Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that mediates the biologic and toxic effects of its xenobiotic ligands. In recent years it has become evident that in the absence of ligand the AHR promotes cell cycle progression and that its activation by high-affinity ligands results in interactions with the retinoblastoma protein (RB) that lead to perturbation of the cell cycle, G0/G1 arrest, diminished capacity for DNA replication and inhibition of cell proliferation. Hence, the AHR has diametrically opposed pro-proliferative and anti-proliferative functions that have yet to be reconciled at the molecular level. Work from our own and from other laboratories suggests that the AHR may function as a tumor suppressor gene that becomes silenced in the process of tumor formation. To develop preliminary support for a more thorough examination of this hypothesis we characterized the expression levels of various tumor suppressor genes, transforming growth factor-β (Tgfb) genes and the Ahr gene in liver tumor samples from mice with a liver-specific RB ablation and their wild-type littermates. In tumors arising in RB-positive livers, Cdkn2d and Tgfb1 were repressed and Cdkn2c, Tgfb2, Tgfb3 and Pai1 were induced, whereas in RB-negative tumors, only Cdkn2c and Tgfb3 were induced. Ahr was significantly repressed in tumors from both sets of mice, supporting the concept that Ahr silencing may be associated with cancer progression.
Keywords: Ah receptor, tumor suppressor genes, TGFβ, hepatocellular carcinoma
INTRODUCTION
The AHR is a ligand-activated member of the bHLH/PAS family of transcription factors that plays an important role in controlling a variety of developmental and physiological events, including induction of drug metabolizing enzymes, xenobiotic detoxification, neurogenesis, tracheal and salivary duct formation, circadian rhythms, response to hypoxia, and hormone receptor function (Schmidt and Bradfield, 1996; Crews and Fan, 1999; Pocar et al., 2005). So far, more than 400 environmental toxicants and naturally occurring compounds have been reported to be AHR ligands (Denison et al., 2002). The cytosolic unliganded AHR is one component of a multi-protein complex containing the immunophilin-like protein XAP2/AIP/ARA9, the 23-kD co-chaperone protein p23 and two molecules of a 80-kD Hsp90 protein (Ma and Whitlock, Jr., 1997; Petrulis et al., 2003). Upon ligand binding, the complex translocates to the nucleus where AHR dissociates from the multi-protein complex and dimerizes with ARNT, a second bHLH/PAS protein, to form a heterodimeric transcription factor. The AHR/ARNT heterodimer binds to DNA recognition sequences, termed AHREs/XREs/DREs, found in the enhancer region of genes coding for many phase I drug metabolizing enzymes, such as the cytochromes P450 CYP1A1, CYP1A2, CYP1B1, and several phase II conjugating enzymes, such as ALDH3A1 and NQO1. Binding leads to chromatin and nucleosome disruption, recruitment of the basal transcriptional machinery and other transcriptional regulators, and initiation of downstream transcription (Hankinson, 1995; Whitlock, Jr., 1999; Schnekenburger, Peng and Puga, 2007; Schnekenburger, Talaska and Puga, 2007). Following nuclear export, the AHR is degraded via the 26S proteasome pathway (Pollenz, 2002).
The AHR has also been recognized as a cell cycle regulator (Ma and Whitlock, 1996; Weiss et al., 1996; Ge and Elferink, 1998; Kolluri et al., 1999; Puga et al., 2000; Strobeck et al., 2000; Marlowe et al., 2004) although the precise molecular mechanisms responsible for this role have not been fully elucidated. In the absence of an exogenous ligand, or after deletion of the ligand-binding PAS-B domain, the AHR has been shown to promote cell cycle progression (Ma and Whitlock, 1996; Elizondo et al., 2000; Chang et al., 2007), whereas exposure to its prototypical ligand, TCDD, inhibits cell cycle proliferation in an AHR-dependent manner (Bauman et al., 1995;Hushka and Greenlee, 1995;Levine-Fridman, Chen, and Elferink, 2004; Marlowe et al., 2004). Mechanistically, at least two separate signaling pathways contribute to the AHR role in cell cycle regulation. First, AHR can repress expression of TGF-β1 (Elizondo et al., 2000) by accelerating TGF-β1 mRNA degradation (Chang et al., 2007) and blocking TGF-β1-dependent inhibition of cell proliferation and promotion of apoptosis. Conversely, overproduction of TGF-β in fibroblasts from AHR knockout mice causes low proliferation rates and increased apoptosis (Elizondo et al., 2000; Chang et al., 2007). Consistent with these observations, livers from AHR knockout mice showed increased levels of TGF-β1 and TGF-β3 proteins and elevated numbers of hepatocytes undergoing apoptosis compared to wild-type mice (Zaher et al., 1998). Second, protein interactions between AHR and RB further repress the RB-dependent repression of the transcription factor E2F and prevent entry of the cells into S-phase (Ge and Elferink, 1998; Puga et al., 2000; Strobeck et al., 2000; Marlowe et al., 2004). Hence, under certain circumstances the AHR could be considered as a pro-proliferative gene with the properties of an oncogene, and under others as a tumor suppressor gene. Consistent with this view, AHR has been found to be silenced by promoter hypermethylation in a significant number of human acute lymphoblastic leukemia cases (Mulero-Navarro et al. 2006).
Epidemiologic studies have shown that the retinoblastoma tumor suppressor protein RB is functionally inactivated in most human neoplasms (Liu et al., 2004) with loss of heterozygosity being frequently observed in hepatocellular carcinomas (Ashida et al., 1997; Kondoh et al., 2001). In good agreement with the epidemiologic observations, mice with a liver-specific ablation of the Rb gene showed aberrant ploidy and increased multiplicity of liver tumors when exposed to DEN (Mayhew et al. 2005; Mayhew et al. 2007; Srinivasan et al. 2007). To explore the role of AHR/RB/TGF-β interactions in liver tumorigenesis, we have examined the expression status of Tgfb genes, tumor suppressor genes, Ahr and several of its downstream targets in DEN-induced liver tumors and normal livers of mice with a liver-specific ablation of the Rb gene and in their wild-type littermates. The central question in these experiments was to determine whether AHR expression would be repressed, favoring the hypothesis that it functioned as a tumor suppressor gene, or would be over-expressed or unaffected, favoring the hypothesis that it functioned as an oncogene. We find distinct patterns of gene expression that for the most part show a significant increase of expression of Tgfb genes and repression of Ahr in RB-positive and RB-negative tumors.
MATERIALS AND METHODS
Mice
Liver-specific Rb conditional knockouts and their normal littermates were derived from Rbfl/fl mice, harboring Rb alleles in which exon 19 is flanked by loxP sites, crossed with Alb-Cre mice to obtain mice homozygous for the floxed Rb locus and hemizygous for Alb-Cre. These mice were then interbred with Rbfl/fl mice to produce Rbfl/fl (wild type) and Rbfl/fl Alb-Cre+/0 (RB-knockout) littermates at a 1:1 ratio (Mayhew et al., 2005). Mice were housed in a pathogen-free animal facility under standard 12-hour light/12-hour dark cycle with ad libitum water and chow. All of the experiments were conducted using the highest standards for humane care in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the University of Cincinnati Institutional Animal Care and Use Committee.
Liver samples
Fifteen-day old Rbfl/fl and Rbfl/fl Alb-Cre+/0 littermates were administered a single i.p. injection of the genotoxic hepatocarcinogen DEN (Sigma) dissolved in saline at a dose of 20 mg/kg body weight; littermates used as controls were inoculated with an equal volume of saline. This protocol resulted in the four groups of mouse livers analyzed in this work: RB-positive, normal, are livers from Rbfl/fl mice inoculated with saline; RB-positive, tumors are livers from Rbfl/fl mice inoculated with DEN; RB-negative, normal are livers from Rbfl/flAlb-Cre+/0 mice inoculated with saline; and RB-negative, tumors are livers from Rbfl/flAlb-Cre+/0 mice inoculated with DEN. Mice were euthanized 9 months post-DEN exposure. Immediately after euthanasia, livers were excised, weighed and photographed to facilitate scoring of surface liver lesions. Visible surface tumors were excised from both Rbfl/fl (normal) and Rbfl/flAlb-Cre+/0 animals, immediately frozen in liquid N2 and stored at −80 °C until use. Genotypes were determined by standard PCR of tail DNA and deletion or retention of the Rb gene was confirmed by genotyping the mouse livers themselves. Characterization of these tumors has been reported elsewhere (Mayhew et al. 2005; Mayhew et al. 2007; Srinivasan et al. 2007).
cDNA sample preparation
RNA was isolated from frozen liver tumor and normal tissues using Triazol. cDNA was synthesized by reverse transcription of 1 µg of total RNA in a total volume of 20 µl containing 1X reverse transcriptase buffer (Invitrogen), 25 µg/ml oligo-dT12–18 primer (Invitrogen), 0.5 mM dNTP mix (GeneChoice), 10 mM dithiothreitol (Invitrogen), 5 mM MgCl2, 20 U of RNase inhibitor (RNasin, Promega), and 100 U of SuperScript™ II RNase H− reverse transcriptase (Invitrogen). Samples were denatured and annealed to the primer for 10 min at 70 °C, and reverse-transcribed for 1 hour at 42 °C. Before amplification, the reverse transcriptase was inactivated by heating to 70 °C for 15 minutes and RNA was hydrolyzed by incubation with 2 U RNase H (Invitrogen) at 37 °C for 20 minutes. The resulting cDNA products were diluted in a final volume of 200 µl and a 2 µl aliquot was used as template for subsequent quantification by real-time PCR amplification.
Real-time PCR
Primers for cDNA amplification are shown in Table 1. Amplification of β-actin cDNA in the same samples was used as an internal control for all real-time PCR amplification reactions. PCR product specificity was confirmed using melting curve analyses and subsequent polyacrylamide gel electrophoresis. PCR reactions were conducted in duplicate in a total volume of 25 µl containing SYBR Green PCR Master Mix (Applied Biosystems), and 0.1 µM of each primer. Amplification was performed on an ABI 7500 (Applied Biosystems) where the reaction was heated to 95 °C for 10 minutes followed by 40 cycles of denaturation at 95 °C for 15 seconds and annealing-elongation at 60 °C for 60 seconds. Detection of the fluorescent product was carried out during the elongation period, and emission data were quantified using threshold cycle (Ct) values. Ct values were determined in duplicate, averaged and normalized to values for β-actin amplification of the same sample (ΔCt).
Table 1.
Primer sequences fo cDNA amplification of selected genes
| Gene | Primer sequences |
|---|---|
| Forward: 5′- CATCCGTAAAGACCTCTATGCC -3′ | |
| β-Actin | Reverse: 5′- ACGCAGCTCAGTAACAGTCC -3′ |
| Forward: 5′- GGCCAAGAGCTTCTTTGATG - 3′ | |
| Ahr | Reverse: 5′- TGCCAGTCTCTGATTTGTGC - 3′ |
| Forward: 5′- GAACAGGGATGGCAGTTAGG - 3′ | |
| Cdkn1a (p21) | Reverse: 5′- AGTATGGGGTGGGGGAAAAG - 3′ |
| Forward: 5′- GGGTTCCAGCTTGTTGTGTT - 3′ | |
| Cdkn1b (p27) | Reverse: 5′- GGCCATTTTCCATCTCTGAA - 3′ |
| Forward: 5′- CCTTCCAAAACTTGAACCCTAC - 3′ | |
| Cdkn2b (p15) | Reverse: 5′- TCCCTTGCTATTTTACACCAC - 3′ |
| Forward: 5′- TGCGCTGCAGGTTATGAAACTTGG - 3′ | |
| Cdkn2c (p18) | Reverse: 5′- AACATCAGCCTGGAACTCCAGCAA - 3′ |
| Forward: 5′- GCCTTGCAGGTCATGATGTTTGGA - 3′ | |
| Cdkn2d (p19) | Reverse: 5′- ATTCAGGAGCTAGGAAGCTGACCA - 3′ |
| Forward: 5′- GTGTCTGGTTACTTTGACAAGTGG-3′ | |
| Cyp1a1 | Reverse: 5′- ACATGGACATGCAAGGACA -3′ |
| Forward: 5′- CTGACCGCTTCTACTTCCTC -3′ | |
| Dnmt1 | Reverse: 5′- TCCCTTTCCCCTTCCCTTTC -3′ |
| Forward: 5′- TTCCAACATGACCAACCAGA -3′ | |
| Hdac1 | Reverse: 5′- GGCAGCATCCTCAAGTTCTC -3′ |
| Forward: 5′- ACCCCACTCTATTTTGCTCC -3′ | |
| Nqo1 | Reverse: 5′- ACTTACTCCTTTTCCCATCCTC -3′ |
| Forward: 5′- GTCTTTCCGACCAAGAGCAG -3′ | |
| Pai1 | Reverse: 5′- GACAAAGGCTGTGGAGGAAG -3′ |
| Forward: 5′- CAACGCCATCTATGAGAAAACC-3′ | |
| Tgfb1 | Reverse: 5′- AAGCCCTGTATTCCGTCTCC-3′ |
| Forward: 5′- CTCAACACACCAAAGTCCTC-3′ | |
| Tgfb2 | Reverse: 5′- ATCAAAACTCCCTCCCTCC-3′ |
| Forward: 5′- CAGCCTACATAGGTGGCAAGAAT-3′ | |
| Tgfb3 | Reverse: 5′- ACCCAAGTTGGACTCTCTCCTCAA -3′ |
| Forward: 5′- TGGAAGACTCCAGTGGGAAC - 3′ | |
| Trp53 | Reverse: 5′- TCTTCTGTACGGCGGTCTCT - 3′ |
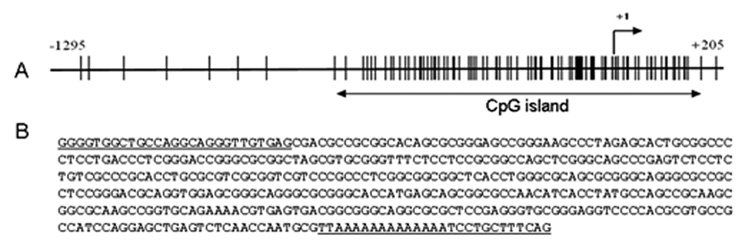
CpG island analysis in the mouse Ahr promoter
A 1,500-nucleotide segment of the mouse Ahr promoter, from coordinates −1295 to +205 and containing the transcription start site, was analyzed with the Methprimer software program (http://www.urogene.org/methprimer) for the presence of CpG islands. A 705-nucleotide segment was identified as a CpG island using an observed/expected CpG ratio of 0.6, a minimum length island of 200 nucleotides and a minimum G+C content of 50%. A 395-nucleotide fragment was used for further analysis.
DNA methylation analysis
The methylation status of the mouse Ahr promoter was determined by PCR amplification of sodium bisulfite-treated genomic DNA followed by cloning and DNA sequencing, as described (Schnekenburger, Peng, and Puga, 2007). Genomic DNA was extracted using standard procedures and 750 ng of each DNA sample underwent sodium bisulfite modification using a bisulfite modification kit (Active Motif MethylDetector™) following the manufacturer’s recommendations. Specific primers for PCR amplification were designed using the same MethPrimer software program. Several primer sets were designed such that the target sequences did not contain any CpG dinucleotide, thus allowing for amplification of both unmethylated and methylated DNA. Primers were tested for their ability to yield high-quality sequencing reactions. PCR products were quality controlled by agarose gel electrophoresis. Inclusion of restriction enzyme sites in the PCR primers allowed for cloning of the amplification products in the pGEM4Z vector (Promega). For each DNA sample, a minimum of 10 clones were analyzed by sequencing.
Statistical analyses
Statistical analyses of RT-PCR data was performed using SigmaStat 2.03. Group comparisons were made using a two-tailed Student’s t-test. A p-value less than 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Comparison of mRNA levels of RB-positive tumors (n = 4) with RB-positive normal livers (n = 3) showed significant changes in the expression of a number of genes. mRNA levels of Ahr, Cdkn2d (p19) and Tgfb1 in tumor tissue were decreased to 30%, 50% and 40%, respectively, of their levels in normal liver. On the other hand, mRNA levels of Cdkn1a (p21), Cdkn2c (p18), Pai1 (Serpine-1), Tgfb2 and Tgfb3 were elevated in RB-positive tumors by 4.9-, 4.3-, 3.2-, 10.5- and 11.3-fold, respectively, relative to their levels in normal liver (Table 2). In contrast, RB-negative tumors (n = 8) and RB-negative normal livers (n = 3) showed fewer differences, with Ahr mRNA being the only one showing significantly reduced levels (60%) in tumors and Cdkn2c (p18) and Tgfb2 mRNAs increased by 6.5- and 8.6-fold, respectively, relative to their levels in RB-negative normal liver tissues. Expression of several other genes, including Cdkn2d, Cyp1a1, Pai1 and Tgfb3, was also increased in RB-negative tumors, but the changes did not reach statistical significance (Table 2). Expression of Trp53 (p53) was unchanged in tumors relative to normal tissues and Dnmt1and Hdac1, which were assayed for their potential role in repression of AHR-regulated genes (Schnekenburger, Peng and Puga, 2007), also showed no significant changes between tumors and normal livers (Table 2).
Table 2. Fold change of the expression of selected genes in tumor vs normal liver tissues of mice.
ΔCT values were calculate using mRNA levels of β-actin as the normalization standard. The values shown are the mean ± S.D. of RB positive normal (n = 3) and tumor (n =4) liver samples and RB-negative normal (n = 3) and tumor (n = 8) liver samples. Fold-change was calculated by raising 2 to the power of ΔCTnormal minus ΔCTtumor; a value <1 indicates less mRNA in tumors than in normal tissues, whereas a value >1 indicates higher mRNA levels in tumors than in normal tissues. P-values were calculated using Student’s t test and are considered significant if ≤ 0.05.
| RB Positive Tumors | RB Negative Tumors | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | ΔCT Normal | ΔCT Tumor | Fold Change | p value | ΔCT Normal | ΔCT Tumor | Fold Change | p value |
| Ahr | 8.2 ± 0.1 | 10.0 ± 0.6 | 0.3 ± 0.2 | 0.004 | 8.8 ± 0.2 | 9.5 ± 0.2 | 0.6 ± 0.1 | <0.001 |
| Cdkn1a (p21) | 12.5 ± 0.8 | 10.2 ± 1.4 | 4.9 ± 0.7 | 0.01 | 8.8 ± 3.1 | 8.3 ± 0.5 | 1.4 ± 1.0 | N.S. |
| Cdkn1b (p27) | 9.0 ± 0.3 | 9.7 ± 0.7 | 0.6 ± 0.3 | N.S. | 10.2 ± 1.0 | 9.4 ± 0.2 | 1.7 ± 0.3 | N.S. |
| Cdkn2b (p15) | 19.7 ± 3.6 | 19.7 ± 3.2 | 1.0 ± 2.0 | N.S. | 18.8 ± 2.0 | 19.1 ± 1.4 | 0.8 ± 0.8 | N.S. |
| Cdkn2c (p18) | 11.6 ± 0.2 | 9.5 ± 1.0 | 4.3 ± 0.4 | 0.02 | 12.2 ± 1.5 | 9.5 ± 0.6 | 6.5 ± 0.5 | 0.001 |
| Cdkn2d (p19) | 10.3 ± 0.3 | 11.4 ± 0.4 | 0.5 ± 0.2 | 0.01 | 12.4 ± 1.8 | 11.1 ± 0.4 | 2.5 ± 0.6 | N.S. |
| Cyp1a1 | 14.2 ± 0.8 | 20.1 ± 4.7 | 0.0 ± 2.0 | N.S. | 20.3 ± 1.4 | 19.0 ± 2.3 | 2.5 ± 0.9 | N.S. |
| Dnmt1 | 11.8 ± 0.2 | 12.2 ± 0.7 | 0.8 ± 0.3 | N.S. | 11.2 ± 0.5 | 11.0 ± 0.4 | 1.1 ± 0.2 | N.S. |
| Hdac1 | 8.0 ± 0.4 | 8.4 ± 0.3 | 0.8 ± 0.2 | N.S. | 8.6 ± 0.4 | 8.2 ± 0.3 | 1.3 ± 0.2 | N.S. |
| Nqo1 | 8.8 ± 0.7 | 9.4 ± 2.1 | 0.7 ± 0.9 | N.S. | 10.4 ± 1.4 | 10.6 ± 1.6 | 0.9 ± 0.7 | N.S. |
| Pai1 | 12.6 ± 0.4 | 10.9 ± 1.0 | 3.2 ± 0.4 | <0.001 | 12.1 ± 2.2 | 10.6 ± 1.4 | 2.8 ± 0.8 | N.S. |
| Tgfb1 | 8.2 ± 0.1 | 9.4 ± 0.2 | 0.4 ± 0.1 | <0.001 | 8.8 ± 0.5 | 8.4 ± 0.3 | 1.3 ± 0.2 | N.S. |
| Tgfb2 | 14.4 ± 1.4 | 11.0 ± 0.5 | 10.6 ± 0.6 | 0.006 | 14.5 ± 1.1 | 11.4 ± 0.8 | 8.6 ± 0.4 | <0.001 |
| Tgfb3 | 18.6 ± 1.2 | 15.1 ± 1.0 | 11.3 ± 0.6 | 0.008 | 16.3 ± 1.3 | 15.3 ± 1.2 | 2.0 ± 0.6 | N.S. |
| Trp53 | 6.7 ± 0.2 | 6.5 ± 0.6 | 1.1 ± 0.3 | N.S. | 6.3 ± 0.6 | 6.1 ± 0.3 | 1.1 ± 0.2 | N.S. |
Changes in DNA methylation have been directly linked to genomic instability and play an important role in tumorigenesis. In tumors and in cancer cell lines, aberrant methylation usually occurs at CpG islands, unmethylated in normal somatic cells and hypermethylated in tumor suppressor gene promoters, representing a major mechanism of gene inactivation in primary human tumors (Fraga et al., 2004; Esteller, 2007). A CpG island in the promoter of the human AHR gene has recently been found to be methylated in a panel of 19 tumor cell lines and in lymphocytes of one-third of acute lymphoblastic leukemia patients (Mulero-Navarro et al., 2006). This observation suggested the possibility that promoter hypermethylation might be a potential mechanism responsible for the down-regulation of AHR expression in our liver tumor samples. Accordingly, we used DNA sequencing of bisulfite-modified cloned PCR products to test this hypothesis. We sequenced a minimum of 10 clones from each tumor DNA sample but found no evidence of methylation (data not shown) in the corresponding CpG island of the mouse Ahr promoter (Fig. 1), indicating that hypermethylation was not the main cause of the decrease in Ahr expression in the mouse tumors analyzed.
Fig. 1.
(A). A 705-nucleotide segment of the mouse Ahr promoter from −556 to + 148 was identified as a CpG island using the Methprimer software program. (B). A 395-nucleotide DNA fragment within this CpG island, from coordinates −245 to +148 and containing 56 CpG dinucleotides was selected for methylation analysis after bisulfite modification followed by PCR amplification with specific oligonucleotide primers (underlined), cloning and sequencing of a minimum of 10 clones from each liver DNA sample.
Because of their role as tumor suppressors, the members of the two families of CDK inhibitors, the INK4 (p16, p15, p18 and p19) and the CIP/KIP (p21, p27 and p57) are often found in a silenced state in tumors of many different origins (Damo, Snyder, and Franklin 2005; Bai et al. 2007); however, recent evidence in pancreatic and lung tumors shows up-regulation, rather than repression of p18 (Cdkn2c) and argues against a pure tumor suppressor role for this gene (Joshi et al., 2007; Lindberg, Akerstrom, and Westin, 2007; Pei et al., 2007), whereas p19/ARF (Cdkn2d) is almost exclusively repressed in tumors (Lowe and Sherr, 2003; Canepa et al., 2007). Our results are consistent with those findings, showing increased expression of Cdkn2c in both RB-positive and RB-negative tumors, and decreased expression of Cdkn2d in RB-positive but not in RB-negative tumors. Possibly, in the absence of RB, repression of one of the main inhibitors of its phosphorylation is less essential for tumor suppression.
The TGF-β proteins are growth modulators involved in cell proliferation, apoptosis, differentiation, adhesion and migration with their growth inhibitory effects resulting from their ability to arrest cells in the G1 phase of the cell cycle (Massague, Blain, and Lo, 2000; Siegel and Massague, 2003; Massague and Gomis, 2006). Reduced expression of TGF-β proteins or loss of their inhibitory effects have been linked to cell hyperproliferation and tumor progression (Massague, Blain, and Lo, 2000; Siegel and Massague, 2003; Massague and Gomis, 2006). In fact, TGF-β appears to have a dual role in tumorigenesis, acting as a tumor suppressor in the earlier tumor phases, and as a tumor promoter in the later phases (Derynck, Akhurst and Balmain, 2001; Ten Dijke et al., 2002). At that time, TGF-β is produced in high amounts in the tumor and it stimulates tumorigenesis by allowing tumor cells to escape immune surveillance (Hazelbag et al., 2002) and promoting angiogenesis (Zhang et al., 2006; Ten Dijke et al., 2002). Consistent with this dual role of TGF-β, we find a significant decrease of Tgfb1 expression in the tumors and very large increases of Tgfb2 and Tgfb3 mRNA levels, with a concomitant increase in Pai1 expression, in agreement with the regulatory role of Tgfb in Pai1 transcription (Kutz et al., 2006). The Pai1 gene encodes the plasminogen activator inhibitor serpine-1, whose expression increases filopodia formation and migration and is highly elevated in many invasive tumors, including breast, brain and gastric cancers (Rao et al., 1993; Chazaud et al., 2002; Lei et al., 2007).
Ahr expression was significantly down-regulated in both RB-positive and RB-negative liver tumors, although not by promoter hypermethylation. These findings are consistent with the hypothesis that the AHR may have tumor suppressor gene properties in these tumors and is in good agreement with recent observations that AHR functions as a tumor suppressor in prostate carcinogenesis in mice (Fritz et al., 2007). A hypothesis to explain mechanistically these results is that in the absence of RB, cells with lower levels of AHR will have a greater probability of survival because they will escape RB/AHR repression of S-phase entry. On the other hand, in both RB-positive and RB-negative cells, the lower AHR levels will promote and increase in TGFβ, as we have recently shown (Chang et al. 2007), allowing tumor cells to escape immune surveillance and promoting angiogenesis. An important limitation of these studies is that they offer a snapshot of a dynamic process that began 9 months before, hence, the data may represent events happening at relatively late stage liver tumors, providing no clues about potential events that may have taken place early in the process and contributed to tumor promotion, as for example, reversal of Tgfb expression consistent with its dual role in tumorigenesis. Future work in this area will of necessity include analyses of earlier time points.
ACKNOWLEDGEMENTS
This research was supported by NIEHS grants R01 ES06273, R01 ES10807 and the NIEHS Center for Environmental Genetics grant P30 ES06096.
ABBREVIATIONS
- ALDH3A1
aldehyde dehydrogenase-3A1
- AHR
aryl hydrocarbon receptor
- AHRE
aryl hydrocarbon receptor response element
- AIP
AHR-interacting protein
- ALL
acute lymphoblastic leukemia
- ARA9
Ah receptor associated protein 9
- ARNT
aryl hydrocarbon receptor nuclear translocator
- bHLH
basic-region helix-loop-helix
- CDK
cyclin-dependent kinase
- DEN
diethylnitrosamine
- DRE
dioxin response element
- HCC
hepatocellular carcinoma
- Hsp90
heat shock protein 90
- MEM-α
minimal essential medium-α
- NQO1
NAD(P)H-dependent quinone oxidoreductase
- PAI-1
plasminogen activator inhibitor type-1
- PAS
Period-Aryl hydrocarbon nuclear translocator-Simple-minded
- RB
retinoblastoma
- TCDD
2, 3, 7, 8-tetrachlorodibenzo-p-dioxin
- TGFβ
transforming growth factor-beta
- XAP2
hepatitis virus X-associated protein-2
- XRE
xenobiotic response element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashida K, Kishimoto Y, Nakamoto K, Wada K, Shiota G, Hirooka Y, Kamisaki Y, Itoh T, Kawasaki H. Loss of heterozygosity of the retinoblastoma gene in liver cirrhosis accompanying hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 1997;123:489–495. doi: 10.1007/BF01192203. [DOI] [PubMed] [Google Scholar]
- Bai F, Pei XH, Nishikawa T, Smith MD, Xiong Y. p18Ink4c, but not p27Kip1, collaborates with Men1 to suppress neuroendocrine organ tumors. Mol Cell Biol. 2007;27:1495–1504. doi: 10.1128/MCB.01764-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman JW, Goldsworthy TL, Dunn CS, Fox TR. Inhibitory effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on rat hepatocyte proliferation induced by 2/3 partial hepatectomy. Cell Prolif. 1995;28:437–451. doi: 10.1111/j.1365-2184.1995.tb00084.x. [DOI] [PubMed] [Google Scholar]
- Canepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB. Life. 2007;59:419–426. doi: 10.1080/15216540701488358. [DOI] [PubMed] [Google Scholar]
- Chang X, Fan Y, Karyala S, Schwemberger S, Tomlinson CR, Sartor MA, Puga A. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Mol Cell Biol. 2007;27:6127–6139. doi: 10.1128/MCB.00323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud B, Ricoux R, Christov C, Plonquet A, Gherardi RK, Barlovatz-Meimon G. Promigratory effect of plasminogen activator inhibitor-1 on invasive breast cancer cell populations. Am. J. Pathol. 2002;160:237–246. doi: 10.1016/S0002-9440(10)64367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr. Opin. Genet. Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- Damo LA, Snyder PW, Franklin DS. Tumorigenesis in p27/p53- and p18/p53-double null mice: functional collaboration between the pRb and p53 pathways. Mol Carcinog. 2005;42:109–120. doi: 10.1002/mc.20068. [DOI] [PubMed] [Google Scholar]
- Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chem. Biol. Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Elizondo G, Fernandez-Salguero P, Sheikh MS, Kim GY, Fornace AJ, Lee KS, Gonzalez FJ. Altered cell cycle control at the G(2)/M phases in aryl hydrocarbon receptor-null embryo fibroblast. Mol. Pharmacol. 2000;57:1056–1063. [PubMed] [Google Scholar]
- Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum. Mol Genet. 2007;16(Spec No 1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Herranz M, Espada J, Ballestar E, Paz MF, Ropero S, Erkek E, et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004;64:5527–5534. doi: 10.1158/0008-5472.CAN-03-4061. [DOI] [PubMed] [Google Scholar]
- Fritz WA, Lin TM, Cardiff RD, Peterson RE. The aryl hydrocarbon receptor inhibits prostate carcinogenesis in TRAMP mice. Carcinogenesis. 2007;28:497–505. doi: 10.1093/carcin/bgl179. [DOI] [PubMed] [Google Scholar]
- Ge N-L, Elferink CJ. A direct interaction between the aryl hydrocarbon receptor and retinoblatoma protein. J. Biol. Chem. 1998;273:22708–22713. doi: 10.1074/jbc.273.35.22708. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hazelbag S, Gorter A, Kenter GG, van den BL, Fleuren G. Transforming growth factor-beta1 induces tumor stroma and reduces tumor infiltrate in cervical cancer. Hum. Pathol. 2002;33:1193–1199. doi: 10.1053/hupa.2002.130109. [DOI] [PubMed] [Google Scholar]
- Hushka DR, Greenlee WF. 2,3,7,8-Tetrachlorodibenzo-p-dioxin inhibits DNA synthesis in rat primary hepatocytes. Mutat. Res. 1995;333:89–99. doi: 10.1016/0027-5107(95)00135-2. [DOI] [PubMed] [Google Scholar]
- Joshi PP, Kulkarni MV, Yu BK, Smith KR, Norton DL, Veelen W, Hoppener JW, Franklin DS. Simultaneous downregulation of CDK inhibitors p18(Ink4c) and p27(Kip1) is required for MEN2A-RET-mediated mitogenesis. Oncogene. 2007;26:554–570. doi: 10.1038/sj.onc.1209811. [DOI] [PubMed] [Google Scholar]
- Kolluri SK, Weiss C, Koff A, Göttlicher M. p27kip1 induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 1999;13:1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh N, Wakatsuki T, Hada A, Shuda M, Tanaka K, Arai M, Yamamoto M. Genetic and epigenetic events in human hepatocarcinogenesis. Int. J. Oncol. 2001;18:1271–1278. doi: 10.3892/ijo.18.6.1271. [DOI] [PubMed] [Google Scholar]
- Kutz SM, Higgins CE, Samarakoon R, Higgins SP, Allen RR, Qi L, Higgins PJ. TGF-beta 1-induced PAI-1 expression is E box/USF-dependent and requires EGFR signaling. Exp. Cell Res. 2006;312:1093–1105. doi: 10.1016/j.yexcr.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Lei H, Hemminki K, Johansson R, Altieri A, Enquist K, Henriksson R, Lenner P, Forsti A. PAI-1 -675 4G/5G polymorphism as a prognostic biomarker in breast cancer. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9635-3. [DOI] [PubMed] [Google Scholar]
- Levine-Fridman A, Chen L, Elferink CJ. Cytochrome P4501A1 promotes G1 phase cell cycle progression by controlling aryl hydrocarbon receptor activity. Mol. Pharmacol. 2004;65:461–469. doi: 10.1124/mol.65.2.461. [DOI] [PubMed] [Google Scholar]
- Lindberg D, Akerstrom G, Westin G. Mutational analysis of p27 (CDKN1B) and p18 (CDKN2C) in sporadic pancreatic endocrine tumors argues against tumor-suppressor function. Neoplasia. 2007;9:533–535. doi: 10.1593/neo.07328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Dibling B, Spike B, Dirlam A, Macleod K. New roles for the RB tumor suppressor protein. Curr. Opin. Genet. Dev. 2004;14:55–64. doi: 10.1016/j.gde.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Ma Q, Whitlock JP., Jr. A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Chem. 1997;272:8878–8884. [PubMed] [Google Scholar]
- Ma Q, Whitlock JPJ. The aromatic hydrocarbon receptor modulates the Hepa 1c1c7 cell cycle and differentiated state independently of dioxin. Molecular & Cellular Biology. 1996;16:2144–2150. doi: 10.1128/mcb.16.5.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe JL, Knudsen ES, Schwemberger S, Puga A. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S-phase specific gene expression. J. Biol. Chem. 2004;279:29013–29022. doi: 10.1074/jbc.M404315200. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, et al. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–984. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Mulero-Navarro S, Carvajal-Gonzalez JM, Herranz M, Ballestar E, Fraga MF, Ropero S, Esteller M, Fernandez-Salguero PM. The dioxin receptor is silenced by promoter hypermethylation in human acute lymphoblastic leukemia through inhibition of Sp1 binding. Carcinogenesis. 2006;27:1099–1104. doi: 10.1093/carcin/bgi344. [DOI] [PubMed] [Google Scholar]
- Pei XH, Bai F, Smith MD, Xiong Y. p18Ink4c collaborates with Men1 to constrain lung stem cell expansion and suppress non-small-cell lung cancers. Cancer Res. 2007;67:3162–3170. doi: 10.1158/0008-5472.CAN-06-4517. [DOI] [PubMed] [Google Scholar]
- Petrulis JR, Kusnadi A, Ramadoss P, Hollingshead B, Perdew GH. The hsp90 Co-chaperone XAP2 alters importin beta recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity. J. Biol. Chem. 2003;278:2677–2685. doi: 10.1074/jbc.M209331200. [DOI] [PubMed] [Google Scholar]
- Pocar P, Fischer B, Klonisch T, Hombach-Klonisch S. Molecular interactions of the aryl hydrocarbon receptor and its biological and toxicological relevance for reproduction. Reproduction. 2005;129:379–389. doi: 10.1530/rep.1.00294. [DOI] [PubMed] [Google Scholar]
- Pollenz RS. The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem. Biol. Interact. 2002;141:41–61. doi: 10.1016/s0009-2797(02)00065-0. [DOI] [PubMed] [Google Scholar]
- Puga A, Barnes SJ, Dalton TP, Chang C, Knudsen ES, Maier MA. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J. Biol. Chem. 2000;275:2943–2950. doi: 10.1074/jbc.275.4.2943. [DOI] [PubMed] [Google Scholar]
- Rao JS, Rayford A, Morantz RA, Festoff BW, Sawaya R. Increased levels of plasminogen activator inhibitor-1 (PAI-1) in human brain tumors. J. Neurooncol. 1993;17:215–221. doi: 10.1007/BF01049977. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Schnekenburger M, Peng L, Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim. Biophys. Acta. 2007;1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnekenburger M, Talaska G, Puga A. Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol Cell Biol. 2007;27:7089–7101. doi: 10.1128/MCB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- Srinivasan SV, Mayhew CN, Schwemberger S, Zagorski W, Knudsen ES. RB loss promotes aberrant ploidy by deregulating levels and activity of DNA replication factors. J. Biol. Chem. 2007;282:23867–23877. doi: 10.1074/jbc.M700542200. [DOI] [PubMed] [Google Scholar]
- Strobeck MW, Fribourg AF, Puga A, Knudsen ES. Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene. 2000;19:1857–1867. doi: 10.1038/sj.onc.1203510. [DOI] [PubMed] [Google Scholar]
- Ten Dijke P, Goumans MJ, Itoh F, Itoh S. Regulation of cell proliferation by Smad proteins. J. Cell Physiol. 2002;191:1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- Weiss C, Kolluri SK, Kiefer F, Gottlicher M. Complementation of Ah receptor deficiency in hepatoma cells: negative feedback regulation and cell cycle control by the Ah receptor. Exp. Cell Res. 1996;226:154–163. doi: 10.1006/excr.1996.0214. [DOI] [PubMed] [Google Scholar]
- Whitlock JP., Jr. Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- Zaher H, Fernandez-Salguero PM, Letterio J, Sheikh MS, Fornace AJ, Jr., Roberts AB, Gonzalez FJ. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Mol. Pharmacol. 1998;54:313–321. doi: 10.1124/mol.54.2.313. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]