using a stringent statistical False Discovery Rate (FDR) of 1%] (Fig.S1 and S2). In comparison, the ERα cistrome in MCF7 cells (Carroll et al., 2006) reanalyzed using the MAT algorithm (Johnson et al., 2006) and updated to the most recent human genome sequence (Hg18) revealed 5782 high confidence sites (FDR 1%) (Fig.S3). Interestingly, the genomic distribution of FoxA1 binding sites was reminiscent of that of ERα (Carroll et al., 2005; Lin et al., 2007). Indeed, the majority of the sites (96.9%) were found distant from the proximal 1 kilobase (kb) promoter regions of genes (Fig.S4B). Accordingly, this distribution contrasted with that of RNA Polymerase II (RNA PolII) (Carroll et al., 2005), which is found primarily at proximal promoters (Fig.S4C). Comparing the FoxA1 and ERα cistromes, revealed a highly significant overlap with ∼50-60% ERα binding sites occurring on FoxA1 occupied sites (Fig.1A and S5A-B). To determine the functional significance of this co-binding, we subsequently determined the distribution of FoxA1 and ERα binding sites with regards to E2 regulated genes in MCF7 cells (Carroll et al., 2006). Hence, we compared the fraction of E2 regulated versus non-regulated genes in MCF7 cells with at least one binding site specific to ERα, FoxA1 or shared by the two factors (as defined in Fig. S5) within 20 kb of their transcription start site (TSS). Importantly, E2-upregulated genes were significantly enriched compared to non-regulated genes near sites of overlapping ERα/FoxA1 recruitment (Fig.1B). Strikingly, this was also the case for E2-downregulated genes (Fig.1B). These results demonstrate that genes having enhancers within 20 kb of the TSS that bind both ERα and FoxA1 together compared to ERα or FoxA1 separately are much more likely to be regulated in response to E2 treatment in breast cancer cells. A role for FoxA1 in E2-downregulated genes independently of its association with ERα was also revealed through the enrichment for this category of genes near sites recruiting FoxA1 only (Fig.1B). In fact, FoxA1 silencing in MCF7 cells reduced the basal expression of these genes to levels equivalent to the reduction seen after E2 treatment (Fig.S6A-B). This is most likely a consequence of FoxA1 role in allowing for the basal activity of enhancers for those genes (Fig.S6C-D). These data indicate that FoxA1 controls the E2 response in breast cancer cells through a combination of mechanisms consisting of maintaining the basal expression of genes repressed following hormone treatment and allowing for the induction of E2-upregulated genes through a direct collaboration with ERα. Interestingly, genes with FoxA1 binding sites within 20 kb of their TSS also had a greater chance to be expressed together with FoxA1 and ERα in primary breast tumors pointing to the biological relevance of the FoxA1 cistrome beyond the MCF7 cell-line (Fig.1C and S7-8).
Figure 1. Genome-wide identification of FoxA1 binding sites reveals its global role in control of E2 signaling in breast cancer cells.

A) Overlap analysis at FDR1% showing the number of binding sites specific to FoxA1 or ERα or shared between the two factors in MCF7 cells. B) Correlation between E2 up-regulated (left panel) or down-regulated (right panel) genes and binding of either ERα only (ERα unique), FoxA1 only (FoxA1 unique), both factors at different sites (ERα+FoxA1) or both factors at a shared site (ERα/FoxA1 overlapping sites) within 20 kb of the TSS of genes. Fold change is presented for instances where significant differences are observed between regulated (t-test p-value ≤ 10-3) versus non-regulated genes (t-test p-value ≥ 10-3). C) Correlation between ERα and FoxA1 binding sites and genes co-expressed with FoxA1 in primary breast tumors (Wang et al., 2005) were analyzed as in B. Fold change is presented for instances where significant differences are observed.
FoxA1 cell type-specific activity depends on differential recruitment to chromatin
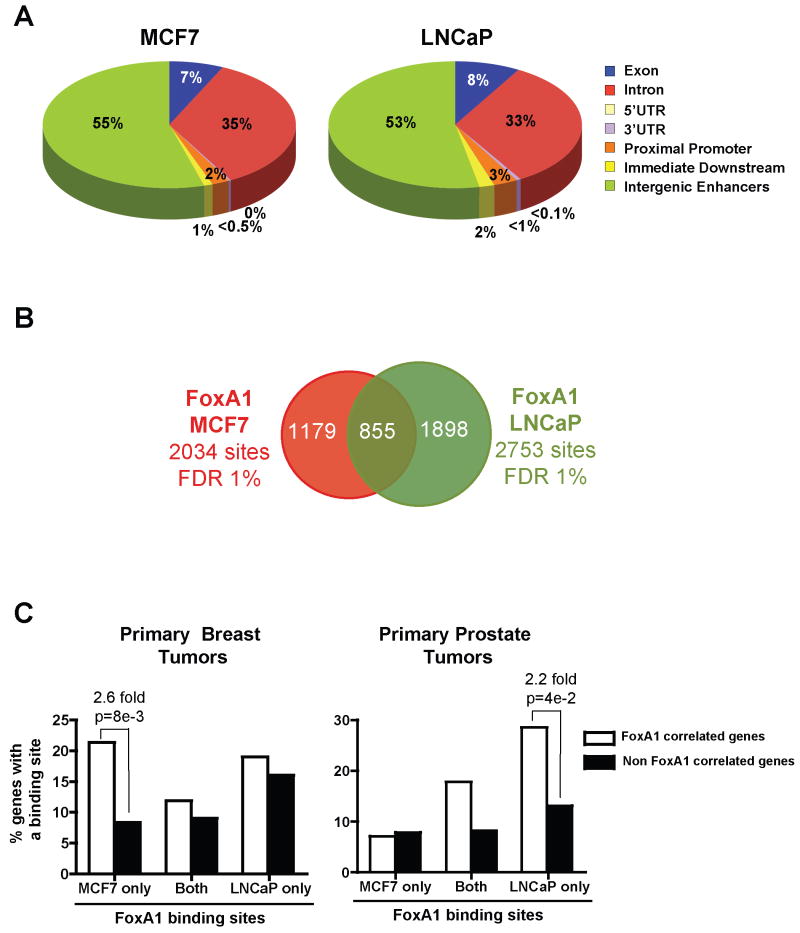
Having shown that FoxA1 recruitment to the chromatin within the MCF7 cell-line was correlated with the regulation of the transcriptional program specific to ERα-positive breast tumors, we investigated how FoxA1 binding to the chromatin relates to its cell-specific functions. This was accomplished by comparing the FoxA1 cistromes originating from cell-types of different lineages, namely the MCF7 breast cancer and LNCaP prostate cancer cell-lines. Through genomic-scale studies performed across the non-repetitive regions of human chromosomes 8, 11 and 12 using ChIP-chip assays, we identified over 2000 high confidence sites of FoxA1 recruitment (FDR 1%) in both cell-lines. As in MCF7 cells, these sites were predominantly found at enhancer positions in LNCaP cells (Fig.2A and S9). Importantly, comparison of the FoxA1 partial cistromes in these two cell-lines revealed both a significant number of shared sites and an even greater number of cell type-specific regions (Fig.2B). Indeed, comparisons of the datasets using various cut-offs indicated that the overlap did not exceed 55% and 40% of the MCF7 and LNCaP binding sites, respectively (Fig.S10A-C). Therefore, of all sites identified in both cell lines (3932 sites total), over 65 % of them correspond to regions of cell type-specific recruitment (886 sites specific to MCF7 cells and 1654 sites specific to LNCaP cells). The accuracy of these predictions was validated by ChIP-qPCR experiments (Fig.S10D). Hence, on a genomic-scale the majority of FoxA1 recruitment sites within the chromatin of two distinct cellular lineages are cell-type specific. These results strongly suggested that FoxA1 might regulate differential transcriptional programs as a result of its cell type-specific recruitment pattern in MCF7 and LNCaP cells.
Figure 2. Cell type-specific recruitment of FoxA1 correlates with differential gene expression patterns.
A) Cis-regulatory element annotation system (CEAS) (Ji et al., 2006) was used to determine the distribution of FoxA1 binding regions identified within chromosomes 8, 11 and 12 in MCF7 and LNCaP cells regarding known genes. B) Overlap analysis at FDR1% showing the number of FoxA1 binding sites specific to MCF7 or LNCaP or shared between the two cell-lines. C) Correlation between cell type-specific or shared FoxA1 binding sites and genes co-expressed with FoxA1 in primary breast (Wang et al., 2005) or prostate (Setlur, SR., Mertz, KD., Hoshida,Y., Demichelis, F., Lupien, M., Perner, S., Sboner, A., Pawitan, Y., Andren, O., Johnson, LA., et al. unpublished results) tumors. The occurrence of FoxA1 binding sites within 20 kb of the TSS of FoxA1 co-expressed genes was compared to that of non co-expressed genes. Fold change is presented for instances where significant differences are observed.
We next investigated the association of FoxA1 binding sites unique to MCF7 or LNCaP, or sites shared between the two cell lines with genes co-expressed with FoxA1 in primary breast or prostate tumors. This revealed a significant enrichment of genes co-expressed with FoxA1 in primary breast tumors over non-co-expressed genes near FoxA1 specific binding sites unique to MCF7 breast cancer cells (Fig.2C and Fig.S11) (van de Vijver et al., 2002; Wang et al., 2005). Reciprocally, genes co-expressed with FoxA1 in primary prostate tumors were significantly enriched over non-co-expressed genes near FoxA1 binding sites unique to LNCaP prostate cancer cells (Fig.2C) (Setlur, SR., Mertz, KD., Hoshida,Y., Demichelis, F., Lupien, M., Perner, S., Sboner, A., Pawitan, Y., Andren, O., Johnson, LA., et al. unpublished results). Altogether, these results demonstrate that differential recruitment is the primary mechanism responsible for the differential function of FoxA1 in these two different cell-lineages.
FoxA1 alternatively collaborates with ERα or AR at cell-specific enhancers
In order to further characterize the functional mechanisms involved in FoxA1 regulation of the breast and prostate cancer specific transcriptional programs, we monitored the transcription factor binding motifs enriched within the common FoxA1 recruitment sites, as well as those unique to each cell-line. As expected, the Forkhead motif (FKHR) was enriched in all three subsets of FoxA1 binding regions (Fig.3A). Conversely, we found that the recognition motifs for the nuclear receptors ERα (ERE and ERE half-site) and AR (ARE and ARE half-site) were specifically enriched in FoxA1 binding sites unique to MCF7 or to LNCaP cells, respectively (Fig.3A). This suggested that the differential FoxA1 recruitment between MCF7 and LNCaP was correlated with cell-specific transcriptional collaborations with ERα or AR. This hypothesis was tested by comparing the FoxA1 cistrome on chromosomes 8, 11 and 12 from both cell-lines to that of AR in LNCaP cells (Q.W. and M.B. unpublished results) and to that of ERα in MCF7 cells (Carroll et al., 2006). Interestingly, as was the case for ERα, we found that more than half of AR binding sites in LNCaP cells occurred on sites where FoxA1 was also present (Fig.3B). These data strongly suggest that the functional relationship between FoxA1 and AR previously demonstrated at a few model genes (Gao et al., 2003) in fact extend to a large fraction of regions used by this nuclear receptor. Accordingly, FoxA1 silencing modulated the transcriptional response to dihydroxytestosterone (DHT) of several studied target genes (Fig.S12). Importantly, the majority of FoxA1 binding sites overlapping with ERα were sites specific to MCF7 cells, while the majority of FoxA1 binding sites overlapping with AR were sites specific to LNCaP cells (Fig.3B). These data suggest that the cell type-specific recruitment of FoxA1 to the chromatin is linked to breast and prostate cancer transcriptional programs through specific collaborations with ERα in breast cells and AR in prostate cells. Indeed, these nuclear receptors are known to be master regulators of the behavior of a large subset of breast and prostate tumors through transmission of estrogenic and androgenic signals. Hence, we investigated the association of the different classes of sites with genes regulated by E2 in MCF7 cells or those regulated by DHT in LNCaP cells (Carroll et al., 2006; Wang et al., 2007). Only genes regulated by E2 were significantly enriched over non-regulated genes near ERα sites overlapping with FoxA1 in MCF7 cells (Fig.3C). In contrast, genes regulated by DHT were specifically significantly enriched over non-regulated genes near AR sites overlapping with FoxA1 in LNCaP cells (Fig.3C). Importantly, E2 or DHT regulated genes were mostly associated with the cell type-specific FoxA1 binding sites overlapping with ERα or AR and not those common to both cell lines (100% for AR/FoxA1 sites and 70% for ERα˜/FoxA1 sites). Overall, these data clearly implicate a role for FoxA1 in the regulation of breast and prostate specific transcriptional programs through cell-specific recruitment and subsequent differential collaboration with the sex steroid nuclear receptors ERα and AR.
Figure 3. FoxA1 cell type-specific binding sites also recruit nuclear receptors ERα or AR and correlate with regulation of sex steroid signaling in breast and prostate cancer cells.
A) Enrichment for the ERE, ERE half-site, FKHR, ARE and ARE half-site in the center of the binding sites specific to MCF7 cells (MCF7-only) or LNCaP cells (LNCaP-only) or shared between the two cell-lines (Both). The occurrence of the motifs (N motifs) was normalized to the number of sites in each subset (N binding sites). B) Venn diagrams depicting the overlap between FoxA1 (red) and ERα (blue) binding sites from MCF7 cells together with FoxA1 (green) and AR (orange) binding sites from LNCaP cells. C). Correlation between E2 or DHT regulated genes and binding sites for FoxA1 and ERα in MCF7 cells or for FoxA1 and AR in LNCaP cells. Analyses were performed as in Fig.1B using hormone regulated or non-regulated genes from chromosomes 8,11 and 12. Fold change is presented for instances where significant differences are observed between regulated versus non-regulated genes.
Differential recruitment to the chromatin extends to other transcription factors present in both MCF7 and LNCaP cells. Indeed, AP-1, whose recognition motif was enriched within the FoxA1 binding sites from MCF7 and LNCaP cells (Fig.S13A), was found to be co-recruited together with FoxA1 at a subset of its cell-specific binding sites (Fig.S13B). Hence, these data demonstrate that cell-specific recruitment also extends to ubiquitously expressed transcription factors such as AP-1 and suggest that this differential recruitment could also play an important role in its well-known cell-lineage differential activities (Jochum et al., 2001).
A cell type-specific histone signature correlates with differential FoxA1 recruitment
The functional importance of FoxA1 cell-specific recruitment described above raises the question as to how FoxA1 is able to bind to distinct regions within the genome of the MCF7 and LNCaP cells. Accordingly, we first considered the possibility that the sequence recognized by FoxA1 could be different between the two cell-lines. However, de novo motif analysis revealed that the Forkhead factor recognition sequence enriched within the FoxA1 binding sites did not show any significant difference between shared and cell-specific binding regions though it varied somewhat from the previously established consensus motif (Fig.4A). Therefore, we investigated whether the differential FoxA1 binding could rather be linked to specific epigenetic modifications. First, we looked at several repressive histone marks (Bernstein et al., 2007; Kouzarides, 2007) and found that H3K9me2 was more highly enriched on sites not recruiting FoxA1 in both cell lines, although not exclusively found on sites not recruiting FoxA1 (Fig.4B-C and S14A). We then sought to determine if FoxA1 recruitment was on the other hand associated with the presence of active histone marks. Recently, a genomic-scale study demonstrated the occurrence of mono (me1) and dimethylation (me2) of H3K4 at active enhancers (Heintzman et al., 2007). Analyzing the presence of these specific histone modifications at the FoxA1 recruitment sites revealed significant enrichment for H3K4me1 and me2 in a cell type-specific manner (Fig.4D-G). Indeed, in MCF7 cells, FoxA1 binding sites unique to MCF7 cells as well as sites common to both cell lines were significantly mono and dimethylated on H3K4 compared to the LNCaP unique FoxA1 binding sites (Fig.4D,F). On the other hand, in LNCaP cells, the LNCaP specific FoxA1 binding sites together with the common sites were significantly enriched for these histone modifications compared to MCF7 specific sites (Fig.4E-G). To confirm this correlation between H3K4 methylation and FoxA1 occupancy on a genomic scale we performed a ChIP-chip analysis of H3K4me2 levels in MCF7 cells across chromosomes 8, 11 and 12. These data revealed that on a genomic scale levels of H3K4me2 in MCF7 cells were indeed significantly greater on MCF7 specific or shared FoxA1 recruitment sites than on LNCaP specific ones (Fig.4H). H3K4me2 levels were also significantly higher on regions with FoxA1 recognition motifs bound by FoxA1 compared to an equivalent number of randomly selected unbound regions with FoxA1 recognition motifs in MCF7 cells (Fig. 4H). Importantly as less than 3.7% of sites harboring FoxA1 recognition motifs actually recruit FoxA1 in MCF7 cells (Fig. S14C), these data derived from the analysis of thousands of sites reveals a strong correlation between the presence of H3K4me2 and FoxA1 binding. Of the FoxA1 recruitment sites tested, as expected, very few demonstrated enrichment for H3K4me3 in accordance with the predominant occurrence of this modification at promoters rather than enhancers (Heintzman et al., 2007) (Fig.S14B). Overall, these results suggest a link between FoxA1 recruitment with the presence of H3K4me1 and me2.
Figure 4. Methylation pattern of histone H3 lysine 4 correlates with cell type-specific FoxA1 recruitment.
A) De novo determination of the sequence recognized by FoxA1 within its cell type-specific or shared binding sites. Logos show the consensus sequences of the enriched Forkhead motifs found by de novo analyses within the FoxA1 binding sites specific to MCF7 (MCF7-only) or LNCaP (LNCaP-only) cells or common to the two cell-lines (Both) in comparison to the Transfac FoxA1 matrix (http://www.gene-regulation.com/pub/databases.html#transfac). B-G) Levels of H3K9me2 (B-C) H3K4me1 (D-E) and H3K4me2 (F-G) on FoxA1 recruitment sites specific to MCF7 cells (MCF7-only) or LNCaP cells (LNCaP-only) or shared between the two cell-lines (Both) were determined by ChIP-qPCR. Box plots were generated from data obtained from three independent experiment testing 11 sites specific to MCF7 cells, 12 to LNCaP cells and 8 common to both cell-types. Statistical analyses of the difference between the non cell type-specific sites and the other sites are presented, *: p≤0.05 and **: p≤0.01. H) ChIP-chip analyses of H3K4me2 levels across chromosomes 8,11 and 12 in MCF7 cells. Two independent ChIP-chip experiments were combined and analyzed using the MAT algorithm. The signals given by the probes localized in the 200 bp central regions of the FoxA1 binding sites unique to MCF7 (MCF7-only) or LNCaP (LNCaP-only) or shared (Both) by the two cell-lines were compared (left graph). Similarly, H3K4me2 levels at 200bp regions containing the FoxA1 recognition motif bound by FoxA1 were compared to randomly selected FoxA1 unbound FoxA1 recognition motif containing regions (right graph). Means +/- S.E. of H3K4me2 levels given by MAT are shown as well as statistically significant differences with *** corresponding to p≤0.001.
FoxA1 is required for chromatin remodeling but not for H3K4 methylation
In MCF7 cells, H3K4me1 and me2 are detected at enhancers prior to E2 stimulation and ERα binding, reminiscent of FoxA1 recruitment (Fig.S15). Accordingly, ERα silencing in these cells did not dramatically affect H3K4 methylation levels or FoxA1 recruitment at most sites where these two factors are recruited (Fig.5A and S16). Moreover, the vast majority (∼80%) of FoxA1 sites specific to MCF7 cells do not recruit ERα (Fig.3B). Hence, while we cannot entirely rule out a potential role for ERα in stabilizing FoxA1 binding at a small subset of sites, these results suggest that in general cell-specific FoxA1 recruitment occurs independently of ERα action in MCF7 cells. This raises the issue of whether H3K4me1 and me2 are required for FoxA1 recruitment or are induced as a result of FoxA1 binding to the chromatin. This question was first addressed by investigating whether FoxA1 silencing would affect H3K4 methylation, chromatin remodeling or both in MCF7 and LNCaP cells. Consistent with its cell type-specific recruitment, FoxA1 silencing impacted the DNAse I sensitivity only at those sites to which it was recruited (Fig.5B). Under these conditions, however, these sites did not in general show a significant reduction in the levels of H3K4me1 or me2 in either MCF7 or LNCaP cells (Fig.5C). In fact, a significant increase in H3K4me1 was detectable at most sites tested in LNCaP cells. Similarly, levels of H3K9me2 were unaffected by FoxA1 silencing (Fig.S17). Overall, these data do not favor a model whereby FoxA1 recruitment leads to the induction of these modifications but rather suggest an important contribution of FoxA1 in opening genomic regions marked by H3K4me1 and me2. Accordingly, even though FoxA1 silencing did not modulate H3K4 methylation levels at enhancers (Fig.5D), it affected the transcriptional regulation of their target genes (Fig.5E and Fig.S18). Considering that H3K4me2 is typically associated with gene transcription (Bernstein et al., 2005), these results highlight the critical interplay between the pioneer factor FoxA1 and H3K4me2 at enhancers for efficient gene regulation.
Figure 5. FoxA1 silencing decreases chromatin accessibility of enhancers but not H3K4 methylation levels.

A) Effect of ERα silencing on FoxA1 recruitment. Eight sites recruiting both ERα and FoxA1 in MCF7 cells were used to monitor the effect of ERα silencing on ERα and FoxA1 recruitment by ChIP-qPCR. Reduction in ERα protein levels by siERα was also demonstrated by western blot (Fig.S16A). B) DNaseI sensitivity assays were performed in both MCF7 and LNCaP cells and the percent change triggered by FoxA1 silencing from at least three independent experiments is reported. C) Effect of FoxA1 silencing on the levels of H3K4me1 and me2 at binding sites used in the DNaseI sensitivity assays in both MCF7 and LNCaP cells from three experiments is presented, * p≤0.05 and **: p≤0.01. D-E) Presence of H3K4me1/me2 at enhancer is not sufficient for transcriptional regulation of BIK and CCND1 in MCF7 cells. H3K4me1/2 levels at FoxA1 recruiting enhancers localized within or nearby FoxA1 target genes were determined by ChIP-qPCR in MCF7 cells transfected with siLuc or siFoxA1 (C). Even though FoxA1 silencing did not modulate the levels of H3K4 methylation, the expression of the target genes was significantly reduced (D).
Cell type-specific FoxA1 recruitment depends on H3K4 methylation
To establish the capacity of H3K4 mono or dimethylation to define the cell type-specific recruitment of FoxA1, we overexpressed the H3K4me1 and me2 specific demethylase KDM1 (also known as LSD1/BHC110) in MCF7 cells and established its impact on FoxA1 recruitment (Shi et al., 2004). Under these conditions, H3K4me1 was slightly reduced (Fig.S19A) and H3K4me2 was significantly lowered on FoxA1 binding sites (Fig.6A). The level of H3K9me2 remained unchanged at these sites (Fig.6C). Although FoxA1 protein levels were unaffected by KDM1 over-expression (Fig.6D), its recruitment to the chromatin was significantly impaired (Fig.6B). Importantly, no global alteration in ChIP efficiency was observed upon KDM1 over-expression (Fig.S20B-C). Hence, these results suggest that H3K4me2 is required to define the cell type-specific regions competent for recruitment of FoxA1. The correlation between the presence of histone marks and FoxA1, ERα or AR recruitment is shown for specific examples of hormone-regulated genes (Fig.6E).
Figure 6. Role of H3K4me2 in FoxA1 recruitment to the chomatin.
A-C) Effect of KDM1 over-expression on H3K4 methylation (A), FoxA1 recruitment (B) and H3K9 methylation (C). H3K4me2 and H3K9me2 levels as well as FoxA1 recruitment were determined in control or KDM1 over-expressing cells by ChIP-qPCR. Box plots were generated from data obtained for 16 sites. Results from one representative experiment are presented with the statistical analyses of the difference between control and KDM1 over-expressing cells, **: p≤0.01. D) Western blots showing KDM1, FoxA1 and Calnexin (Control) levels in MCF7 cells transfected with an empty control plasmid or a plasmid coding for KDM1. E) Specific examples of genes regulated by E2, DHT or by both hormones. One gene specifically regulated by E2 in MCF7 cells (MCF7-only), by DHT in LNCaP cells (LNCaP only) and by both hormones in MCF7 and LNCaP cells respectively (both) are shown. E2 and DHT regulated genes were identified using expression array analyses performed in MCF7 and LNCaP cells, respectively. Significantly regulated genes were determined using a t-test and a p-value cut-off of 5×10-3. ERα, AR and FoxA1 binding sites from ChIP-chip are indicated together with the occurrence of histone modifications derived from ChIP-qPCR at these sites. Enrichment for the various factors is presented by green and red blocks in LNCaP and MCF7 cells, respectively. White blocks indicate the absence of enrichment for the ChIPed factors or a decrease of more than two-fold for histone marks in MCF7 cells following KDM1 over-expression. A 4 kb wide view of the probe signals obtained by ChIP-chip for FoxA1, ERα and AR at the analyzed binding sites is also shown. Complete probe signal across the 3 genes selected is presented in Fig.S21.
Discussion
Networks of transcription factors are known to be at the center of cell type-specific transcriptional programs that characterize different cell lineages (Olson, 2006; Schrem et al., 2002). However, how a particular transcription factor manages to regulate gene expression in a cell type-specific fashion within the context of different transcription factor networks is still poorly understood. In particular, it is still elusive how a pioneer factor, such as FoxA1, able to bind condensed chromatin structures in vitro can mediate differential gene regulation in vivo (Cirillo et al., 2002; Eeckhoute et al., 2006). Here, we show that FoxA1 differential transcriptional activities in breast and prostate cells relies primarily on its differential recruitment to the chromatin and alternative collaboration with the lineage-specific factors ERα or AR at cell-specific enhancers (Fig.6E, 7 and S21). These findings indicate that alternative transcriptional programs depend both on the orchestrated expression of a particular set of collaborating transcription factors together with their ability to bind cell-specific enhancer elements in the vicinity of their target genes. Alternatively, other transcription factor networks may primarily target gene promoters (Bieda et al., 2006; Geles et al., 2006). This may allow for a tight regulation of gene expression both at basal levels and in response to stimuli through combined activities of promoter and enhancer bound regulatory complexes (Hatzis and Talianidis, 2002; Marr et al., 2006). Importantly, we found that even ubiquitous transcription factors, such as AP-1, show differential recruitment to cell type-specific enhancers. Combined with other recent studies (So et al., 2007), this suggests that cell-specific binding to the chromatin represents a general mechanism for differential transcription factor regulatory activities. Cell-specific recruitment of AP-1 to FoxA1 sites could have important functional implications in breast cells especially for E2 downregulated genes where FoxA1 binding sites are enriched for AP-1 and Sp1 motifs (p≤0.05) that can tether ERα to mediate gene repression (Carroll et al., 2006; Stossi et al., 2006). Other important candidates for a global role in control of sex steroid signaling through collaborations with FoxA1 and ERα or AR include GATA family members (Eeckhoute et al., 2007) (Wang et al., 2007), c-myc (Cheng et al., 2006) and NFIC (Eeckhoute et al., 2006).
Figure 7. Model of the cell type-specific interplay between the epigenetic signature and FoxA1 for the establishment of lineage specific transcriptional programs.

Schematic representation of how FoxA1 recruitment occurs primarily on H3K9me2-poor but H3K4me1/me2-rich regions. H3K4me1/me2 could guide FoxA1 cell type-specific recruitment through direct physical interactions. FoxA1 regulation of differential transcriptional programs is subsequently achieved through transcriptional collaborations with cell type-specific (ERα and AR) as well as ubiquitously expressed (AP-1) transcription factors.
The occurrence of specific histone modifications at cis-regulatory elements commonly characterizes transcriptionally active or inactive regions (Bernstein et al., 2007; Kouzarides, 2007). Recently, the balance between the presence of active or repressive histone modifications (trimethylation of H3K4 and H3K27) has been shown to correlate with promoter activity (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007). Here, we show that the cell type-specific activity of enhancers correlates with the presence of the positive mark H3K4me2, previously shown to be distributed in a cell type-specific manner (Bernstein et al., 2005), while inactive enhancers lack H3K4me2 and harbor higher levels of the repressive mark H3K9me2. Interestingly, even though FoxA1 silencing does not modulate levels of H3K4 and K9 methylation at enhancers (Fig.5 and S17) it is required for their activity and consequently for their target gene transcriptional regulation (Fig.5, S6 and S18). Therefore, H3K4me1/2 appear to correlate with competent enhancers but not necessarily with transcriptional activation of target genes that requires factors such as FoxA1 to activate the functionality of these enhancers.
The capacity of FoxA1 to bind unique binding sites in reconstituted chromatin has been studied extensively in vitro (Cirillo et al., 2002; Cirillo et al., 1998; Sekiya and Zaret, 2007). Under these conditions, no histone modifications appear to be required for FoxA1 recruitment. However, our results demonstrate that in vivo FoxA1 actually occupies only a very small fraction of all its potential recognition motifs found in the genome (less than 3.7%). Moreover, this limited number of occupied sites is significantly different between two different cell-types. Therefore, although FoxA1 can act as a pioneer factor able to bind to condensed chromatin, we show here that in vivo its pioneer function is limited to a small subset of sites that are largely cell-type specific. Our data further defines on a genomic-scale the chromatin components involved in directing FoxA1 recruitment to this subset of its potential binding sites. Indeed, our results point to an important role of active and repressive histone marks notably H3K4me2 and H3K9me2, respectively, in guiding FoxA1 recruitment. These data indicate that a better understanding of cell-lineage transcriptional commitment will require the study of how these marks are established and how they regulate recruitment of pioneer transcription factors such as FoxA1. Altogether, our data reveal an additional layer of complexity in the regulation of FoxA1 recruitment to chromatin in vivo that goes beyond the mere presence of its recognition motif. Indeed, FoxA1 translates an epigenetic signature into functional cell-type specific enhancers leading to the establishment of cell type-specific transcriptional programs.
Experimental Procedures
ChIP-chip and ChIP-qPCR
ChIP-chip experiments using Affymetrix Human Tiling 2.0R Array Set were performed as previously described (Carroll et al., 2005; Carroll et al., 2006). For each ChIP-chip experiment, at least three independent assays were performed. Analyses were performed using MAT (Johnson et al., 2006), whose probe mapping had been updated to the latest human genomic sequence (Hg18). We used statistical False Discovery Rate (FDR) as cut-off in those analyses. All ChIP-chip data used in this study can be accessed at http://research.dfci.harvard.edu/brownlab/datasets/. ChIP-qPCR experiments were performed as in (Carroll et al., 2005). Statistical analyses were performed using Student's t-test comparison for unpaired data. Primer sequences can be found in Supplementary Table 1.
Antibodies used for ChIP experiments were FoxA1 (Ab5089 and Ab23738 from Abcam, FOX1 from CeMines) ERα (Ab-10 from Neomarkers, HC-20 from Santa Cruz), pan-jun (D from Santa-Cruz), pan-fos (K-25 from Santa-Cruz) (Schwartz et al., 2007), AR (N20 from Santa-Cruz), H3K4me1, me2, me3, H3K9me1, me2, me3, H4K20me1, me2, me3 (Ab8895, Ab7766, Ab8580, Ab9045 Ab1220, Ab8898, Ab9051, Ab9052 and Ab9053, respectively from Abcam) (Mikkelsen et al., 2007) (Barski et al., 2007), H3K27me1, me2, me3 (07-448, 07-449 and 07-452 from Upstate Biotechnology Inc) (Barski et al., 2007; Mikkelsen et al., 2007; Vakoc et al., 2006) RNA PolII (H-224 from Santa-Cruz and Ab5408 from Abcam), H3 (Ab1791 from Abcam) and AcH4 (from Cell Signaling).
Genomic distribution and binding site overlap
Genomic distribution of binding sites identified by ChIP-chip was performed using cis-regulatory element annotation system (CEAS) (Ji et al., 2006). Two binding sites were considered to overlap as long as they had one base pair in common. The average size of the ChIP-chip regions being 1 kb, this means that the center of the two binding sites had to be in average within 1 kb of each other to be considered overlapping.
Transcription factor recognition motif enrichment analysis
Known DNA motifs that are enriched relative to the center of ChIP-chip sites were identified using the following statistic. All sites were trimmed or expanded to 600 bps centered at the middle point of the identified ChIP-enriched regions. All sub-sequences within the trimmed regions were scored by a TRANSFAC motif (Matys et al., 2006) and the genomic background sequence composition to identify hits above certain relative entropy cutoff t. Letting xi, a value between 0 and 1, denote the relative location of motif hit i on the ChIP-regions (0 and 1 representing the center and edge of a ChIP-region, respectively) out of N total motif hits, we define a z-score, to assess the positional bias of a motif towards the center of the regions. Different integer cutoffs t ≥ 3 were tested for each motif, and the cutoff resulting in the highest z was selected. This statistic is based on the assumptions that insignificant DNA motifs will be uniformly distributed across the ChIP-regions and the null distribution of Σxi can be estimated as the N-fold convolution of uniform density functions. In figure 3A, a Gaussian kernel was used to smooth the curves in case too few motif hits appear at particular positions.
Association of trends in gene expression with transcription factor binding sites
Gene expression data was normalized and summarized using RMA (Irizarry et al., 2003) and updated RefSeq probeset definitions (Dai et al., 2005). Where multiple transcripts were associated with a single gene expression index the transcript with the transcription start site closest to a ChIP-enriched region was selected. “Differentially expressed” genes were denoted as those genes with a t-test p-value ≤ 10-3. Genes “close” to a ChIP region were defined as those having such a region within 20 kb of the transcription start site. Fisher's exact test was used to assess the statistical significance of the association between “close” genes and “differentially expressed” genes.
De novo motif search
De novo motif searches were performed on sequences ±100 bps from the center of FoxA1 ChIP regions in MCF7 cells or LNCaP cells by using LeitMotif (Song, J. and X.S.L. unpublished results), a modified MDscan (Liu et al., 2002) with ninth-order Markov dependency for the genome background. Motif logos were generated by enoLOGOS (Workman et al., 2005).
RNA interference
FoxA1 was silenced using the following small interfering RNA duplexes: siFoxA1 #1 sense 5′-GAGAGAAAAAAUCAACAGC-3′; antisense 5′-GCUGUUGAUUUUUUCUCUC-3′ (Carroll et al., 2005; Eeckhoute et al., 2006) and siFoxA1 #2 5′-GGACUUCAAGGCAUACGAAUU-3′; 5′-UUCGUAUGCCUUGAAGUCCUU-3′ (Fig.S17). SMARTpool siRNA directed against ERα was purchased from Dharmacon. Small interfering RNA against Luciferase was used as a negative control (Carroll et al., 2005).
DNase I hypersensitivity assays
DNase I hypersensitivity assays were performed as in (Eeckhoute et al., 2006).
KDM1 overexpression experiments
A total of 15mg of pCMX-KDM1 construct or the control empty vector were transfected in MCF7 cells using lipofectamine 2000 (Invitrogen) according to the manufacturers instructions. Following 76 hours of expression, cells were processed for ChIP-qPCR as previously described.
Real-Time RT-PCR
RNA was isolated from MCF7 and LNCaP cells using RNeasy mini kit (Qiagen), with on-column DNase treatment to remove contaminating genomic DNA. Real-time reverse transcription-PCR (RT-PCR) was done as in (Keeton and Brown, 2005). Primers used in RT-qPCR are listed in Supplementary Table 2.
Western Blots
Western blots were processed as described in (Lupien et al., 2007) using antibodies against KDM1 kindly provided by R. Schule (Universitäts-Frauenklinik und Zentrum für Klinische Forschung, Freiburg, Germany), FoxA1 (Abcam) and Calnexin (Stressgen Biotechnologies).
Supplementary Material
Supplemental Data
Supplement Data include twenty one figures, two tables and Supplemental References and can be found with this article online at www.cell.com
Acknowledgments
We thank Dr. Roland Schüle (Universitäts-Frauenklinik und Zentrum für Klinische Forschung, Freiburg, Germany) for pCMX-Flag-KDM1 as well as KDM1 antibodies. We also thank Drs. Mark A. Rubin (Dana Farber Cancer Institute, Massachusetts, USA) and Andrea Sboner (Yale University, Connecticut, USA) for help in analyzing the primary prostate tumor expression datasets. We acknowledge Dr. Shannon T. Bailey for his help with the validation of the siRNA targeting ERα. This work was supported by grants from the NIDDK (R01DK074967 to M.B.), the NCI (P01 CA8011105 and the DF/HCC Breast Cancer SPORE Grant to M.B.), and the DFCI Women's Cancers Program. This study was designed by J.E., M.L., C.A.M, X.S.L and M.B. The experimental procedures were primarily carried out by M.L. and J.E. with assistance from J.S.C. The AR cistrome was provided by Q.W. Biostatistical and computational support and data analysis was provided primarily by C.A.M. with the assistance of Y.Z. and W.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Badve S, T D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H. FOXA1 expression in breast cancer - Correlation with luminal subtype A and survival. Clinical cancer research. 2007;13:4415–4421. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome research. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. Repressive and restrictive mesodermal interactions with gut endoderm: possible relation to Meckel's Diverticulum. Development. 2000;127:4915–4923. doi: 10.1242/dev.127.22.4915. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Molecular cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Molecular cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim EY, Clark KL, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. The EMBO journal. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic acids research. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes & development. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. doi: 10.1158/0008-5472.CAN-07-0746. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Zhang J, Rao MA, Case TC, Mirosevich J, Wang Y, Jin R, Gupta A, Rennie PS, Matusik RJ. Molecular endocrinology. Vol. 17. Baltimore, Md: 2003. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes; pp. 1484–1507. [DOI] [PubMed] [Google Scholar]
- Geles KG, Freiman RN, Liu WL, Zheng S, Voronina E, Tjian R. Cell-type-selective induction of c-jun by TAF4b directs ovarian-specific transcription networks. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2594–2599. doi: 10.1073/pnas.0510764103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes & development. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Hatzis P, Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Molecular cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic acids research. 2006;34:W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2001;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton EK, Brown M. Molecular endocrinology. Vol. 19. Baltimore, Md: 2005. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT; pp. 1543–1554. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 Maintains the Differentiation of the Luminal Cell Fate in the Mammary Gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. Whole-Genome Cartography of Estrogen Receptor alpha Binding Sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Miller CT, Contreras JI, Prescott MS, Dagenais SL, Wu R, Yee J, Orringer MB, Misek DE, Hanash SM, et al. The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 2002;62:5273–5279. [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nature biotechnology. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Lupien M, Jeyakumar M, Hebert E, Hilmi K, Cotnoir-White D, Loch C, Auger A, Dayan G, Pinard GA, Wurtz JM, et al. Molecular endocrinology. Vol. 21. Baltimore, Md: 2007. Raloxifene and ICI182,780 increase estrogen receptor-alpha association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12; pp. 797–816. [DOI] [PubMed] [Google Scholar]
- Marr MT, 2nd, Isogai Y, Wright KJ, Tjian R. Coactivator crosstalk specifies transcriptional output. Genes & development. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic acids research. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirosevich J, Gao N, Gupta A, Shappell SB, Jove R, Matusik RJ. Expression and role of Foxa proteins in prostate cancer. The Prostate. 2006;66:1013–1028. doi: 10.1002/pros.20299. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev. 2002;54:129–158. doi: 10.1124/pr.54.1.129. [DOI] [PubMed] [Google Scholar]
- Schwartz B, Melnikova VO, Tellez C, Mourad-Zeidan A, Blehm K, Zhao YJ, McCarty M, Adam L, Bar-Eli M. Loss of AP-2alpha results in deregulation of E-cadherin and MMP-9 and an increase in tumorigenicity of colon cancer cells in vivo. Oncogene. 2007;26:4049–4058. doi: 10.1038/sj.onc.1210193. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Molecular cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of Cell- and Gene-Specific Transcriptional Regulation by the Glucocorticoid Receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son CG, Bilke S, Davis S, Greer BT, Wei JS, Whiteford CC, Chen QR, Cenacchi N, Khan J. Database of mRNA gene expression profiles of multiple human organs. Genome research. 2005;15:443–450. doi: 10.1101/gr.3124505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear BT, Jin L, Ramasamy S, Dobierzewska A. Transcriptional control in the mammalian liver: liver development, perinatal repression, and zonal gene regulation. Cell Mol Life Sci. 2006;63:2922–2938. doi: 10.1007/s00018-006-6258-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. The Journal of biological chemistry. 2006;281:16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Molecular and cellular biology. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen recepor dependent prostate cancer growth. Molecular cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic acids research. 2005;33:W389–392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data
Supplement Data include twenty one figures, two tables and Supplemental References and can be found with this article online at www.cell.com