Summary
Little is known of the control of gene expression in the animal hemisphere of the Xenopus embryo. Here we show that expression of FoxI1e, a gene essential for normal ectoderm formation, is expressed regionally within the animal hemisphere, in a highly dynamic fashion. In situ hybridization shows that FoxI1e is expressed in a wave-like fashion that is initiated on the dorsal side of the animal hemisphere, extends across to the ventral side by the mid-gastrula stage, and is then turned off in the dorsal ectoderm, the neural plate, at the neurula stage. It is confined to the inner layers of cells in the animal cap, and is expressed in a mosaic fashion throughout. We show that this dynamic pattern of expression is controlled by both short and long range signals. Notch signaling controls both the mosaic, and dorsal/ventral changes in expression, and is controlled, in turn, by Vg1 signaling from the vegetal mass. FoxI1e expression is also regulated by nodal signaling downstream of VegT. Canonical Wnt signaling contributes only to late changes in the FoxI1e expression pattern.
These results provide new insights into the roles of vegetally localized mRNAs in controlling zygotic genes expressed in the animal hemisphere by long range signaling. They also provide novel inights into the role of Notch signaling at the earliest stages of vertebrate development.
Introduction
The Xenopus blastula is conventionally divided into three regions, with respect to both cell fate and gene expression. Cells in the vegetal region give rise to the embryonic endoderm. They inherit the maternally encoded transcription factor VegT, which activates the synthesis of nodals. These, in turn, induce mesoderm in the adjacent marginal region. Cells in the animal region form the ectoderm. It is not known how this fate is initiated.In addition to forming the germ layers, the three blastula regions each become patterned to form the axes of the body, by the expression of dorsal and ventral genes.
This picture of gene expression in the blastula tends to be regarded as dynamic with respect to time, but static with respect to each region of the blastula; a linear progression within each region of transcriptional initiation, leading to specification to enter a particular lineage, or to exhibit a particular type of cell behavior.
We show here that in fact, gene expression is highly dynamic within the animal region. The forkhead transcription factor FoxI1e is expressed in the animal hemisphere, starting at the blastula stage. It is required for the expression of genes in both the early neural and ecodermal lineages, and for the later differentiation of the epidermis (Mir et al., 2007). It is also required to repress mesoderm specification in the animal hemisphere (Suri et al., 2005). Here we show that its expression pattern changes rapidly in both time and space within the animal hemisphere. Expression is initiated dorsally at the blastula stage, then spreads to encompass the ventral region of the animal hemisphere by the mid-gastrula stage. At the early neurula stage, expression is lost from the dorsal ectoderm cells that form the neural plate. During this sequence, its expression domain also changes with respect to the layers of the animal cap. First, it is restricted to the inner cells of the animal cap at the blastula stage, and then to a layer of cells between the inner and outer layers of the animal cap at the gastrula stage. Furthermore, throughout this temporal and spatial progression, FoxI1e expression is always in a mosaic pattern, with positive cells interspersed with non-expressing cells.
These observations revealed a previously unsuspected regionalization of the animal hemisphere. The total expression domain of FoxI1e extends across the animal cap, excluding the outer cells at the blastula stage, and both inner and outer cells at the gastrula stage, and all the time is restricted to only some cells, but not others, in this expression domain. During the late blastula to mid-gastrula stages, the expression of FoxI1e moves across this expression domain in a dorsal to ventral direction. This pattern is completely different from the expression patterns of other animally localized transcripts previously reported, such as ectodermin (Dupont et al.,2005), and Xlim5 (Toyama et al., 1995, Houston et al., 2003), which are not mosaic, nor dynamic with respect to the dorsal-ventral axis. We therefore set out to identify some of the factors that might control these spatial and temporal changes.
We found that both long and short range signals control this pattern of expression. First, mosaic expression is controlled by Notch signaling. Both gain- and loss-of-function experiments showed that Notch signaling represses FoxI1e expression, and that loss of Notch signaling causes expression of FoxI1e in all cells of its total expression domain, and eliminates both the temporal and spatial progression of expression within this domain. This raised the issue of how Notch signaling is controlled in the animal hemisphere. It could be intrinsic, or controlled at long range from the other developing germ layer regions. We show that control of FoxI1e expression by Notch is, in turn, controlled by the TGFβ signal, Vg1, whose mRNA is maternally encoded, and inherited only by the vegetal cells of the embryo (Melton, 1987). We also find that the level of FoxI1e, but not its mosaic distribution, is controlled by nodal signaling downstream of VegT, a vegetally localized, maternally encoded transcription factor (Zhang and King, 1996).
These data reveal multiple levels of control of early zygotic genes in Xenopus. First, long range signals from the vegetal cells of the embryo, as well as short range signals through the Notch pathway, combine to control both amount and position of expression of animal-specific genes during the blastula and gastrula stages. Second, none of these signals controls the “total expression domain” of FoxI1e, which remains confined to the group of cells that normally express it during its changing temporal expression at the blastula and gastrula stages. Instead, they control the temporal and spatial sequence of expression within this domain.
MATERIALS AND METHODS
Oocytes and embryos
Oocytes were generated for host transfer experiments by manual defolliculation of surgically-removed ovary as previously described (Heasman et al., 1991). Culture and injections were carried out in L15-based Oocyte Culture Medium (OCM). The length of the culture period between injection and host transfer varied by experiment. All mRNA injections were cultured overnight. VegT-depleted embryos were generated using a morpholino oligo [5′-CCCGACAGCAGTTTCTCATTCCACG-3′], and cultured for 3 days after injection. Vg1 depletions were carried out using the oligo Vg1c as previously described (Birsoy et al., 2006), with 4 days of culture after injection. Xotch was depleted using AS14MP [5′-GGAAGGGCTCAGCGCTAC-3′], with 3 days of culture. For rescue experiments, mRNA was injected on the last day of culture, before progesterone treatment, or at the 2-cell stage after fertilization. Eggs were collected in high-salt solution, fertilized in vitro with isolated testis, and cultured in 0.1X MMR. Staging was according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1967). Dissections were performed at stage 9 or stage 10 on agarose-coated dishes in 1x MMR and then cultured in OCM. β-Catenin-depleted embryos were generated using a morpholino as previously described (Heasman et al., 2000). Synthetic mRNAs encoding β-Catenin, BMP4, cmBMP7, β-Galactosidase, NICD, Su(H)-DBM, and Vg1 were generated using Ambion mMessage mMachine kits.
Real-time RT-PCR
Total RNA was extracted as previously described (Zhang et al., 1998). Unless otherwise indicated, input was 2 whole embryos, 3 marginal zones, or 5 vegetal masses per sample. cDNA was synthesized using oligo dT primers, and semi-quantitative real-time RT-PCR was carried out using the LightCycler system as described (Kofron et al., 2001). Ornithine decarboxylase (ODC) was used as a loading control, and all values were normalized to ODC levels. In all cases, water-only and reverse transcriptase-negative controls failed to produce specific products. Each experiment was repeated a minimum of three times in independent experiments to verify reproducibility of results. Primer sequences were: Siamois: U: 5′-CTGTCCTACAAGAGACTCTG-3′, D: 5′-TGTTGACTGCAGACTGTTGA-3′. Xnr3: U: 5′-CTTCTGCACTAGATTCTG-3′, D: 5′-CAGCTTCTGGCCAAGACT-3′. FoxI1e: U-5′-GCACCTGCTGTGGTTCATAA-3′, D-5′-CACCACTGTAGTGCGTCAGAA-3′. Xotch: U: 5′-AGTAACCCGTGCAAAAATGG-3′, D: 5′-AGCTTCCGGTAAATCCAGGT-3′. ESR-1: U: 5′-TGGCAAAACTGGAACAGGAT-3′, D: 5′-TGGGATACAACAGGGAGCTT-3′.
Whole-mount in situ hybridization, membrane staining, and Red-Gal staining
In situ hybridizations were carried out as described by Harland (Harland, 1991), with a probe concentration for FoxI1e of 5 μg/ml, and using BMB Purple (Roche) as the alkaline phosphatase substrate. For membrane staining, embryos were stained for FoxI1e, sectioned, and stained with Alexa-488-conjugated Wheat Germ Agglutinin (Molecular Probes) at 0.01 μg/μl in PBS+0.1% Tween-20 for 30 minutes. For lineage labeling, embryos injected with 50 pg of nuclear β-Galactosidase mRNA in the A1 blastomere were fixed for 1 hour at room temperature in MEMFA, stained with Red-Gal (Research Organics) for 20 minutes at 37°C, and fixed for another hour at room temperature before in situ hybridization for FoxI1e.
RESULTS
FoxI1e is expressed in a dorsal to ventral wave, in a subset of cells in the animal cap
In previous work, we found that FoxI1e expression was mosaic in the early embryo (Mir et al., 2007). This could be due either to an asynchronous onset of expression, leading to a more stable homogeneous expression pattern, or it could be that gene expression in the animal hemisphere is not uniform, but controlled in more complex ways than previously thought. To distinguish between these possibilities we carried out in situ hybridization on embryos from the late blastula stage (stage 9) to the mid-gastrula stage (stage 11). We found that expression in all embryos began in a localized manner above the equator on one side of the embryo and then spread across the animal cap (Fig. 1A). The site of onset of expression was identified as the dorsal side by injection of β-galactosidase mRNA into the 2 dorsal animal cells at the 8-cell stage. The β-Gal signal consistently co-localized with FoxI1e expression (Fig. 1B, 92%, n=71), showing that FoxI1e expression begins on the dorsal side of the embryo, and spreads ventrally. To confirm this finding, we dissected early gastrulae into dorsal and ventral halves, and used real-time RT-PCR to compare the amount of FoxI1e expression. We found that FoxI1e mRNA was indeed expressed at higher levels in dorsal halves at the late blastula stage (Fig. 1C).
Fig. 1. FoxI1e is expressed in a dorsal to ventral wave in the blastula and gastrula stages.
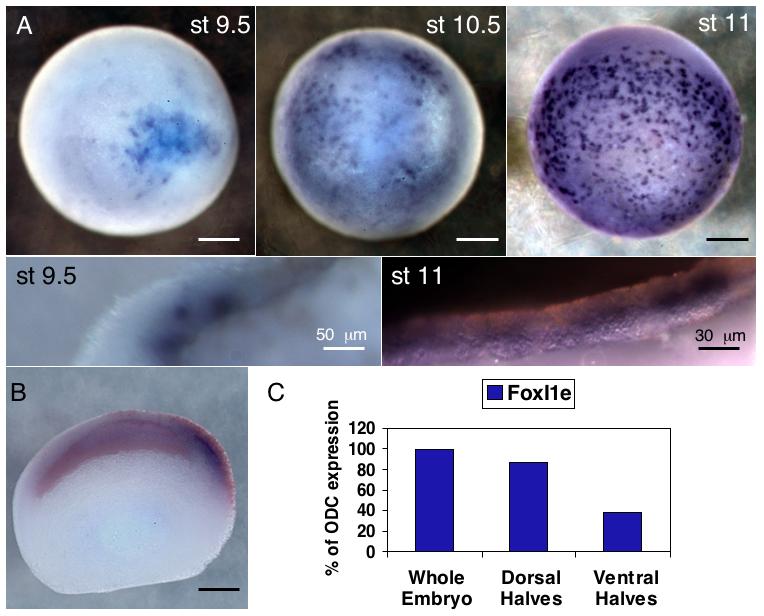
(A) In situ hybridization for FoxI1e shows initial staining at stage 9.5 on one side of the embryo, and then spreading across the embryo. Its expression is always mosaic. (B) Embryos injected in the two dorsal, animal blastomeres at the 4-cell stage with 50 pg of β-Gal mRNA were stained with Red-Gal before in situ hybridization, showing the initial expression is on the dorsal side of the embryo. (C) Embryos were dissected into dorsal and ventral halves at stage 10 and frozen for real-time PCR. FoxI1e expression is enriched on the dorsal side at stage 10. Results are normalized to ODC expression levels. (D) Stage 11 embryos were stained for FoxI1e and sectioned. Staining with Wheat Germ Agglutinin defines a small population of FoxI1e positive cells between the sensorial and epithelial layers of the ectoderm. Scale bars represent 200 μm, unless otherwise noted.
The animal cap consists of two cell layers—an epithelial, or outer layer, and an inner, sensorial layer (Chalmers et al., 2003). From late blastula to early gastrula stages, FoxI1e was expressed only in the sensorial layer. By the mid-gastrula stage, however, when expression had spread to the ventral side of the embryo, there was no signal in either the inner or outer cell layers of the cap. Instead, there was expression in between the two layers.
These observations raise the question of how such an unexpected pattern of gene expression in the animal hemisphere is controlled.
Late, but not early expression of FoxI1e is controlled by Wnt-dependent dorsal axis specification
Since FoxI1e expression begins dorsally, we wanted to know if its initial expression is controlled by the Wnt signal transduction pathway, which is known to activate dorsal axis formation in Xenopus. To test this, we either depleted β-Catenin (β-Cat) on the dorsal side of the embryo (by injecting the two dorsal cells at the 4-cell stage with 20 ng morpholino oligo (Heasman et al., 2000), or overexpressed β-Cat on the ventral side (by injection of 100 pg β-Cat mRNA into the two ventral cells at the 4-cell stage). Embryos were dissected at the early gastrula stage into dorsal and ventral halves, and FoxI1e mRNA levels measured by real-time RT-PCR. In embryos injected with β-Cat MO, there was a reduction in the expression of the direct targets Xnr3 and Siamois in dorsal halves, and in mRNA injected embryos, Xnr3 and Siamois were upregulated in ventral halves, indicating that the manipulations affected the embryos as expected (Fig. 2A, B). However, we found that regardless of the experimental treatment, FoxI1e was always expressed at higher levels in dorsal halves than in ventral halves. Also, the overall level of FoxI1e expression in whole embryos was not consistently affected by β-Cat MO or mRNA (Fig. 2C-F). These results show that the spatial regulation of FoxI1e expression at the blastula and gastrula stages is independent of canonical Wnt signaling.
Fig. 2. Wnt-dependent dorsal axis formation controls late, but not early, FoxI1e expression.
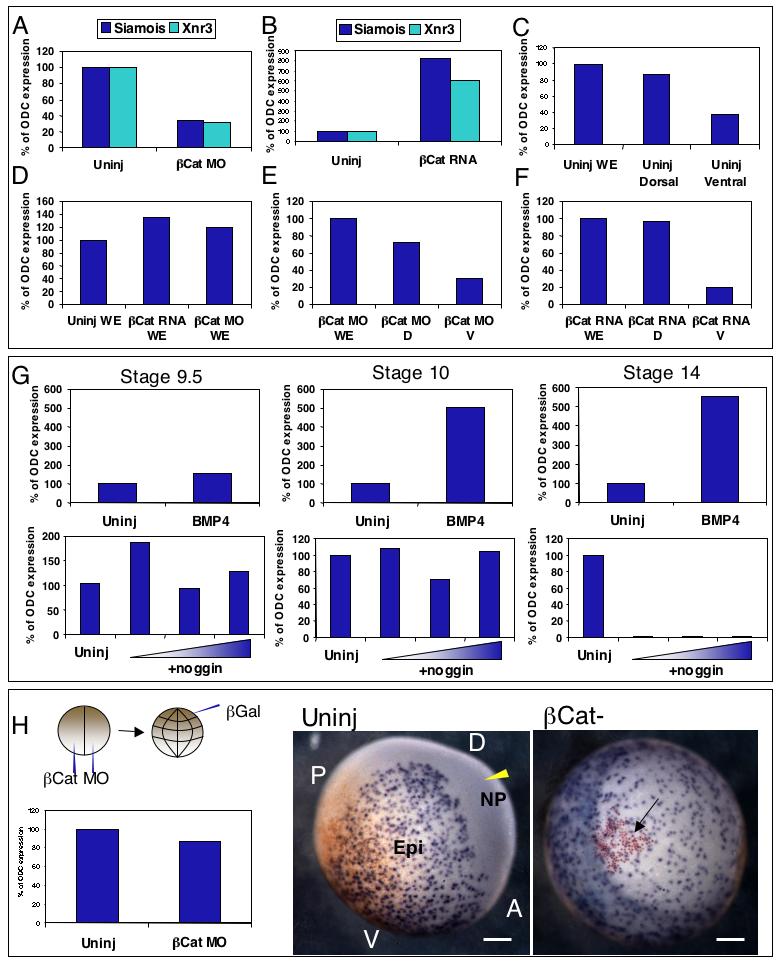
Embryos injected with 40ng β-Cat MO had reduced levels of direct targets Siamois and Xnr3 at stage 10 (A), and embryos injected with 50pg β-Cat mRNA had increased levels (B). In control explants, the level of FoxI1e was higher in dorsal halves than ventral halves (C). The total level and distribution of FoxI1e was unchanged by β-Cat MO or mRNA (D-F). (G) shows levels of FoxI1e mRNA at stages 9.5, 10, and 14 (compared to ODC mRNA levels at each stage) in embryos injected with either 100pg BMP4 mRNA (upper panels), or 10, 40, or 160pg of noggin mRNA (lower panels). Neither BMP4 overexpression, nor inhibition using Noggin, consistently affected the level of FoxI1e expression at the late blastula stage (st. 9.5). By the early gastrula stage (st. 10), BMP4 overexpression increased FoxI1e expression, but Noggin still had little effect. Expression of FoxI1e in the early neurula (st. 14) was increased by BMP4 overexpression and completely ablated by Noggin. This indicates that the early expression of FoxI1e is BMP-independent, but that the restriction of FoxI1e to the epidermis at neurulation is BMP-dependent. (H) Overall level of FoxI1e expression at stage 14 is unaffected by β-Cat MO. Embryos injected with 40ng β-Cat MO at the 2-cell stage were injected with β-Gal at the 32-cell stage in the A1 blastomere. Red-gal staining, and FoxI1e in situ hybridization were carried out at stage 14. The anterior (A), posterior (P), dorsal (D), and ventral (V) regions of the embryo are marked in the control embryo, with the derivatives of the dorsal animal blastomere (A1), marked by the yellow arrowhead in the neural plate. Embryos lacking β-Cat (right panel) lack axes altogether. The red-gal positive cells (arrowed) mark the derivatives of the A1 blastomere. FoxI1e expression in β-Cat-depleted embryos persists in that clone of cells, indicating the β-Cat dependence of restriction of FoxI1e from the prospective CNS. The reddish cast toward the posterior end of the uninjected embryo is residual maternal pigment, not affected by bleaching. Scale bars represent 200 μm.
One of the downstream effects of Wnt signaling on the dorsal side of the embryo is inhibition of BMP signaling (Baker et al., 1999). We therefore assayed the effects of altered BMP signaling on FoxI1e expression. To test the effect of increased BMP signaling on FoxI1e, 500 pg of mRNA encoding BMP4 were injected into manually defolliculated oocytes, which were fertilized using the host transfer method after overnight culture. The effect of blocking BMP signaling was tested by injection of mRNA encoding the endogenous BMP inhibitor Noggin, at doses between 10 and 500pg (Zimmerman et al., 1996) into embryos at the two cell stage. In both groups, the levels of FoxI1e mRNA were measured by real-time RT-PCR.
Fig. 2G shows that, at the late blastula stage (st. 9.5), neither increased nor decreased BMP signaling consistently affected the level of FoxI1e expression, suggesting that the initiation, and the initial level of FoxI1e expression on the dorsal side is not controlled by BMP signaling. At the early gastrula stage, loss of BMP signaling did not change FoxI1e expression levels, but increased BMP signaling dramatically upregulated FoxI1e expression. At the neurula stage (St. 14), loss of BMP signaling caused complete loss of FoxI1e expression and increased BMP signaling enhanced its expression (Fig. 2G). These data show that initiation of expression on the dorsal side, and the control of its initial level of expression, is not controlled by BMP inhibition. However, the subsequent increase in expression on the ventral side, and loss on the dorsal side, in the neural plate, is controlled by BMP signaling.
Next, we analyzed the spatial expression of FoxI1e in β-catenin-depleted embryos. Embryos were injected at the 2-cell stage with 40 ng β-catenin MO, and dorsal cells marked by the injection of 50 pg of mRNA encoding nuclear β-galactosidase (β-gal) in a dorsal animal cell (cell A1) at the 32-cell stage. At Stage 14, real-time RT-PCR showed similar levels of expression between β-catenin-depleted and control whole embryos. Localization of FoxI1e expression was assayed by in situ hybridization, combined with Red-Gal staining for the β-gal marker of dorsal cells. In uninjected embryos, FoxI1e expression was visible in the epidermis, but absent from the presumptive neural plate. In β-Cat MO-injected embryos however, cells marked by Red-Gal staining that would normally have become part of the CNS and switched off FoxI1e, still had positive FoxI1e staining (Fig. 2H).
Taken together, these data show that neurula stage FoxI1e expression is dependent on dorsal-ventral embryonic patterning initiated by canonical Wnt signaling, and propagated through the ectoderm by modulation of BMP signaling, but that the initial dorsal expression of FoxI1e is independent of this pathway.
VegT and downstream Nodals, as well as the maternal TGF-β ligand Vg1, control the level of FoxI1e expression in the animal hemisphere
FoxI1e was initially identified in this project by its upregulation in embryos depleted of VegT, in which a 12-fold upregulation of expression was seen in the vegetal mass (Mir et al., 2007). Although consistent, this upregulation comprises only a small fraction of the increase in overall expression in the whole embryo, suggesting that VegT may play a greater role in controlling expression in the animal hemisphere. To test this, we carried out in situ hybridization for FoxI1e expression in embryos depleted of VegT using a morpholino oligo injected into the oocyte. The increase in FoxI1e expression in VegT-depleted embryos was found to be much more dramatic in the animal than in the vegetal hemisphere (Fig. 3A), supporting the hypothesis that a major role of VegT in the embryo is to repress the level of FoxI1e in its normal expression domain, rather than inhibit expression elsewhere in the embryo.
Fig. 3. VegT and Nodal signaling act at long-range to affect FoxI1e expression in the animal cap.
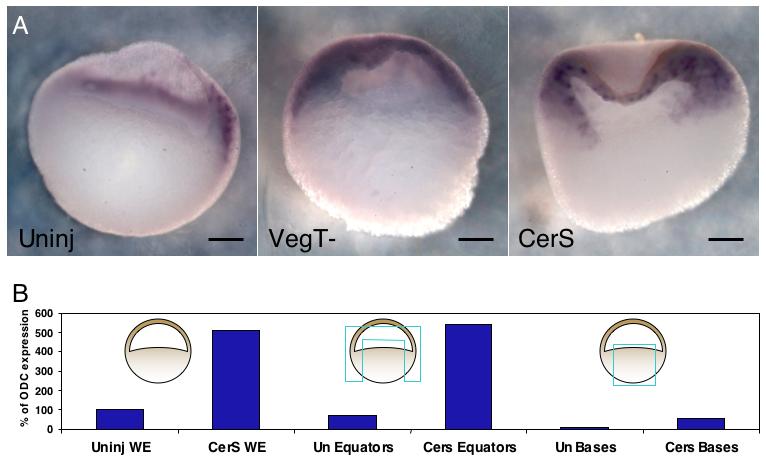
(A) In situ hybridization in VegT-depleted and CerS-injected embryos shows FoxI1e is most upregulated in the animal cap rather than the vegetal mass at stage 10. Dorsal is the to the right in the bisected uninjected embryo. The VegT and CerS embryos did not dorsal axes. (B) Vegetal masses stripped of all mesoderm contamination dissected from control and CerS-injected embryos confirm that the vast majority of increase in FoxI1e expression is derived from non-endodermal tissue. Scale bars represent 200 μm.
This role for VegT must be non cell-autonomous, since its transcripts are vegetally localized. It has been shown previously that Nodal signaling is downstream of VegT, and activates mesoderm gene expression in the adjacent marginal zone. In addition, Nodal signaling has previously been shown to repress FoxI1e expression, as indicated by an upregulation in embryos injected with the Nodal inhibiting construct CerS (Agius et al., 2000). We therefore analyzed this effect more carefully by dissecting vegetal masses which had all potential mesodermal contamination removed, from embryos injected with 1 ng of CerS mRNA at the 2-cell stage. Real-time RT-PCR analysis at stage 10 confirmed that although there was an upregulation of FoxI1e in the vegetal mass relative to controls, a much greater increase was found outside the vegetal mass (Fig. 3B). In situ hybridizations for FoxI1e mirrored the pattern of upregulation caused by loss of VegT, with a very large increase of expression in the animal cap, but little in the vegetal mass. Interestingly, the increased expression in the animal cap did not include the outer layer of cells (Fig. 3A). These results indicate that the VegT-Nodal pathway regulates the level of FoxI1e expression within its normal expression domain, rather than determining the expression domain itself.
Since VegT and Nodal signaling do not seem to be the main factors involved in repression of FoxI1e in the vegetal mass, we next analyzed the role of the maternally-encoded, vegetally-localized TGF-β ligand, Vg1. Vg1 has recently been shown to function as an essential endogenous mesoderm inducing signal in Xenopus (Birsoy et al., 2006), and so we wanted to know if it also controls FoxI1e expression. Vg1-depleted embryos were generated by injecting 4 ng of Vg1-specific thioate-modified DNA oligo (Vg1c) into oocytes as previously described, and then fertilizing them by the host transfer procedure. Vegetal masses were dissected from blastula-stage embryos, and frozen at stage 10. Real-time RT-PCR analysis showed a dramatic upregulation of FoxI1e expression in Vg1-depleted whole embryos compared to controls, again, with a disproportionate upregulation in the animal hemisphere (Fig. 4A). In situ hybridization reveals a massive upregulation of FoxI1e within the inner layer of the animal cap, with all cells in the inner layer expressing FoxI1e in the most strongly affected embryos (Fig. 4B, 20%, n=55).
Fig. 4. Vg1 is a long-range inhibitor of FoxI1e expression.
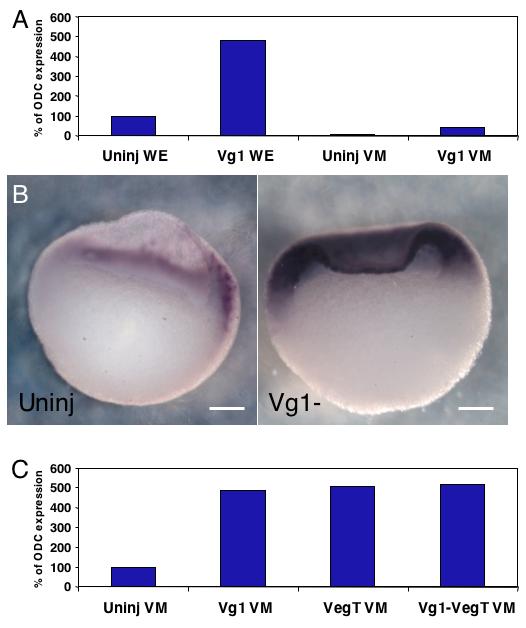
(A,B) Depletion of Vg1 results in a 5-fold increase in FoxI1e expression at stage 10, resulting largely from an increase in expression in non-endodermal tissues. (C) Co-depletion of VegT and Vg1 does not increase the expression of FoxI1e in the vegetal mass over either one alone, indicating the presence of an unidentified inhibitor in the vegetal mass, or the absence of an activator. Dorsal is to the right in the bisected embryos shown. Scale bars represent 200 μm.
To address the possibility that Vg1 and VegT cooperate to repress FoxI1e in the vegetal mass, embryos lacking both Vg1 and VegT were generated. Depletion of each mRNA alone caused a modest increase in FoxI1e expression in cultured vegetal masses. Depletion of both mRNAs did not show an increased effect (Fig. 4C).
Taken together, these data indicate that VegT and TGF-β signaling from the vegetal mass act in a long-range, non cell-autonomous manner to control FoxI1e expression in the animal cap. They also suggest the presence in the early embryo of either animally localized activators of FoxI1e, or vegetally localized repressors.
Notch represses FoxI1e expression
To determine if the Notch signaling pathway is responsible for the mosaic expression of FoxI1e, we designed antisense DNA oligonucleotides against the Xenopus Notch homolog (Xotch) coding sequence. These were tested for efficiency by injection into full-grown oocytes. After 48 hours in culture, the level of depletion was analyzed by real-time RT-PCR. Of the oligos tested, oligo 14 was most effective at reducing endogenous levels of Xotch mRNA. To stabilize the oligo, we used a phosphorothioate-modified version, which was able to reduce Xotch mRNA to 15-25% of wildtype. Xotch-depleted embryos were generated using the host transfer method, and Fig. 5A shows that the mRNA remained depleted through the gastrula stage.
Fig. 5. Notch signaling is responsible for the initial dorsal restriction and mosaic expression of FoxI1e.
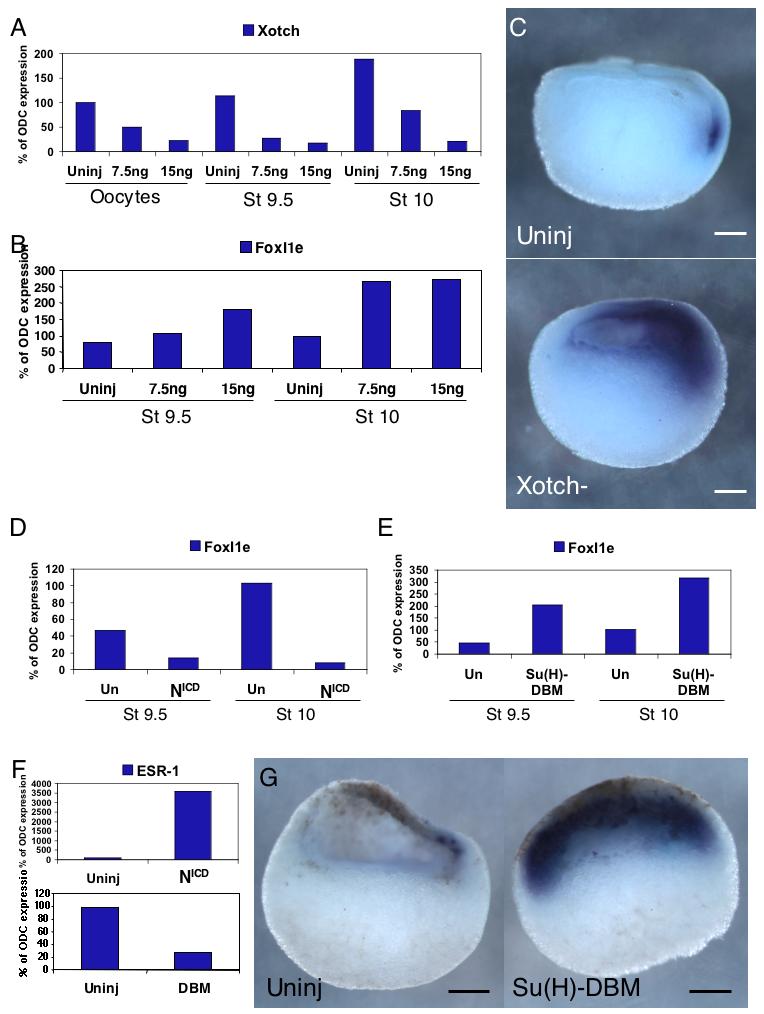
(A) Maternal Xotch mRNA is depleted to 20-25% with 15 ng of thioate-modified DNA oligo. The level of Xotch remains low in the blastula and gastrula. (B) FoxI1e expression is 2-3-fold upregulated by depletion of Xotch. In situ hybridization shows an expansion of FoxI1e expression (C). (D) Injection of the constitutively active Notch Intracellular Domain (NICD) causes downregulation of FoxI1e relative to controls, and injection of the dominant negative construct Su(H)-DBM causes an upregulation of FoxI1e (E). (F) NICD upregulates the Notch target ESR-1, and Su(H)-DBM downregulates it. (G) In situ hybridization for FoxI1e comparing control and Su(H)-DBM injected embryos at stage 9.5 indicates that more, and in the most severe cases, all of the sensorial-layer animal cap cells express FoxI1e. Dorsal is to the right in all embryos shown. Scale bars represent 200 μm.
At the late blastula stage in Xotch-depleted embryos, there was a reproducible increase in FoxI1e mRNA expression (Fig. 5B). We further analyzed these embryos by in situ hybridization, and found that whereas in the controls, FoxI1e expression was restricted to a small number of cells on the dorsal side of the embryo, in Xotch-depleted embryos, expression of FoxI1e extended further across the animal cap towards the ventral side (Fig. 5C, 90%, n=46).
To test whether an increase in Notch signaling would have the reciprocal effect, we injected oocytes with 500 pg of synthetic mRNA encoding the intracellular domain of Notch (NICD), which is constitutively translocated to the nucleus and activates Notch signaling (Coffman et al., 1993; Deblandre et al., 1999). At the late blastula and early gastrula stages, FoxI1e mRNA expression was dramatically decreased in the NICD - injected embryos compared to uninjected controls (Fig. 5D). The Notch target; enhancer of split-related-1 (ESR-1); was dramatically upregulated in these embryos, indicating the expected activity of the construct (Fig. 5F). These results show that Notch signaling is responsible for the restriction of FoxI1e expression to its mosaic pattern in the animal hemisphere, and for its initial dorsal expression.
We next wanted to determine if the core Notch signaling pathway was involved in regulating FoxI1e expression. In Xenopus, the core pathway is mediated by the transcription factor Suppressor of Hairless [Su(H)]. We injected oocytes with 500 pg of mRNA encoding a mutated version of Su(H) that is missing the DNA binding domain [Su(H)-DBM], and acts in a dominant negative manner (Deblandre et al., 1999). Su(H)-DBM-injected oocytes were fertilized by the host transfer method, as above, and analyzed by RT-PCR and in situ hybridization. Real-time RT-PCR analysis revealed a reproducible upregulation of FoxI1e (Fig. 5E). ESR-1 expression was downregulated in Su(H)-DBM-injected embryos, confirming the inhibition of Notch signaling (Fig. 5F). In situ hybridization showed expression of FoxI1e in all cells of the inner layer of the ectoderm (Fig. 5G, 85%, n=52), rather than the dorsal, mosaic pattern seen in control embryos. The results are similar to the Xotch knockdown, confirming the requirement for Notch signaling to restrict FoxI1e to a salt-and-pepper expression pattern.
Maternal Vg1 activates Notch signaling
Depletions of Vg1 and Notch have similar effects of FoxI1e expression. This could be because they act in parallel, or because they act in series in a single pathway to control FoxI1e expression. It has been suggested previously that Activin can activate Notch signaling in the blastula (Abe et al., 2004). To test the possibility that Vg1 controls Notch signaling in the animal hemisphere, cultured oocytes were depleted of Vg1 mRNA by injection of 4ng Vg1C oligo, and fertilized. They were then injected at the 2-cell stage with 50 to 500pg NICD mRNA. Embryos were harvested during the late blastula and early gastrula stages for real-time RT-PCR analysis, and during the early gastrula stage for in situ hybridization. Introduction of the activated Notch construct rescued FoxI1e expression levels in the Vg1-depleted embryos (Fig. 6A,B). This suggests that maternal Vg1 activates Notch signaling in the blastula. To confirm this, we showed that expression of the Notch target gene ESR-1 is reduced by depletion of Vg1 in the early embryo (Fig. 6C), and that Vg1 overexpression could not rescue the FoxI1e overexpression caused by reduction of Notch signaling caused by injection of 500pg Su(H)-DBM mRNA (Fig. 6D).
Fig. 6. Maternal Vg1 activates Notch signaling in the blastula to control FoxI1e expression.
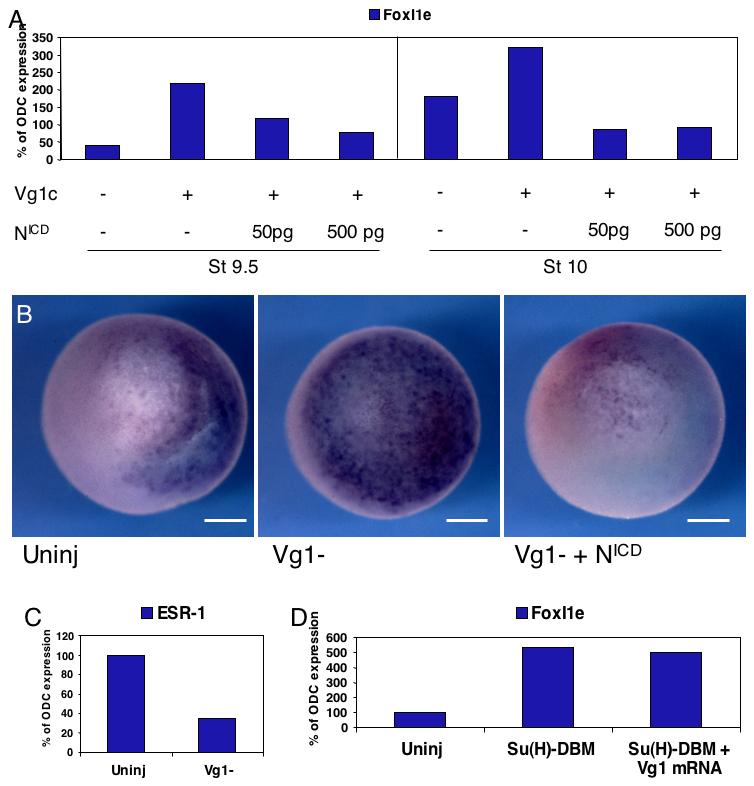
(A) Vg1-depleted embryos were injected with 50 or 500 pg of NICD mRNA at the 2-cell stage. NICD rescued the increase in FoxI1e expression caused by Vg1 depletion. (B) These results were confirmed by in situ hybridization for FoxI1e at Stage 10, which shows an upregulation of FoxI1e in Vg1-depleted embryos, and a reversal of this upregulation by subsequent injection with NICD. The control embryo is oriented with dorsal to the right. Depletion of Vg1 results in a delay of gastrulation, and so the orientations of both the Vg1-depleted and the NICD-rescued embryos is indeterminate. Scale bars represent 200 μm. (C) Real-time PCR at stage 10 shows that the Notch target ESR-1 is downregulated in Vg1-depleted embryos relative to controls, indicating that Notch signaling depends on Vg1 at this stage. (D) 200 pg of Vg1 mRNA was unable to rescue the increase in FoxI1e expression induced by loss of Notch signaling by injection of 500pg Su(H)-DBM mRNA.
Discussion
In this work we have shown that expression of FoxI1e, a gene expressed in the animal hemisphere, which controls ectoderm formation, is subject to multiple levels of control. First is the surprising observation that it is expressed in a mosaic fashion throughout its whole period of expression in the embryo. This is particularly interesting because no other genes have been shown to be expressed in such a pattern at this early stage in development. In a previous paper, we showed that expression of genes in both neural and epidermal branches of ectodermal differentiation are downregulated in FoxI1e-depleted embryos. These target genes are not expressed in mosaic fashions, indicating that FoxI1e-expressing cells in the blastula and gastrula are probably controlling expression of downstream targets in a non-cell-autonomous manner.
The mosaic expression of FoxI1e could be accounted for in 3 possible ways. First, it could be cell cycle dependent, so that at any given time, only a subset of cells at a particular point in the cell cycle express it. The second possibility is that FoxI1e-expressing cells could originate from a few cells and then disperse by migration across the animal cap. Finally, it could be activated or repressed in a mosaic pattern by intercellular signaling. We show in this paper that blockade of Notch signaling abrogates the mosaic expression of FoxI1e, suggesting that the third possibility is correct. This does not preclude the first two. However, it is unlikely that FoxI1e mRNA is turned over during part of each cell cycle, and lineage analysis excludes that possibility that there is large-scale migration of inner animal cells from the dorsal to the ventral side of the embryo. This work also confirms that Notch signaling is active in the blastula, a fact that was previously underappreciated.
Second, we show that the normal expression domain of FoxI1e extends across the whole animal cap, but excludes the most superficial layer of cells throughout its expression period. Cells in this expression domain turn on FoxI1e in a temporal sequence, from the dorsal to the ventral side, so that at the gastrula stage, cells all across the expression domain are expressing FoxI1e. Such a progression could be controlled by local factors in the animal cap, or at longer range by factors that control dorsal/ventral patterning in the rest of the embryo. We show that Notch, and thus short-range signaling, is involved. However, we find that longer range signaling, originating in vegetal cells, also controls the expression pattern of FoxI1e. We show that the shifting expression of FoxI1e within its expression domain results from different signaling pathways acting at different times. For example, FoxI1e expression in the blastula and gastrula is unaffected by manipulation of the Wnt signaling that establishes the dorsal axis. However, the later downregulation of FoxI1e in the neural plate requires the inhibition of BMP signaling, which is downstream of Wnt signaling. In the late blastula and early gastrula ectoderm, vegetal pathways initiated by VegT and Vg1 influence the spatial pattern of FoxI1e expression. In the absence of either Vg1, VegT, or nodal signaling, early expression of FoxI1e is initiated all across the animal cap, instead of gradually spreading from dorsal to ventral.
Finally, we have shown that Vg1 activates the Notch signaling pathway to restrict FoxI1e expression to a mosaic pattern. The mechanism by which Vg1 activates Notch signaling remains unclear, but the most likely possibilities are through interaction of phospho-smad2 with Notch or by upregulating transcription of zygotic components of Notch signaling. Our attempts to identify these components have been unsuccessful, thus far. Though it has been shown that activin can induce Delta-1 and Delta-2 in animal caps (Abe et al., 2004), our analysis of their expression in Vg1-depleted embryos has not confirmed this relationship. Previously, Vg1 has been shown to be a maternal inducer of mesoderm (Birsoy et al., 2006). The effect of Vg1 on Notch could be downstream of mesoderm induction, or through release of signaling molecules into the blastocoel fluid, which would then act on the inner surface of the animal cap. The mechanisms by which vegetal pathways influence animal patterning require further study.
Previous studies have shown that FoxI1e is upregulated in VegT-depleted and in CerS-injected embryos. However, it has not, until now, been fully appreciated that the increase in expression of FoxI1e and other ectodermal genes in the vegetal mass is minor compared to the increase in the animal half of the embryo. This strengthens the hypothesis that there is an animally localized maternal activator of ectoderm formation. Additionally, there must be either an activator(s) specific to the deep layer of the ectoderm, or a repressor(s) of FoxI1e in the superficial layer. Indeed, a number of differentially expressed transcription factors have been identified, and the candidate may be among them (Chalmers et al., 2006).
This work represents the first description of regulation of animally expressed zygotic genes. Previous work has focused primarily on the exclusion of mesodermal gene expression from the ectoderm. FoxI1e has been shown to inhibit FGF-mediated mesoderm induction (Suri et al., 2005), and the Smad4 ubiquitin ligase ectodermin attenuates mesoderm induction in the animal cap by inihibiting all TGF-β signaling, both activin-type and BMP-type (Dupont et al., 2005). Additionally, the MADS box transcription factor SRF disrupts the interaction of Smad2 and FoxH1, thereby preventing activin-type TGF-β signaling (Yun et al., 2007). In the absence of ectodermin, SRF, or FoxI1e, mesodermal gene expression expands animally. It is clear from these studies that the inhibition of mesoderm induction in the animal cap keeps the stage clear for ectoderm specification, and offers a mechanism to control its boundaries. However, they also provide evidence that signals originating from vegetal hemisphere can reach the ectoderm. Although we have not shown that the Vg1 pathway or nodal signaling directly affect FoxI1e expression, these data do allow for this possibility.
FoxI1e is not the first zygotic gene identified that is expressed in the entire early ectoderm. The trancription factors Xlim5 (Toyama et al., 1995, Houston et al., 2003) and AP-2 (Luo et al., 2002) are restricted to the CNS and epidermis, respectively, late during the gastrula stage, but are both broadly expressed throughout the ectoderm before this restriction. However, this early expression is generally ignored. In an unpublished study, we have shown that both of these genes are upregulated in VegT-depleted embryos, indicating overlapping regulation. It will be important to analyze the expression of these genes when testing animally localized maternal factors for their role in ectoderm specification.
We have begun to assemble a pathway from maternal control, to intercellular signaling, to ectoderm patterning, but the question of why FoxI1e is expressed in a mosaic pattern remains. Although it is not expressed in every ectodermal cell, and later becomes confined to the epidermis, FoxI1e is important for formation of the ectoderm germ layer, before it divides into epidermis and CNS. There must be signaling events downstream of FoxI1e that allow it to activate gene expression in a non-cell autonomous fashion. It has been show that Notch signaling prolongs mesodermal competence in the animal cap (Abe et al., 2005; Abe et al., 2004; Coffman et al., 1993). It has also been shown that ectoderm determination is a gradual process that begins during the late blastula stage and continues through gastrulation (Heasman et al., 1984; Snape et al., 1987). It is possible that Notch signaling represses FoxI1e expression in the animal cap in the mid-late blastula, but as animal Notch signaling weakens, FoxI1e+ cells begin to appear in the ectoderm, forcing the cells around them to activate other ectodermal genes. The gradual activation of FoxI1e coincides with the restriction of animal cap cells to ectodermal fates. This represents a model integrating Vg1, Notch, VegT, Nodals, and FoxI1e into the specification and patterning of the early ectoderm.
Fig. 7. Model of control of FoxI1e expression in the early embryo.
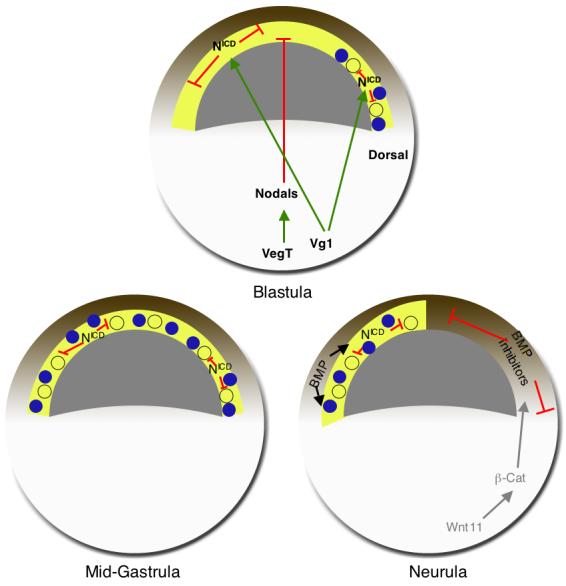
(A) At the blastula stage, signals from the vegetal hemisphere restrict the expression of FoxI1e to the dorsal side of the embryo within the subset of prospective ectodermal cells that are competent to express FoxI1e. Vg1 activates Notch signaling to maintain mosaic expression. The competent tissue, in yellow, is adjacent to the blastocoel in the sensorial layer of the animal cap. The molecular nature of this region that permits FoxI1e expression is unknown. It could be the inheritance of an intrinsic transcription factor, or signaling from other tissues. (B) As signaling from the vegetal hemisphere weakens, FoxI1e expression spreads to the ventral side of the embryo. The appearance and spread of FoxI1e expression coincides temporally with the restriction of animal cells to ectodermal lineages. The inner layer of cells is still competent to express FoxI1e. (C) At the neurula stage, FoxI1e expression is BMP-dependent. Activation of dorsal axis formation earlier in development leads to the release of BMP inhibitors from the dorsal mesoderm, resulting in a restriction of FoxI1e expression potential to the ventral ectoderm.
Acknowledgements
The authors would like to thank Chris Kintner () for his generous gift of the NICD and Su(H)-DBM constructs, and for his feedback on this manuscript. The authors would also like to acknowledge the financial support of the Cincinnati Children’s Hospital Research Foundation, and the National Institutes of Health (1R01HD045737).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe T, Furue M, Kondow A, Matsuzaki K, Asashima M. Notch signaling modulates the nuclear localization of carboxy-terminal-phosphorylated smad2 and controls the competence of ectodermal cells for activin A. Mech Dev. 2005;122:671–80. doi: 10.1016/j.mod.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Abe T, Furue M, Myoishi Y, Okamoto T, Kondow A, Asashima M. Activin-like signaling activates Notch signaling during mesodermal induction. Int J Dev Biol. 2004;48:327–32. doi: 10.1387/ijdb.041838ta. [DOI] [PubMed] [Google Scholar]
- Agius E, Oelgeschlager M, Wessely O, Kemp C, De Robertis EM. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127:1173–83. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JC, Beddington RS, Harland RM. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes and Development. 1999;13:3159–59. doi: 10.1101/gad.13.23.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy B, Kofron M, Schaible K, Wylie C, Heasman J. Vg 1 is an essential signaling molecule in Xenopus development. Development. 2006;133:15–20. doi: 10.1242/dev.02144. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Lachani K, Shin Y, Sherwood V, Cho KW, Papalopulu N. Grainyhead-like 3, a transcription factor identified in a microarray screen, promotes the specification of the superficial layer of the embryonic epidermis. Mech Dev. 2006;123:702–18. doi: 10.1016/j.mod.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Strauss B, Papalopulu N. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development. 2003;130:2657–68. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- Coffman CR, Skoglund P, Harris WA, Kintner CR. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993;73:659–71. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development. 1999;126:4715–28. doi: 10.1242/dev.126.21.4715. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–95. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hawley SH, Wunnenberg-Stapleton K, Hashimoto C, Laurent MN, Watabe T, Blumberg BW, Cho KW. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes Dev. 1995;9:2923–35. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- Heasman J, Holwill S, Wylie CC. Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Cell Biol. 1991;36:213–30. doi: 10.1016/s0091-679x(08)60279-4. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev Biol. 2000;222:124–34. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Heasman J, Wylie CC, Hausen P, Smith JC. Fates and states of determination of single vegetal pole blastomeres of X. laevis. Cell. 1984;37:185–94. doi: 10.1016/0092-8674(84)90314-3. [DOI] [PubMed] [Google Scholar]
- Houston DW, Wylie C. The Xenopus LIM-homeodomain protein Xlim5 regulates the differential adhesion properties of early ectoderm cells. Development. 2003;130:2695–2704. doi: 10.1242/dev.00509. [DOI] [PubMed] [Google Scholar]
- Kofron M, Klein P, Zhang F, Houston DW, Schaible K, Wylie C, Heasman J. The role of maternal axin in patterning the Xenopus embryo. Dev Biol. 2001;237:183–201. doi: 10.1006/dbio.2001.0371. [DOI] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Thomas ML, Weeks DL, Sargent TD. Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev Biol. 2002;245:136–44. doi: 10.1006/dbio.2002.0621. [DOI] [PubMed] [Google Scholar]
- Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328:80–2. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- Mir A, Kofron M, Zorn AM, Bajzer M, Haque M, Heasman J, Wylie CC. FoxI1e activates ectoderm formation and controls cell position in the Xenopus blastula. Development. 2007;134:779–88. doi: 10.1242/dev.02768. [DOI] [PubMed] [Google Scholar]
- Snape A, Wylie CC, Smith JC, Heasman J. Changes in states of commitment of single animal pole blastomeres of Xenopus laevis. Dev Biol. 1987;119:503–10. doi: 10.1016/0012-1606(87)90053-4. [DOI] [PubMed] [Google Scholar]
- Suri C, Haremaki T, Weinstein DC. Xema, a foxi-class gene expressed in the gastrula stage Xenopus ectoderm, is required for the suppression of mesendoderm. Development. 2005;132:2733–42. doi: 10.1242/dev.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama R, Curtiss PE, Otani H, Kimura M, Dawid IB, Taira M. The LIM class homeobox gene lim5: implied role in CNS patterning in Xenopus and zebrafish. Dev Biol. 1995;170:583–93. doi: 10.1006/dbio.1995.1238. [DOI] [PubMed] [Google Scholar]
- Yun CH, Choi SC, Park E, Kim SJ, Chung AS, Lee HK, Lee HJ, Han JK. Negative regulation of Activin/Nodal signaling by SRF during Xenopus gastrulation. Development. 2007;134:769–77. doi: 10.1242/dev.02778. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–24. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–29. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- Zimmerman LB, De Jesús-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]


